Published online Feb 7, 2015. doi: 10.3748/wjg.v21.i5.1595
Peer-review started: July 18, 2014
First decision: October 14, 2014
Revised: October 28, 2014
Accepted: December 8, 2014
Article in press: December 8, 2014
Published online: February 7, 2015
Processing time: 207 Days and 0.2 Hours
AIM: To investigate gene mutations and DNA mismatch repair (MMR) protein abnormality in Chinese colorectal carcinoma (CRC) patients and their correlations with clinicopathologic features.
METHODS: Clinical and pathological information for 535 patients including 538 tumors was reviewed and recorded. Mutation analyses for exon 2 of KRAS gene and exon 15 of BRAF gene were performed by Sanger sequencing except that in 9 tumors amplification refractory mutation system PCR was used. Expression of MMR proteins including MHL1, MSH2, MSH6 and PMS2 was evaluated by immunohistochemistry. Correlations of KRAS and BRAF mutation status and the expression status of MMR proteins with age, gender, cancer stage, location, and histology were analyzed. Correlations between KRAS or BRAF mutations and MMR protein expression were also explored.
RESULTS: The overall frequencies of KRAS and BRAF mutations were 37.9% and 4.4%, respectively. KRAS mutations were more common in patients ≥ 50 years old (39.8% vs 22% in patients < 50 years old, P < 0.05). The frequencies of BRAF mutants were higher in tumors from females (6.6% vs males 2.8%, P < 0.05), located in the right colon (9.6% vs 2.1% in the left colon, 1.8% in the rectum, P < 0.01), with mucinous differentiation (9.8% vs 2.8% without mucinous differentiation, P < 0.01), or being poorly differentiated (9.5% vs 3.4% well/moderately differentiated, P < 0.05). MMR deficiency was strongly associated with proximal location (20.5% in the right colon vs 9.2% in the left colon and 5.1% in the rectum, P < 0.001), early cancer stage (15.0% in stages I-II vs 7.7% in stages III-IV, P < 0.05), and mucinous differentiation (20.2% vs 9.2% without mucin, P < 0.01). A higher frequency of MLH1/PMS2 loss was found in females (9.2% vs 4.4% in males, P < 0.05), and MSH2/MSH6 loss tended to be seen in younger (<50 years old) patients (12.0% vs 4.0% ≥ 50 years old, P < 0.05). MMR deficient tumors were less likely to have KRAS mutations (18.8% vs 41.7% in MMR proficient tumors, P < 0.05) and tumors with abnormal MLH1/PMS2 tended to harbor BRAF mutations (15.4% vs 4.2% in MMR proficient tumors, P < 0.05).
CONCLUSION: The frequency of sporadic CRCs having BRAF mutation, MLH1 deficiency and MSI in Chinese population may be lower than that in the Western population.
Core tip: Tests for KRAS, BRAF and DNA mismatch repair protein status were important for clinical management of patients with colorectal carcinoma (CRC). Investigations from large samples for these molecular markers were limited in Chinese CRC patients. In the present study, we collected and summarized clinicopathological and molecular data of 535 CRC patients in our institution. These results would help to understand CRC molecular features and guide Lynch syndrome screening, CRC clinical management and individualized therapy in the Chinese population.
-
Citation: Ye JX, Liu Y, Qin Y, Zhong HH, Yi WN, Shi XY.
KRAS andBRAF gene mutations and DNA mismatch repair status in Chinese colorectal carcinoma patients. World J Gastroenterol 2015; 21(5): 1595-1605 - URL: https://www.wjgnet.com/1007-9327/full/v21/i5/1595.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i5.1595
Colorectal carcinoma (CRC) is one of the most common malignancies in the world. Although CRC in China is not as common as that in most Western countries, its incidence has increased steadily in recent years due to living standard improvement and lifestyle change[1].
Based on current knowledge on the carcinogenesis of CRCs, individualized clinical managements have been recommended for CRC patients[2,3]. Testing tumor tissue for predictive or prognostic gene mutations to guide personalized therapy is a rapidly emerging field in pathology.
The epidermal growth factor receptor (EGFR) is a major therapeutic target in CRCs[4]. Activating mutations of the KRAS gene is thought to stimulate the RAS/RAF/MAPK pathway independent of EGFR activation, therefore CRCs with KRAS mutations are resistant to EGFR inhibitors[5]. Although the predictive value of BRAF mutation status for response to EGFR inhibitors is still uncertain[6,7], its prognostic value for CRCs is widely accepted, i.e., patients with the BRAF V600E mutation tend to have a poor prognosis[8-10]. Moreover, the presence of the BRAF V600E mutation in a MLH1 deficient CRC indicates that it is a sporadic rather than a Lynch syndrome associated carcinoma with high level of microsatellite instability (MSI-H)[11]. MSI-H CRCs are either caused by germ line mutations or epigenetic silencing of DNA mismatch repair (MMR) genes[2] and have distinct clinical and pathological features. Detection of mismatch repair protein deficiency or MSI status is not only useful for screening Lynch syndrome but also can serve as a prognostic marker for favorable outcome. In addition, it is also a negative predictive marker for fluoropyrimidine-based chemotherapy in patients with stage II disease[12,13]. From 2010, mutation analysis for KRAS and BRAF as well as MSI/MMR testing has been suggested to be performed for CRC patients by the National Comprehensive Cancer Network (NCCN) clinical practice guidelines[14].
Frequencies of KRAS and BRAF mutations and MSI-H in CRCs have been widely studied in Western populations. Among them, KRAS mutations are the most frequent molecular changes, with a frequency ranging from 22% to 46.7%[15-20], while BRAF mutations are less frequent, with a frequency ranging from 5.0% to 21.8%[15,21-25]. Several studies have reported the frequencies of KRAS and BRAF mutations in Chinese CRC patients in the English literature. However, most of the studies performed with limited sample size and the results were controversial, with a frequency of KRAS mutations ranging from 19.7% to 43.9% and that of BRAF mutations ranging from 1.7% to 25.4%[26-32]. MSI CRCs account for approximately 15-20% of all CRCs in Western countries[17,19,20,33-35]. Limited reports from China show a frequency of MSI CRCs in Chinese patients (ranging from 9.6% to 13%) lower than that in Western populations but close to reports from Korea[36-40].
Information from previous studies raises the possibility that geographic and/or racial differences may present between Chinese and Western populations. Therefore, more data are needed to further clarify the characteristics of these important molecular changes in Chinese CRC patients. In the present study, we collected the data of CRC patients treated from 2010 to 2013 in our department and hope to provide more information about CRC in Chinese patients.
We searched the pathology database of the Department of Pathology of Peking University Third Hospital from 2010 to 2013 for primary or metastatic colorectal adenocarcinomas. Five hundred and thirty-five patients with 538 tumors tested for KRAS and BRAF mutations or MMR protein expression were collected. The pathology records and clinical charts were reviewed to obtain the following information: patient gender, age, anatomic site of tumor, morphological characteristics (histologic type, tumor grade, depth of tumor penetration, lymph node involvement, lymphatic or vascular invasion, perineural invasion), and history of metastasis or other tumors. Base on clinical data, primary locations of tumors were defined as the right colon (from the cecum through the transverse colon), left colon (from the splenic flexure through the rectosigmoid flexure) and rectum (15 cm above the anal verge). Tumors were staged according to the seventh edition of the American Joint Commission on Cancer (AJCC) TNM staging system. Well to moderately differentiated tumors were grouped together and tumors diagnosed as mucinous adenocarcinomas, signet-ring cell carcinomas and adenocarcinomas with mucinous or signet-ring cell differentiation were recorded as mucin-producing tumors.
Data and tissue collection was approved by the Ethics Committee of Peking University Health Science Center, following the ethical guidelines of the 1975 Declaration of Helsinki.
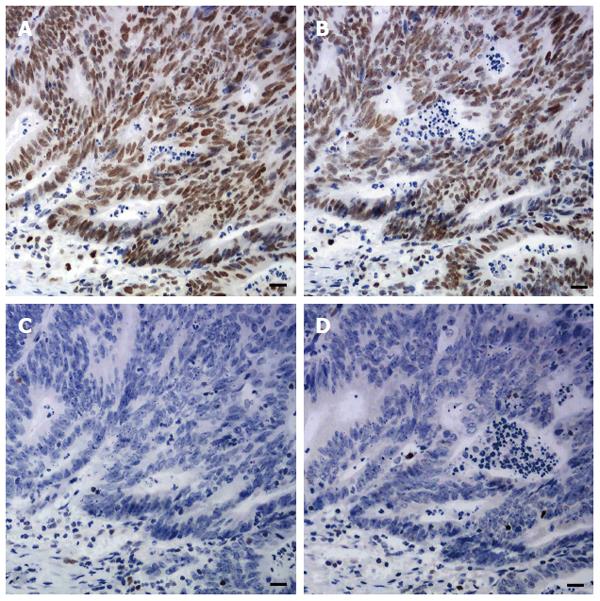
Sections of one representative block of each tumor were incubated with antibodies to MLH1, MSH2, MSH6 and PMS2. Standard heat-induced epitope retrieval in EDTA solution (pH 9.0 for MLH1, PMS2, and MSH2, pH 6.0 for MSH6) and immunostaining and signal detection using the Dako EnVision Detection System and a Dako Autostainer (Dako North America Inc., California, United States) were performed. The sources, dilutions, and incubation time of each primary antibody are listed in Table 1. Any tumor cell with nuclear staining was defined as positive for that marker. Positive staining for all these proteins was regarded as proficient MMR (pMMR). Negative staining for any of these four proteins was regarded as deficient MMR (dMMR) (Figure 1). Since family history and genetic information were unavailable, no attempt was made to further classify patients into with Lynch syndrome or sporadic MSI-H CRCs.
| Antibody | Supplied by | Clone No. | Positive signal localization | Dilution | Incubation temperature (°C) | Incubationperiod (h) |
| MLH1 | Novocastra, UK | 14 | Nuclear | 1:50 | 37 | 2 |
| MSH2 | Novocastra, UK | FE11 | Nuclear | 1:20 | 37 | 2 |
| MSH6 | Novocastra, UK | BC/44 | Nuclear | 1:80 | 37 | 2 |
| PSM2 | Novocastra, UK | EP51 | Nuclear | 1:30 | 37 | 2 |
As previously described by Zhong et al[41] and Liu et al[42], genomic DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tumor tissue sections with the Qiagen Blood and Tissue Kit (QiagenInc, Valencia, CA) according to the manufacturer’s protocol. The KRAS exon 2 and BRAF exon 15 were amplified by polymerase chain reaction (PCR) using PromegaGoTaq Hot Start Colorless Master Mixes (Promega Corporation, Madison, WI, United States). The specific primers and sizes of the expected amplicons are presented in Table 2. Genomic DNA of 50-100 ng was amplified in a 50 μL reaction system containing 25 μL of Hot Start Colorless Master Mix and 5 μL of 10 μmol/L primer mix. The PCR reaction conditions consisted of 2 min at 95 °C, 40 cycles of 94 °C for 30 s, 55 °C for 40 s and 72 °C for 1 min, and 72 °C for 7 min. Five microliters of the PCR product was analyzed on a 1.2% agarose gel with 100 to 600 bp DNA marker. Gels were visualized on a BioRad Gel Doc 2000TM system and Quantity One software (BioRad, Hercules, CA, United States). The resulting PCR amplicons were purified and sequenced in both directions using the BigDye Terminator kit and an ABI Prism 3500 DNA Analyzer (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Mutant cases were validated by a second independent PCR and sequencing. The sequencing results were observed with ABI Sequence Scanner software and compared with the reference sequences of the BRAF and KRAS genes from NCBI database to mark the position of nucleotide change.
| Exon | Primer No. | Primer sequence 5’-3’ | AT (°C) | Product size (bp) |
| KRAS 2 | 2-F | TAGTCACATTTTCATTATTTTTAT | 55 | 160 |
| 2-R | AGATTTACCTCTATTGTTGGAT | |||
| BRAF 15 | 15-F | ATCTACTGTTTTCCTTTACTTACT | 55 | 160 |
| 15-R | ATTCTTACCATCCACAAAATG |
For nine cases, mutations in KRAS exon 2 and BRAF exon 15 were identified by amplification refractory mutation system (ARMS)-PCR. Briefly, FFPE tissues were digested using 20 mg/mL proteinase K in ATL buffer (Qiagen) overnight at 56 °C. DNA isolation was performed with the QIAamp DNA Mini Kit (Qiagen) according to the manufacturer’s protocol. KRAS and BRAF mutation status was assessed with Human KRAS Gene 7 Mutations Fluorescence Polymerase Chain Reaction Diagnostic Kit and Human BRAF Gene V600E Mutations Fluorescence Polymerase Chain Reaction Diagnostic Kit (Amoy Diagnostics Co. Ltd, Xiamen, China) on the Agilent-Stratagene M × 3000P Q-PCR System (Agilent Technologies, Santa Clara, CA), according to the manufacturers’ instructions. The 7 most common KRAS mutations (p.G12D, p.G12V, p.G12A, p.G12C, p.G12S, p.G12R, and p.G13D) in CRCs were detected. The reaction conditions included 1 cycle at 95 °C for 5min; 15 cycles at 95 °C for 25 s, 64 °C for 20 s, 70 °C for 20 s; 31 cycles at 93 °C for 25 s, 60 °C for 35 s, 72 °C for 20 s. Fluorescence signals were collected at 60 °C.
Clinical and pathological characteristics are summarized as percentages. Mutation rates of KRAS and BRAF and overall or each MMR protein expression deficiency were also calculated. Clinical and pathological characteristics were compared across different subgroups and analyzed with χ2 test. Correlations between KRAS or BRAF mutations and dMMR status were also explored. P values < 0.05 were considered statistically significant.
Of these 535 patients, males were slightly more than females with a male to female ratio of 1.34:1. Patient age at presentation ranged from 21 to 95 years (median, 65 years) with 10.7% of patients < 50 years at diagnosis. Most patients (61.2%) presented with stage II or III disease. Almost all patients had solitary primary tumor except that three patients had synchronous tumors (two patients had two tumors confined in the left colon, one had two tumors with one in the right colon and the other in the rectum). In 6 patients, only metastatic lesions were available for testing, and 3 of them received neoadjuvant therapy. Both preoperative biopsy and radical specimens were tested for KRAS and BRAF mutations in two patients, and both primary and metastatic lesions were tested in 5 patients. Since there was no discrepancy between biopsy and radical resection specimens or between primary and metastatic tumors, the results from two different tests were recorded as once. Clinical information of all studied patients is summarized in Table 3.
| Clinical feature | Total cases | |
| Gender | Male | 306 (57.2) |
| Female | 229 (42.8) | |
| Age (yr) | < 50 | 57 (10.7) |
| ≥ 50 | 478 (89.3) | |
| Location | Right colon | 177 (32.9) |
| Left colon | 165 (30.7) | |
| Rectum | 196 (36.4) | |
| Tumor differentiation | Poor | 94 (17.5) |
| Well-moderate | 444 (82.5) | |
| Tumor stage | I | 95 (17.7) |
| II | 168 (31.2) | |
| III | 215 (40.0) | |
| IV | 50 (9.3) | |
| Not available | 10 (1.9) | |
KRAS status was ascertained for 485 patients including 488 tumors. The over-all mutation rate was 37.9% (185/488). KRAS mutations identified in codon 12 included G12D (n = 90, 18.4%), G12V (n = 37, 7.6%), G12C (n = 9, 1.8%), G12A (n = 8, 1.6%), G12S (n = 7, 1.4%), and G12R (n = 1, 0.2%). Mutations in KRAS codon 13 included G13D (n = 31, 6.4%), G13C (n = 1, 0.2%) and G13G (silent mutation) (n = 1, 0.2%). One patient was identified with concomitant KRAS mutations in codon 12 and codon 33 (G12D and D33N). Compared with patients < 50 years old, KRAS mutations in patients ≥ 50 years old were more common (39.8% vs 22%, P < 0.05). The rate of KRAS mutations was not significantly associated with gender, tumor location, tumor differentiation, stage or mucin production (Table 4).
| Clinicopathological feature | KRAS | BRAF | ||
| Mutant/tested cases | P value | Mutant/tested cases | P value | |
| Gender | ||||
| Male | 100/277 (36.1) | 0.336 | 7/254 (2.8) | 0.048 |
| Female | 84/208 (40.4) | 13/196 (6.6) | ||
| Age (yr) | ||||
| < 50 | 11/50 (22) | 0.014 | 3/47 (6.4) | 0.758 |
| ≥ 50 | 173/435 (39.8) | 17/403 (4.2) | ||
| Location | ||||
| Right colon | 67/161 (41.6) | 0.318 | 14/146 (9.6) | 0.001 |
| Left colon | 50/150 (33.3) | 3/167 (2.1) | ||
| Rectum | 68/177 (38.4) | 3/140 (1.8) | ||
| Mucin production | ||||
| With | 41/108 (38) | 0.990 | 10/102 (9.8) | 0.006 |
| Without | 144/380 (37.9) | 10/351 (2.8) | ||
| Tumor differentiation | ||||
| Poor | 25/84 (29.8) | 0.091 | 7/74 (9.5) | 0.045 |
| Well-moderate | 160/404 (39.6) | 13/379 (3.4) | ||
| Tumor stage | ||||
| I-II | 85/232 (36.6) | 0.553 | 10/218 (4.6) | 0.918 |
| III-IV | 97/247 (39.3) | 10/228 (4.4) | ||
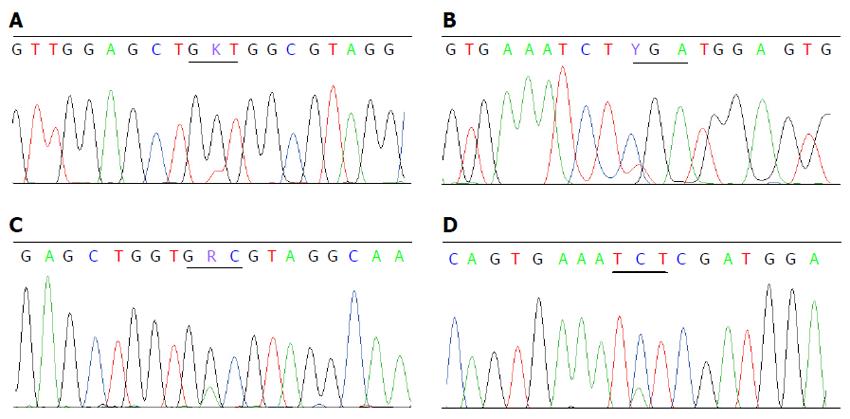
Twenty (4.4%) mutations were detected in 453 CRCs from 450 patients. Sixteen mutations were V600E and the other four were D594G, D594N, R603stop and S602Y. There were no concomitant KRAS and BRAF V600E mutations, but one patient had concomitant mutations of KRAS G12V (Figure 2A) and BRAF R603stop (Figure 2B), and one with KRAS G13D (Figure 2C) and BRAF S602Y (Figure 2D). The proportion of mutant BRAF was higher in females (6.6% vs 2.8% in males, P < 0.05), in tumors located in the right colon (9.6% vs 2.1% in the left colon and 1.8% in the rectum, P < 0.05), with poor differentiation (9.5% vs 3.4% with well-moderate differentiation, P < 0.05) and with mucinous appearance (9.8% vs 2.8% without mucin, P < 0.05). BRAF mutations were not related with patient’s age or tumor stage (Table 4).
Overall, 11.4% of tumors showed loss of expression for at least one MMR protein, and the deficient rates of MLH1, MSH2, MSH6 and PMS2 in studied patients were 5.8% (28/481), 2.7% (13/481), 3.8% (18/480) and 6.5% (19/293), respectively. The proportion of dMMR tumors varied by site with a significantly higher rate (20.5%) in tumors located in the right colon compared to those in the left colon (9.2%) and rectum (5.1%, P < 0.001). There were more stages I-II dMMR tumors than stages III-IV dMMR tumors (15.0% vs 7.7%, P < 0.05) and dMMR tumors tended to show mucinous differentiation. In females, a higher frequency of MLH1/PMS2 deficiency was found (9.2% vs 4.4% in males, P < 0.05). Loss of MSH2/MSH6 expression was more frequent in patients < 50 years old than in those ≥ 50 years old (12.0% vs 4.0%, P < 0.05). Although dMMR tumors were more often in tumors with poor differentiation than in those with well-moderate differentiation, it did not show a significant difference (16.5% vs 10.4%, P > 0.05) (Table 5).
| Clinicopathological feature | dMMR | MLH1/PMS2 | MSH2/MSH6 | ||||
| Defective/tested | P value | Defective/tested | P value | Defective/tested | P value | ||
| Total | 55/481 (11.4) | 31/481 (6.4) | 24/481 (5.0) | ||||
| Gender | Male | 25/272 (9.2) | 0.095 | 12/272 (4.4) | 0.034 | 13/272 (4.8) | 0.970 |
| Female | 29/206 (14.1) | 19/206 (9.2) | 10/206 (4.9) | ||||
| Age (yr) | < 50 | 9/50 (18.0) | 0.114 | 3/50 (6.0)) | 1.000 | 6/50 (12.0) | 0.031 |
| ≥ 50 | 45/428 (10.5) | 28/428 (6.5) | 17/428 (4.0) | ||||
| Location | Right colon | 33/161 (20.5) | 0.000 | 23/161 (14.3) | 0.000 | 10/161 (6.2) | 0.446 |
| Left colon | 13/142 (9.2) | 5/142 (3.5) | 8/142 (5.6) | ||||
| Rectum | 9/178 (5.1) | 3/178 (1.7) | 6/178 (3.4) | ||||
| Mucin production | With | 20/99 (20.2) | 0.002 | 11/99 (11.1) | 0.034 | 9/99 (9.1) | 0.065 |
| Without | 35/382 (9.2) | 20/382 (5.2) | 15/382 (3.9) | ||||
| Tumor differentiation | Poor | 13/79 (16.5) | 0.125 | 6/79 (7.6) | 0.649 | 7/79 (8.9) | 0.148 |
| Well-moderate | 42/402 (10.4) | 25/402 (6.2) | 17/402 (4.2) | ||||
| Tumor stage | I-II | 37/246 (15.0) | 0.012 | 20/246 (8.1) | 0.127 | 17/246 (6.9) | 0.049 |
| III-IV | 18/234 (7.7) | 11/234 (4.7) | 7/234 (3.0) | ||||
Less KRAS mutants were seen in dMMR tumors than in pMMR tumors (18.8% vs 41.7%, P < 0.05). BRAF mutation rate was higher in dMMR tumors than in pMMR tumors, although it did not show a significant difference (9.3% vs 4.2%, P > 0.05). Nevertheless, tumors with defected MLH1/PMS2 tended to harbor BRAF mutations compared with pMMR tumors (15.4% vs 4.2%, P < 0.05). No BRAF mutation was detected in tumors with MSH2/MSH6 deficiency (Table 6).
As predictive and/or prognostic biomarkers, KRAS and BRAF mutation tests are important for predicting response to EGFR-targeted therapy and prognosis of CRC patients. However, regional and racial differences in mutation rates may be present[15-19,38,43,44]. Marked differences between Chinese population and other countries were also observed[26-32], which need to be further confirmed in studies based on large sample size.
In the present study, we found an overall KRAS mutation rate of 37.9% in CRCs, which was close to most of the previous reports either about Chinese or other ethnicities[15-18,20,26-30,43,45-49]. Studies based on clinical practices or CRC cell line models showed that CRCs with KRAS G13D mutation responded partly to cetuximab and panitumumab[50,51]. Therefore, the subtype of KRAS mutations may also have clinical implications. In our cases, the major mutant types were G12D, G12V and G13D, accounting for 85% of all mutations. About 6.4% of CRC patients in our group were found to have KRAS G13D mutation, which was also consistent with other reports[15,16,52]. One uncommon finding in our cases was concomitant KRAS mutations in codon 12 (G12D) and codon 33 (D33N). The patient was a 59-year-old male with a poorly differentiated tumor located in the left colon. To our knowledge, the latter mutation is the first identified mutant in CRCs. Other non-hot mutations were also found in previous reports, however, clinical impact of these mutations is unknown[53-55]. CRCs or colorectal cancer cell lines with special KRAS mutations seemed to have different malignant potential and proliferative ability[50,56], which may lead to different responses to anti-EGFR agents. However, detailed mechanism needs further exploration.
Correlations between KRAS status and clinicopathological features are controversial. Some of the previous reports showed that the frequency of KRAS mutation was in association with age, gender, tumor grade or stage but some did not[15,16,19,26,28,30]. Reports from four independent Chinese groups[28,30,45,52] showed that KRAS mutations were associated with patient gender, but not with patient age. In a study including 966 CRCs, Gao et al[57] observed that KRAS mutations were not only associated with patient gender and age, but also with tumor differentiation. Similar inconsistent results were also presented in studies on other ethnicities[46,55]. In our series of cases, we could only find that tumors in patients older than 50 years tended to harbor mutant KRAS. These diverse findings suggest that the difference in KRAS mutation rates in different groups is too minute to be declared with limited samples. The different criteria for age division might also be a cause[46,58].
BRAF mutation rates are significantly different in previous studies of non-Chinese population[15,21-25]. The lowest mutation rate came from a study on Japanese CRC patients (3%), followed by reports from Russia and Israel (4.1% and 5%, respectively)[24,43,44]. On the contrary, reports about European and Americans showed significantly higher rates of BRAF mutation, which were mostly around 15%[19,20,23,59]. In most of the studies about Chinese population[30,31,48], low percentage of CRCs were found to harbor mutant BRAF, ranging from 1.7% to 7%. The only exception is a report from Mao et al[32], which showed that the frequency of BRAF mutations was as high as 25.4% in a group of Chinese CRC patients. However, their report also showed that 24% of KRAS mutant cases had concomitant BRAF V600E mutation. This phenomenon was inconsistent with the general opinion that concomitant KRAS and BRAF V600E mutations were rare if not mutually exclusive[15,45,60,61]. Considering the relatively limited number of cases included in their study (69 cases), the result may be not representative. In our study, we also observed a low BRAF mutation rate of 4.4%, which was consistent with most of other reports in Chinese and was similar to that in Japanese[44]. Our finding together with other reports supports the opinion that the frequency of BRAF mutation varies among different races and/or regions[21,31,61]. In addition, no patient was found to have concomitant mutations for KRAS and BRAF V600E in our cases. However, of the four cases with non-V600E BRAF mutation, two had concomitant KRAS mutation. Although the significance for these uncommon mutations was uncertain[22], our findings further confirmed the rarity of concomitant KRAS and BRAF V600E mutations in CRCs. Also in accordance with previous reports[21,61], we found BRAF mutation to be more common in females, in proximally located poorly differentiated tumors, or in mucin-producing tumors, although no significant association was found between BRAF mutations and patient age or tumor stage. This may be partially because the number of BRAF mutant cases is limited in our study.
CRCs with deficient MMR exhibit high frequency MSI and have distinctive clinicopathological features, biological behavior and clinical treatments compared to CRCs with pMMR, which in most cases are microsatellite stable (MSS)[2]. dMMR CRCs accounted for about 15%-20% of all CRCs in reports from Western countries[12,13,17,19,20,33,34,62], and are more common in stage II tumors (up to 22%) than in stage III tumors (up to 14%)[15,35]. Information about dMMR CRCs in Chinese population is limited. Huang et al[36] and Jin et al[37] using PCR-based MSI testing showed that the frequencies of MSI-H CRCs in their cases from Southeast China were 11.9% to 13%, which were slightly lower than most of the reports from Western populations[15,17,19,20,33-35]. Detecting MMR protein loss by IHC showed similar efficiency to more complex gene analysis for MSI or MMR gene mutations[63,64]. The current study is one of the largest series that analyzed the MMR status by IHC in Chinese CRCs. Again, our data showed a low frequency of dMMR in Chinese CRCs. The overall frequency of dMMR CRCs and the frequencies of dMMR CRCs in early stage (stage I-II) and advanced stage (stage III-IV) CRCs were 11.4%, 15.4% and 7.7%, respectively. Our findings, together with Huang et al [36]’s and Jin et al[37]’s reports, suggest that CRCs in Chinese might have different genetic background from that in Western populations, and were less likely to have MSI.
Of note, the majority of dMMR CRCs are sporadic and caused by inactivation of MLH1 (about 95%). Lynch syndrome, which is caused by germ-line mutations of MMR genes, accounts for about 20% of the dMMR CRCs, and MLH1 mutation is found in approximately 40% of the cases[65]. Therefore, the vast majority of dMMR CRCs should have MLH1 deficiency. By using IHC, our study was able to inform which MMR proteins were lost in an MSI CRC, and provide more information than previous reports in Chinese CRCs. Surprisingly, our data showed only 50.9% of dMMR CRCs with MLH1 loss, which was much lower than expected. Moreover, BRAF mutations have been reported in 33% to 60% of MSI-H tumors particularly in tumors with methylation of the MLH1 promoter[20,35,66,67]. Among our dMMR cases, only 15.4% had BRAF mutations, which was also much lower than previous reports. These findings suggest that the low frequency of dMMR CRCs in our series is likely caused by including less sporadic dMMR CRCs. Our finding further indicated that ethnic and geographic differences might be present in Chinese dMMR CRCs, although further investigations, such as germ-line mutation analysis of MMR genes and/or analysis of MLH1 promoter methylation, are needed to clarify this possibility. Since Hampel et al[68]’s study has showed that the widely used Amsterdam or Bethesda screening guideline may miss as many as 22% of patients with Lynch syndrome, feasible and economic IHC technique was suggested to perform on all newly diagnosed CRCs to screen Lynch syndrome and guide clinical management for MSI patients[69,70]. In this case, there will be more and more data coming out to provide detailed information of Chinese CRCs.
In summary, our results show a low frequency of BRAF mutations and MMR deficiency, especially less MLH1 deficiency, in a large series of Chinese CRC patients. It suggests that CRCs are less likely to have MSI in Chinese populations, and it is probably caused by the fact that there are less sporadic MMR deficient CRCs in Chinese. However, additional epidemiologic data and genetic investigations are needed to confirm the difference.
Mutation analyses for KRAS and BRAF as well as microsatellites instability/DNA mismatch repair (MSI/MMR) testing have been suggested to be performed for patients with colorectal carcinomas (CRCs). Information from previous studies raises the possibility that geographic and/or racial differences may be present between Chinese and Western populations. Investigations from large samples for these molecular markers were limited in Chinese CRC patients.
The current study demonstrated that a low frequency of BRAF mutations and MMR deficiency, especially MLH1 deficiency, in a large series of Chinese CRC patients.
Results in the present study suggest that CRCs are less likely to have MSI in Chinese populations, and it is probably caused by the fact that there are less sporadic MMR deficient CRCs in Chinese.
These results would help to understand CRC molecular features and guide Lynch syndrome screening, CRC clinical management and individualized therapy in the Chinese population.
The study reports KRAS and BRAF gene mutations and MMR protein expression status in large number of Chinese CRC patients. The results corroborate the earlier findings and show high rates of mutations in the KRAS gene. Associations of these genetic markers with various clinicopathological parameters have also been made. The study also shows less MMR defects in these patients.
P- Reviewer: Roychoudhury S S- Editor: Yu J L- Editor: Wang TQ E- Editor: Wang CH
| 1. | Yang G, Wang Y, Zeng Y, Gao GF, Liang X, Zhou M, Wan X, Yu S, Jiang Y, Naghavi M. Rapid health transition in China, 1990-2010: findings from the Global Burden of Disease Study 2010. Lancet. 2013;381:1987-2015. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1375] [Cited by in RCA: 1446] [Article Influence: 120.5] [Reference Citation Analysis (0)] |
| 2. | Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490-1502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1965] [Cited by in RCA: 2294] [Article Influence: 208.5] [Reference Citation Analysis (1)] |
| 3. | Kelley RK, Van Bebber SL, Phillips KA, Venook AP. Personalized medicine and oncology practice guidelines: a case study of contemporary biomarkers in colorectal cancer. J Natl Compr Canc Netw. 2011;9:13-25. [PubMed] |
| 4. | Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1515] [Cited by in RCA: 1568] [Article Influence: 92.2] [Reference Citation Analysis (0)] |
| 5. | Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626-1634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2504] [Cited by in RCA: 2403] [Article Influence: 141.4] [Reference Citation Analysis (0)] |
| 6. | Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023-1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1610] [Cited by in RCA: 1733] [Article Influence: 144.4] [Reference Citation Analysis (0)] |
| 7. | Maughan TS, Adams RA, Smith CG, Meade AM, Seymour MT, Wilson RH, Idziaszczyk S, Harris R, Fisher D, Kenny SL. Addition of cetuximab to oxaliplatin-based first-line combination chemotherapy for treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet. 2011;377:2103-2114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 766] [Cited by in RCA: 763] [Article Influence: 54.5] [Reference Citation Analysis (2)] |
| 8. | Sclafani F, Gullo G, Sheahan K, Crown J. BRAF mutations in melanoma and colorectal cancer: a single oncogenic mutation with different tumour phenotypes and clinical implications. Crit Rev Oncol Hematol. 2013;87:55-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 56] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 9. | Xu Q, Xu AT, Zhu MM, Tong JL, Xu XT, Ran ZH. Predictive and prognostic roles of BRAF mutation in patients with metastatic colorectal cancer treated with anti-epidermal growth factor receptor monoclonal antibodies: a meta-analysis. J Dig Dis. 2013;14:409-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 10. | Pai RK, Jayachandran P, Koong AC, Chang DT, Kwok S, Ma L, Arber DA, Balise RR, Tubbs RR, Shadrach B. BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: an aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. Am J Surg Pathol. 2012;36:744-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 154] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 11. | Loughrey MB, Waring PM, Tan A, Trivett M, Kovalenko S, Beshay V, Young MA, McArthur G, Boussioutas A, Dobrovic A. Incorporation of somatic BRAF mutation testing into an algorithm for the investigation of hereditary non-polyposis colorectal cancer. Fam Cancer. 2007;6:301-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 127] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 12. | Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219-3226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1132] [Cited by in RCA: 1212] [Article Influence: 80.8] [Reference Citation Analysis (0)] |
| 13. | Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1585] [Cited by in RCA: 1636] [Article Influence: 74.4] [Reference Citation Analysis (0)] |
| 14. | NCCN Clinical practice guidelines in oncology. Colon Cancer. Version 2. Accessed: 2 June 2014; Available from: http://www.nccn.org/professionals/physician_gls/pdf/ colon.pdf. |
| 15. | Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 801] [Cited by in RCA: 917] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 16. | Pérez-Ruiz E, Rueda A, Pereda T, Alcaide J, Bautista D, Rivas-Ruiz F, Villatoro R, Pérez D, Redondo M. Involvement of K-RAS mutations and amino acid substitutions in the survival of metastatic colorectal cancer patients. Tumour Biol. 2012;33:1829-1835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Vasovcak P, Pavlikova K, Sedlacek Z, Skapa P, Kouda M, Hoch J, Krepelova A. Molecular genetic analysis of 103 sporadic colorectal tumours in Czech patients. PLoS One. 2011;6:e24114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 18. | Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet. 2010;375:1030-1047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1208] [Cited by in RCA: 1178] [Article Influence: 78.5] [Reference Citation Analysis (0)] |
| 19. | Naguib A, Mitrou PN, Gay LJ, Cooke JC, Luben RN, Ball RY, McTaggart A, Arends MJ, Rodwell SA. Dietary, lifestyle and clinicopathological factors associated with BRAF and K-ras mutations arising in distinct subsets of colorectal cancers in the EPIC Norfolk study. BMC Cancer. 2010;10:99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 20. | Zlobec I, Bihl MP, Schwarb H, Terracciano L, Lugli A. Clinicopathological and protein characterization of BRAF- and K-RAS-mutated colorectal cancer and implications for prognosis. Int J Cancer. 2010;127:367-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 94] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 21. | Chen D, Huang JF, Liu K, Zhang LQ, Yang Z, Chuai ZR, Wang YX, Shi DC, Huang Q, Fu WL. BRAFV600E mutation and its association with clinicopathological features of colorectal cancer: a systematic review and meta-analysis. PLoS One. 2014;9:e90607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 116] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 22. | Tie J, Gibbs P, Lipton L, Christie M, Jorissen RN, Burgess AW, Croxford M, Jones I, Langland R, Kosmider S. Optimizing targeted therapeutic development: analysis of a colorectal cancer patient population with the BRAF(V600E) mutation. Int J Cancer. 2011;128:2075-2084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 186] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 23. | Shaukat A, Arain M, Thaygarajan B, Bond JH, Sawhney M. Is BRAF mutation associated with interval colorectal cancers? Dig Dis Sci. 2010;55:2352-2356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 24. | Rozek LS, Herron CM, Greenson JK, Moreno V, Capella G, Rennert G, Gruber SB. Smoking, gender, and ethnicity predict somatic BRAF mutations in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:838-843. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 60] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | Phipps AI, Buchanan DD, Makar KW, Burnett-Hartman AN, Coghill AE, Passarelli MN, Baron JA, Ahnen DJ, Win AK, Potter JD. BRAF mutation status and survival after colorectal cancer diagnosis according to patient and tumor characteristics. Cancer Epidemiol Biomarkers Prev. 2012;21:1792-1798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 26. | Shen Y, Wang J, Han X, Yang H, Wang S, Lin D, Shi Y. Effectors of epidermal growth factor receptor pathway: the genetic profiling ofKRAS, BRAF, PIK3CA, NRAS mutations in colorectal cancer characteristics and personalized medicine. PLoS One. 2013;8:e81628. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 85] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 27. | Yunxia Z, Jun C, Guanshan Z, Yachao L, Xueke Z, Jin L. Mutations in epidermal growth factor receptor and K-ras in Chinese patients with colorectal cancer. BMC Med Genet. 2010;11:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 28. | Liou JM, Wu MS, Shun CT, Chiu HM, Chen MJ, Chen CC, Wang HP, Lin JT, Liang JT. Mutations in BRAF correlate with poor survival of colorectal cancers in Chinese population. Int J Colorectal Dis. 2011;26:1387-1395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 29. | Hsieh LL, Er TK, Chen CC, Hsieh JS, Chang JG, Liu TC. Characteristics and prevalence of KRAS, BRAF, and PIK3CA mutations in colorectal cancer by high-resolution melting analysis in Taiwanese population. Clin Chim Acta. 2012;413:1605-1611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 30. | Shen H, Yuan Y, Hu HG, Zhong X, Ye XX, Li MD, Fang WJ, Zheng S. Clinical significance of K-ras and BRAF mutations in Chinese colorectal cancer patients. World J Gastroenterol. 2011;17:809-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 27] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 31. | Liao W, Liao Y, Zhou JX, Xie J, Chen J, Huang W, Luo R. Gene mutations in epidermal growth factor receptor signaling network and their association with survival in Chinese patients with metastatic colorectal cancers. Anat Rec (Hoboken). 2010;293:1506-1511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 32. | Mao C, Zhou J, Yang Z, Huang Y, Wu X, Shen H, Tang J, Chen Q. KRAS, BRAF and PIK3CA mutations and the loss of PTEN expression in Chinese patients with colorectal cancer. PLoS One. 2012;7:e36653. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 33. | Kim GP, Colangelo LH, Wieand HS, Paik S, Kirsch IR, Wolmark N, Allegra CJ. Prognostic and predictive roles of high-degree microsatellite instability in colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project Collaborative Study. J Clin Oncol. 2007;25:767-772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 268] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 34. | Sinicrope FA, Foster NR, Yoon HH, Smyrk TC, Kim GP, Allegra CJ, Yothers G, Nikcevich DA, Sargent DJ. Association of obesity with DNA mismatch repair status and clinical outcome in patients with stage II or III colon carcinoma participating in NCCTG and NSABP adjuvant chemotherapy trials. J Clin Oncol. 2012;30:406-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 35. | Sinicrope FA, Foster NR, Thibodeau SN, Marsoni S, Monges G, Labianca R, Kim GP, Yothers G, Allegra C, Moore MJ. DNA mismatch repair status and colon cancer recurrence and survival in clinical trials of 5-fluorouracil-based adjuvant therapy. J Natl Cancer Inst. 2011;103:863-875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 406] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 36. | Huang YQ, Yuan Y, Ge WT, Hu HG, Zhang SZ, Zheng S. Comparative features of colorectal and gastric cancers with microsatellite instability in Chinese patients. J Zhejiang Univ Sci B. 2010;11:647-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 37. | Jin HY, Liu X, Li VK, Ding Y, Yang B, Geng J, Lai R, Ding S, Ni M, Zhao R. Detection of mismatch repair gene germline mutation carrier among Chinese population with colorectal cancer. BMC Cancer. 2008;8:44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 38. | Jeon CH, Lee HI, Shin IH, Park JW. Genetic alterations of APC, K-ras, p53, MSI, and MAGE in Korean colorectal cancer patients. Int J Colorectal Dis. 2008;23:29-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 39. | Jung SB, Lee HI, Oh HK, Shin IH, Jeon CH. Clinico-pathologic Parameters for Prediction of Microsatellite Instability in Colorectal Cancer. Cancer Res Treat. 2012;44:179-186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 40. | Xiao XY, Zhou XY, Sun MH, Yan G, Du X. [Microsatellite instability of sporadic colorectal carcinomas and its clinicopathological significance]. Zhonghua Zhong Liu Za Zhi. 2006;28:289-293. [PubMed] |
| 41. | Zhong H, Liu Y, Talmor M, Wu B, Hui P. Deparaffinization and lysis by hydrothermal pressure (pressure cooking) coupled with chaotropic salt column purification: a rapid and efficient method of DNA extraction from formalin-fixed paraffin-embedded tissue. Diagn Mol Pathol. 2013;22:52-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 42. | Liu Y, Wu BQ, Zhong HH, Hui P, Fang WG. Screening for EGFR and KRAS mutations in non-small cell lung carcinomas using DNA extraction by hydrothermal pressure coupled with PCR-based direct sequencing. Int J Clin Exp Pathol. 2013;6:1880-1889. [PubMed] |
| 43. | Yanus GA, Belyaeva AV, Ivantsov AO, Kuligina ESh, Suspitsin EN, Mitiushkina NV, Aleksakhina SN, Iyevleva AG, Zaitseva OA, Yatsuk OS. Pattern of clinically relevant mutations in consecutive series of Russian colorectal cancer patients. Med Oncol. 2013;30:686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 44. | Umeda Y, Nagasaka T, Mori Y, Sadamori H, Sun DS, Shinoura S, Yoshida R, Satoh D, Nobuoka D, Utsumi M. Poor prognosis of KRAS or BRAF mutant colorectal liver metastasis without microsatellite instability. J Hepatobiliary Pancreat Sci. 2013;20:223-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 45. | Zhu XL, Cai X, Zhang L, Yang F, Sheng WQ, Lu YM, Du X, Zhou XY. [KRAS and BRAF gene mutations in correlation with clinicopathologic features of colorectal carcinoma in Chinese]. Zhonghua Bing Li Xue Za Zhi. 2012;41:584-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 46. | Samowitz WS, Curtin K, Schaffer D, Robertson M, Leppert M, Slattery ML. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: a population-based study. Cancer Epidemiol Biomarkers Prev. 2000;9:1193-1197. [PubMed] |
| 47. | Xu XM, Qian JC, Cai Z, Tang T, Wang P, Zhang KH, Deng ZL, Cai JP. DNA alterations of microsatellite DNA, p53, APC and K-ras in Chinese colorectal cancer patients. Eur J Clin Invest. 2012;42:751-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 48. | Li HT, Lu YY, An YX, Wang X, Zhao QC. KRAS, BRAF and PIK3CA mutations in human colorectal cancer: relationship with metastatic colorectal cancer. Oncol Rep. 2011;25:1691-1697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 49. | Molinari F, Felicioni L, Buscarino M, De Dosso S, Buttitta F, Malatesta S, Movilia A, Luoni M, Boldorini R, Alabiso O. Increased detection sensitivity for KRAS mutations enhances the prediction of anti-EGFR monoclonal antibody resistance in metastatic colorectal cancer. Clin Cancer Res. 2011;17:4901-4914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 140] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 50. | Kumar SS, Price TJ, Mohyieldin O, Borg M, Townsend A, Hardingham JE. KRAS G13D Mutation and Sensitivity to Cetuximab or Panitumumab in a Colorectal Cancer Cell Line Model. Gastrointest Cancer Res. 2014;7:23-26. [PubMed] |
| 51. | Messner I, Cadeddu G, Huckenbeck W, Knowles HJ, Gabbert HE, Baldus SE, Schaefer KL. KRAS p.G13D mutations are associated with sensitivity to anti-EGFR antibody treatment in colorectal cancer cell lines. J Cancer Res Clin Oncol. 2013;139:201-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 52. | Wang Q, Zhong M, Lü YL, Yuan J, Wei LX. [Correlation of KRAS gene mutations and clinicopathologic parameters in colorectal carcinoma]. Zhonghua Bing Li Xue Za Zhi. 2012;41:603-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 53. | de Macedo MP, de Lima LG, Begnami MD, de Melo FM, Andrade LD, Lisboa BC, Soares LM, Soares FA, Carraro DM, da Cunha IW. KRAS insertions in colorectal cancer: what do we know about unusual KRAS mutations? Exp Mol Pathol. 2014;96:257-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 54. | Palmirotta R, Savonarola A, Formica V, Ludovici G, Del Monte G, Roselli M, Guadagni F. A novel K-ras mutation in colorectal cancer. A case report and literature review. Anticancer Res. 2009;29:3369-3374. [PubMed] |
| 55. | Brink M, de Goeij AF, Weijenberg MP, Roemen GM, Lentjes MH, Pachen MM, Smits KM, de Bruïne AP, Goldbohm RA, van den Brandt PA. K-ras oncogene mutations in sporadic colorectal cancer in The Netherlands Cohort Study. Carcinogenesis. 2003;24:703-710. [PubMed] |
| 56. | Guerrero S, Casanova I, Farré L, Mazo A, Capellà G, Mangues R. K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer Res. 2000;60:6750-6756. [PubMed] |
| 57. | Gao J, Sun ZW, Li YY, Shen L. [Mutations of KRAS and BRAF in Chinese patients with colorectal carcinoma: analyses of 966 cases]. Zhonghua Bing Li Xue Za Zhi. 2012;41:579-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 58. | Rosty C, Young JP, Walsh MD, Clendenning M, Walters RJ, Pearson S, Pavluk E, Nagler B, Pakenas D, Jass JR. Colorectal carcinomas with KRAS mutation are associated with distinctive morphological and molecular features. Mod Pathol. 2013;26:825-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 59. | Toon CW, Chou A, DeSilva K, Chan J, Patterson J, Clarkson A, Sioson L, Jankova L, Gill AJ. BRAFV600E immunohistochemistry in conjunction with mismatch repair status predicts survival in patients with colorectal cancer. Mod Pathol. 2014;27:644-650. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 60. | Wójcik P, Okoń K, Osuch C, Klimkowska A, Tomaszewska R. BRAF mutations in sporadic colorectal carcinoma from polish patients. Pol J Pathol. 2010;61:23-26. [PubMed] |
| 61. | Hanna MC, Go C, Roden C, Jones RT, Pochanard P, Javed AY, Javed A, Mondal C, Palescandolo E, Van Hummelen P. Colorectal cancers from distinct ancestral populations show variations in BRAF mutation frequency. PLoS One. 2013;8:e74950. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 62. | Koopman M, Kortman GA, Mekenkamp L, Ligtenberg MJ, Hoogerbrugge N, Antonini NF, Punt CJ, van Krieken JH. Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br J Cancer. 2009;100:266-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 291] [Cited by in RCA: 379] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 63. | Kheirelseid EA, Miller N, Chang KH, Curran C, Hennessey E, Sheehan M, Kerin MJ. Mismatch repair protein expression in colorectal cancer. J Gastrointest Oncol. 2013;4:397-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 35] [Reference Citation Analysis (0)] |
| 64. | Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, Walsh-Vockley C, Petersen GM, Walsh MD, Leggett BA. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043-1048. [PubMed] |
| 65. | French AJ, Sargent DJ, Burgart LJ, Foster NR, Kabat BF, Goldberg R, Shepherd L, Windschitl HE, Thibodeau SN. Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin Cancer Res. 2008;14:3408-3415. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 192] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 66. | Vilkin A, Niv Y, Nagasaka T, Morgenstern S, Levi Z, Fireman Z, Fuerst F, Goel A, Boland CR. Microsatellite instability, MLH1 promoter methylation, and BRAF mutation analysis in sporadic colorectal cancers of different ethnic groups in Israel. Cancer. 2009;115:760-769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 67. | Parsons MT, Buchanan DD, Thompson B, Young JP, Spurdle AB. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet. 2012;49:151-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 225] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 68. | Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Nakagawa H, Sotamaa K, Prior TW, Westman J. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med. 2005;352:1851-1860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1013] [Cited by in RCA: 1019] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 69. | Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med. 2009;11:35-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 468] [Cited by in RCA: 514] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 70. | Zhang X, Li J. Era of universal testing of microsatellite instability in colorectal cancer. World J Gastrointest Oncol. 2013;5:12-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 45] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |