Published online Oct 14, 2015. doi: 10.3748/wjg.v21.i38.10776
Peer-review started: April 22, 2015
First decision: June 23, 2015
Revised: July 7, 2015
Accepted: September 14, 2015
Article in press: September 14, 2015
Published online: October 14, 2015
Processing time: 176 Days and 9.5 Hours
It is well established that hepatitis C virus (HCV) infection and replication relies on host lipid metabolism. HCV proteins interact and associate with lipid droplets to facilitate virion assembly and production. Besides, circulating infective particles are associated with very low-density lipoprotein. On the other hand, higher serum lipid levels have been associated with sustained viral response to pegylated interferon and ribavirin therapy in chronic HCV infection, suggesting a relevant role in viral clearance for host proteins. Host and viral genetic factors play an essential role in chronic infection. Lipid metabolism is hijacked by viral infection and could determine the success of viral replication. Recently development of direct acting antiviral agents has shown a very high efficacy (> 90%) in sustained viral response rates even for cirrhotic patients and most of the viral genotypes. HCV RNA clearance induced by Sofosbuvir has been associated with an increased concentration and size of the low-density lipoprotein particles. In this review, host genetic factors, viral factors and the interaction between them will be depicted to clarify the major issues involved in viral infection and lipid metabolism.
Core tip: Hepatitis C virus (HCV) is known to be closely related and associated with host lipid metabolism. Recently development of direct acting antiviral agents has shown a very high efficacy (> 90%) in sustained viral response rates even for cirrhotic patients and most of the viral genotypes. HCV RNA clearance induced by Sofosbuvir has been associated with an increased concentration and size of the low-density lipoprotein particles. Host and viral genetic factors play an essential role in chronic infection. Lipid metabolism is hijacked by viral infection and could determine the success of viral replication.
- Citation: Del Campo JA, Romero-Gómez M. Modulation of host lipid metabolism by hepatitis C virus: Role of new therapies. World J Gastroenterol 2015; 21(38): 10776-10782
- URL: https://www.wjgnet.com/1007-9327/full/v21/i38/10776.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i38.10776
Hepatitis C virus (HCV) infection is a relevant public health problem, infecting approximately 170 million people worldwide[1]. About 70% of infected patients will develop chronic HCV infection. One third of them have a significant increased risk of advanced liver fibrosis, cirrhosis development and finally, hepatocellular carcinoma. With the recent emergence of first generation direct acting antivirals (DAAs), and the development of a second generation DAAs, They have been a near-final step towards the eradication of HCV infection[2-5].
HCV is known to be closely related and associated with host lipid metabolism. HCV proteins interact and associate with lipid droplets to facilitate virion assembly and production[6]. Besides, circulating infective particles are associated with very low-density lipoprotein (VLDL)-like particles, referred as lipoviral particles (LVP)[7]. A proposed mechanism to facilitate HCV entry has been postulated based on the incorporation of host apolipoproteins into the LVP[7-9]. It has been shown that several apolipoproteins are necessary for viral assembly and the production of infective particles[10-12]. Moreover, elevated serum lipid levels have been associated with the rate of sustained viral response to pegylated interferon and ribavirin (Peg-IFN + RBV) therapy for chronic HCV infected patients, suggesting a key role for host proteins in the eradication of viral infection[13,14]. In this review, host genetic factors, viral factors and the interaction between them will be depicted to clarify the major issues involved in viral infection and lipid metabolism.
All viruses, as obligate intracellular parasites, are implicitly dependent on host cell functions for their survival and propagation. There is an emerging understanding of the possible role played by lipid droplets (LDs) in the life cycle of a growing number of viruses, including HCV[15,16]. In the establishment of HCV infection, LDs occupy a central role in the generation of infectious virions and are specifically targeted by viral proteins for this purpose[17]. Diacylglycerol acyltransferase-1 (DGAT1) catalyses the final stage in triglyceride synthesis, and also plays a central role in formation of LDs. It has been shown that DGAT1 interacts with both core and NS5A to facilitate their recruitment to LDs[18]. DGAT1 also appears to facilitate interaction between core and NS5A, thereby functioning as a molecular bridge between the two proteins to ensure that they are targeted to the same LD[19].
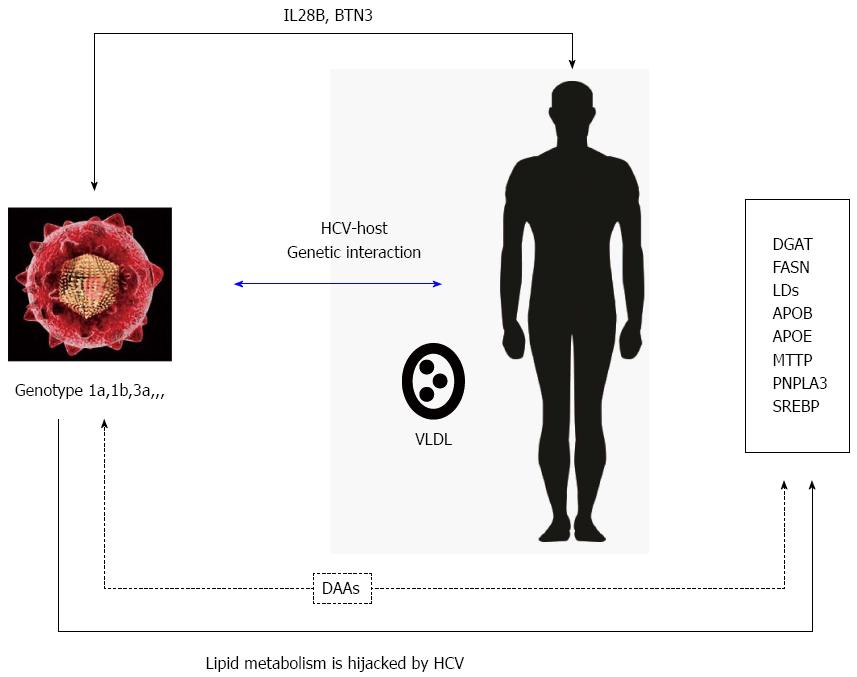
The close relationship between serum LDL-cholesterol (LDL-C) concentration and the chance of achieving sustained viral response has been reported largely in patients under Peg-IFN + RBV therapy[20] as well as with direct-acting antiviral-based triple therapy[21]. Lipid-conforming LVPs are released after HCV eradication, thus increasing concentration can be found in plasma and their concentrations increase in plasma. As previously pointed out, the higher the baseline LDL-C serum level, the greater the chance of curing hepatitis C. This finding is especially relevant in patients in patients bearing non-favourable IL28B genotype, together with previous non-responders patients to Peg-IFN + RBV when treated with triple therapy using telaprevir[22]. Some works have analyzed several genes implicated in lipid transport, such as APOB, APOC-III, APO-L3, and lipid-signaling leptin receptor, MTTP together with liver X receptor/retinoid X receptor pathways. Several changes in these genes have corroborated the link between HCV infection and lipid metabolism and could also identify these genes as therapeutic targets for HCV infection, like FASN inhibition or DGAT activity blockage for inhibition of viral particles production, together with the prevention of the viral entry in the cell[23,24] (Figure 1).
The liver is the main organ for lipid homeostasis in the entire body, through production and uptake of lipoproteins. Lipid homeostasis is a complex mechanism that involves a large amount of genes. Several genetic analysis, including Genome-Wide analysis have been performed to shed some light on this process. This type of analysis has identified a strong association between single nucleotide polymorphisms (SNPs) near the IL28B locus and the chance of achieving sustained virologic response (SVR) to Peg-IFN + RBV therapy in HCV patients, as well as spontaneous viral clearance[25,26]. Moreover, higher plasma levels of ApoB have been associated with sustained virological response in HCV patients bearing the rs8099917 responder genotype (located proximal to rs12979860) in the IL28B gene[27]. Besides, Duggal et al[28] described the association of SNP rs4273729 related to the HLA class II genes on Chromosome 6 with spontaneous HCV clearance independently of IL28B genotype. Nowadays, the role of the IL28B genotype on SVR is attenuated - non significant - in the setting new therapies with NS3 protease, NS5A or NS5B polymerase inhibitors.
Adiponutrin or patatin-like phospholipase domain containing 3 (PNPLA3) is a member of the patatin-like phospholipase family. It is expressed in several human tissues with highest expression in the liver[29]. PNPLA3 acts as a transacylase, which synthesises intracellular triglycerides by transferring acyl groups from monoglycerides to mono- and diglycerides[30]. A study by Trépo et al[31] found, in Caucasian chronic hepatitis C (CHC) patients, a strong and independent association between PNPLA3 and liver damage. Patients with homozygosity of the risk allele had a 2.5-fold higher risk for hepatic steatosis and an over three-fold higher risk for fibrosis as well as for fibrosis progression.
HCV interacts with several proteins of the VLDL secretion pathway for the production of infectious particles. Circulating LVP in an infected patient indicate that HCV virions are associated with hepatically derived triglyceride-rich lipoproteins (TRL) containing apoB-100. These lipo-viro-particles are also associated with gut related lipoproteins containing apoB[8,32]. HCV infection also leads to TRL accumulation through transcriptional activation of lipogenic genes, thus stimulating synthesis of lipids in patients[33]. Besides, several studies on HCV patients have indicated that the virus induced lipogenic genes over-expression. This process may exert a strong influence on inflammation and fibrosis progression in HCV patients, rather than causing the lipid accumulation observed in hepatic steatosis[34].
ApoE plays a relevant role in the assembly and production of viral particles during HCV infection. ApoE depletion has a significant effect in HCV particles production compared to apoB or apoA1 in the same model. This effect may be related to the role of apoE in HCV assembly and interaction with the viral protease NS5A, as previously described[11,12,35]. The interplay NS5A-apoE is a key factor for the building of the viral assembly machinery.
A previous work performed by our group demonstrated a relationship between IL28B polymorphism and lipid profile in patients with hepatitis C genotype 1[20]. This association was not present in patients with hepatitis C genotype 3 or 4 and in the non-infected control group. LDL and total cholesterol levels were higher in patients infected with HCV genotypes 1 and 4 harbouring the favourable (CC) genotype for IL28B gene. HCV directly causes the appearance of large lipid droplets in hepatocytes. Remarkably, HCV replication rates are higher in patients infected with genotype 3, concomitant with more frequent and severe hepatic steatosis[36]. In addition, HCV-induced steatosis related to genotype 3 infections is abolished when antiviral therapy is achieved. Moreover, studies performed in vitro, where cells are transfected with HCV core protein from different genotypes show that core protein is sufficient for lipid droplets induction in the hepatocytes, which is especially relevant - more efficient - in the case of genotype 3a core protein[37]. Lack of understanding for these mechanisms still hamper the characterizarion of these processes, including the appearance of very large lipid droplets in genotype 3. The reasons to explain why genotype 3 is more efficient in steatosis development are still unknown, since very limited studies have been performed using different genotypes in the same model[38].
HCV (including genotype 3a) has been reported to activate in vitro the sterol regulatory element binding proteins 1c and 2, two transcription factors involved in the control of neolipogenesis[39]. However, the evidence obtained in patients with different viral genotypes is inconclusive[34,40] and thus it is unclear whether steatosis in genotype 3 is favoured by an increased fatty acid and/or cholesterol synthesis.
HCV belongs to the Flaviviridae family. These viruses use the secretory pathway of the cell for their way out. Lipoprotein metabolism is tightly associated to the secretory pathway. For this reason, it has been suggested that in HCV infection, the virus uses for its own benefit the VLDL synthesis mechanism of the host cell. Based on an extensive siRNA analysis, it has been shown that most of the host proteins involved in HCV secretion belongs to the classical trafficking pathway, including microtubules, Golgi recycling endosomes, VAMP1 secretory vesicles and the lipoprotein apoE, which is linked to the core protein in the trafficking pathway[41].
High frequency of chronic infection reflects the fact that HCV has evolved several mechanisms to evade and suppress innate immunity, resulting in HCV progression to chronicity[42]. The viral NS3/4A protease is a central component of the HCV innate immune evasion strategy. The multifunctional NS3/4A protease is required for HCV replication, during which it processes the HCV polyprotein at several sites to liberate the viral NS proteins[43]. NS3/4A also targets and cleaves mitochondrial antiviral signaling protein (MAVS) from intracellular membranes to prevent signal transduction[44,45] thus, MAVS cleavage by the HCV NS3/4A protease disrupts RIG-I signaling of innate antiviral immunity and attenuates IFN production[46].
The interaction host-virus resulted on clone selection, immune response modulation and induction/inhibition of proteins involved in the viral entry into the hepatocyte. Recent insights into how HCV regulates innate immune signaling within the liver reveal a complex interaction of patient genetic background with viral and host factors of innate immune triggering and control that imparts the outcome of HCV infection and immunity[47]. Host immune responses, both innate[48] and adaptive[49] together with factors regulating HCV entry into the cell and viral quasispecies, have been explored[50]. In a previous analysis, we identified BTN3A2 (rs9104) to be associated with the selection of viral genotype[51]. Our group is currently exploring HCV susceptibility and to determine the influence of butyrophilin (BTN) family on the selection of HCV genotype. An association between BTN3A2 SNP rs9104 and HCV infection by genotype 1 has been recently described, where genetic variants play a relevant role in selecting a HCV genotype and influencing disease progression[52].
Sofosbuvir is one of the most relevant drugs for hepatitis C therapy. It is a nucleotide analogue inhibitor of the NS5B polymerase which has been recently approved by the Food and Drug Administration and European Medicines Agency for HCV treatment and is currently used in combination with other antivirals like daclatasvir and ledipasvir (NS5A inhibitors). Other combinations include a protease inhibitor such as simeprevir or even with the formerly defined as Standard of Care for hepatitis C (peg-IFN + RBV). Sofosbuvir has demonstrated a consistently potent antiviral activity across several HCV genotypes, and has been found to be safe and well tolerated, showing a very high efficacy (> 90%) in sustained viral response rates even for cirrhotic patients. HCV RNA clearance induced by Sofosbuvir has been associated with an increased concentration and size of the LDL particles. Recently, Meissner et al[53] have demonstrated rapid changes in serum lipoprotein particle concentration during treatment of chronic HCV, genotype 1-infected patients with an IFN-free regimen of SOF and RBV. This likely reflects an altered balance of lipogenesis subsequent to removal of host lipid metabolism perturbation induced by HCV. This fact could be due to differential regulation of genes associated with lipid transport (APOC3 and APOL3) and lipid assembly and signaling (LEPR and MTTP) that has been observed in patients with paired liver biopsies available for analysis[54,55].
Several studies have suggested that statins [3-hydroxy-3-methylglutaryl CoA reductase (HMG Co-A) inhibitors] that inhibit de novo cholesterol synthesis, can block HCV replication[56]. Statins appear to inhibit HCV replication via inhibition of geranylgeranylation of a host protein FBL2 which is required for HCV replication[57]. Rao et al[58] have demonstrated that statin use was associated with an improved SVR among both diabetic patients and non-diabetic patients receiving combination antiviral therapy. Hence, poor diabetes control leads to a lower SVR rate.
Host and viral genetic factors play an essential role in chronic infection. Lipid metabolism is hijacked by viral infection and could determine the success of viral replication. Mechanisms of treatment relapse with DAA therapy are nuclear and differential regulation of host lipid metabolic pathways may be associated with treatment relapse and support further investigation of lipid metabolites as predictors of treatment response to DAA-therapy.
P- Reviewer: Fanning LJ, Ho SB, Nakajima H, Panduro A, Shimizu Y S- Editor: Ma YJ L- Editor: A E- Editor: Ma S
| 1. | Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect. 2011;17:107-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 919] [Cited by in RCA: 945] [Article Influence: 67.5] [Reference Citation Analysis (2)] |
| 2. | Feld JJ, Kowdley KV, Coakley E, Sigal S, Nelson DR, Crawford D, Weiland O, Aguilar H, Xiong J, Pilot-Matias T. Treatment of HCV with ABT-450/r-ombitasvir and dasabuvir with ribavirin. N Engl J Med. 2014;370:1594-1603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 681] [Cited by in RCA: 656] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 3. | Kowdley KV, Gordon SC, Reddy KR, Rossaro L, Bernstein DE, Lawitz E, Shiffman ML, Schiff E, Ghalib R, Ryan M. Ledipasvir and sofosbuvir for 8 or 12 weeks for chronic HCV without cirrhosis. N Engl J Med. 2014;370:1879-1888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 911] [Cited by in RCA: 928] [Article Influence: 84.4] [Reference Citation Analysis (0)] |
| 4. | Kowdley KV, Lawitz E, Poordad F, Cohen DE, Nelson DR, Zeuzem S, Everson GT, Kwo P, Foster GR, Sulkowski MS. Phase 2b trial of interferon-free therapy for hepatitis C virus genotype 1. N Engl J Med. 2014;370:222-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 200] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 5. | Poordad F, Hezode C, Trinh R, Kowdley KV, Zeuzem S, Agarwal K, Shiffman ML, Wedemeyer H, Berg T, Yoshida EM. ABT-450/r-ombitasvir and dasabuvir with ribavirin for hepatitis C with cirrhosis. N Engl J Med. 2014;370:1973-1982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 698] [Cited by in RCA: 683] [Article Influence: 62.1] [Reference Citation Analysis (0)] |
| 6. | Popescu CI, Dubuisson J. Role of lipid metabolism in hepatitis C virus assembly and entry. Biol Cell. 2010;102:63-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 85] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 7. | André P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Bréchot C, Paranhos-Baccalà G, Lotteau V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol. 2002;76:6919-6928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 517] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 8. | Diaz O, Delers F, Maynard M, Demignot S, Zoulim F, Chambaz J, Trépo C, Lotteau V, André P. Preferential association of Hepatitis C virus with apolipoprotein B48-containing lipoproteins. J Gen Virol. 2006;87:2983-2991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 78] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 9. | Meunier JC, Russell RS, Engle RE, Faulk KN, Purcell RH, Emerson SU. Apolipoprotein c1 association with hepatitis C virus. J Virol. 2008;82:9647-9656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 112] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 10. | Chang KS, Jiang J, Cai Z, Luo G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J Virol. 2007;81:13783-13793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 342] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 11. | Jiang J, Luo G. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J Virol. 2009;83:12680-12691. [RCA] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 201] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 12. | Benga WJ, Krieger SE, Dimitrova M, Zeisel MB, Parnot M, Lupberger J, Hildt E, Luo G, McLauchlan J, Baumert TF. Apolipoprotein E interacts with hepatitis C virus nonstructural protein 5A and determines assembly of infectious particles. Hepatology. 2010;51:43-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 177] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 13. | Ramcharran D, Wahed AS, Conjeevaram HS, Evans RW, Wang T, Belle SH, Yee LJ; Virahep-C Study Group. Associations between serum lipids and hepatitis C antiviral treatment efficacy. Hepatology. 2010;52:854-863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 68] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 14. | Ramcharran D, Wahed AS, Conjeevaram HS, Evans RW, Wang T, Belle SH, Yee LJ. Serum lipids and their associations with viral levels and liver disease severity in a treatment-naïve chronic hepatitis C type 1-infected cohort. J Viral Hepat. 2011;18:e144-e152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 15. | Seo JY, Cresswell P. Viperin regulates cellular lipid metabolism during human cytomegalovirus infection. PLoS Pathog. 2013;9:e1003497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 94] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 16. | Boulant S, Montserret R, Hope RG, Ratinier M, Targett-Adams P, Lavergne JP, Penin F, McLauchlan J. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J Biol Chem. 2006;281:22236-22247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 171] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 17. | Filipe A, McLauchlan J. Hepatitis C virus and lipid droplets: finding a niche. Trends Mol Med. 2015;21:34-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 18. | Herker E, Harris C, Hernandez C, Carpentier A, Kaehlcke K, Rosenberg AR, Farese RV, Ott M. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med. 2010;16:1295-1298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 271] [Cited by in RCA: 270] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 19. | Camus G, Herker E, Modi AA, Haas JT, Ramage HR, Farese RV, Ott M. Diacylglycerol acyltransferase-1 localizes hepatitis C virus NS5A protein to lipid droplets and enhances NS5A interaction with the viral capsid core. J Biol Chem. 2013;288:9915-9923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 109] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 20. | Rojas Á, del Campo JA, Maraver M, Aparcero R, García-Valdecasas M, Diago M, Carmona I, Andrade RJ, Solà R, Romero-Gómez M. Hepatitis C virus infection alters lipid metabolism depending on IL28B polymorphism and viral genotype and modulates gene expression in vivo and in vitro. J Viral Hepat. 2014;21:19-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 21. | Serfaty L, Forns X, Goeser T, Ferenci P, Nevens F, Carosi G, Drenth JP, Lonjon-Domanec I, DeMasi R, Picchio G. Insulin resistance and response to telaprevir plus peginterferon α and ribavirin in treatment-naive patients infected with HCV genotype 1. Gut. 2012;61:1473-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 22. | Ogawa E, Furusyo N, Kajiwara E, Nomura H, Dohmen K, Takahashi K, Nakamuta M, Satoh T, Azuma K, Kawano A. Influence of low-density lipoprotein cholesterol on virological response to telaprevir-based triple therapy for chronic HCV genotype 1b infection. Antiviral Res. 2014;104:102-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Yang W, Hood BL, Chadwick SL, Liu S, Watkins SC, Luo G, Conrads TP, Wang T. Fatty acid synthase is up-regulated during hepatitis C virus infection and regulates hepatitis C virus entry and production. Hepatology. 2008;48:1396-1403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 159] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 24. | Sung PS, Murayama A, Kang W, Kim MS, Yoon SK, Fukasawa M, Kondoh M, Kim JS, Kim H, Kato T. Hepatitis C virus entry is impaired by claudin-1 downregulation in diacylglycerol acyltransferase-1-deficient cells. J Virol. 2014;88:9233-9244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 25. | Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2776] [Cited by in RCA: 2723] [Article Influence: 170.2] [Reference Citation Analysis (0)] |
| 26. | Balagopal A, Thomas DL, Thio CL. IL28B and the control of hepatitis C virus infection. Gastroenterology. 2010;139:1865-1876. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 188] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 27. | Yoshizawa K, Abe H, Aida Y, Ishiguro H, Ika M, Shimada N, Tsubota A, Aizawa Y. Serum apolipoprotein B-100 concentration predicts the virological response to pegylated interferon plus ribavirin combination therapy in patients infected with chronic hepatitis C virus genotype 1b. J Med Virol. 2013;85:1180-1190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 28. | Duggal P, Thio CL, Wojcik GL, Goedert JJ, Mangia A, Latanich R, Kim AY, Lauer GM, Chung RT, Peters MG. Genome-wide association study of spontaneous resolution of hepatitis C virus infection: data from multiple cohorts. Ann Intern Med. 2013;158:235-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 173] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 29. | Huang Y, He S, Li JZ, Seo YK, Osborne TF, Cohen JC, Hobbs HH. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc Natl Acad Sci USA. 2010;107:7892-7897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 307] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 30. | Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279:48968-48975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 634] [Cited by in RCA: 676] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 31. | Trépo E, Gustot T, Degré D, Lemmers A, Verset L, Demetter P, Ouziel R, Quertinmont E, Vercruysse V, Amininejad L. Common polymorphism in the PNPLA3/adiponutrin gene confers higher risk of cirrhosis and liver damage in alcoholic liver disease. J Hepatol. 2011;55:906-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 32. | Felmlee DJ, Sheridan DA, Bridge SH, Nielsen SU, Milne RW, Packard CJ, Caslake MJ, McLauchlan J, Toms GL, Neely RD. Intravascular transfer contributes to postprandial increase in numbers of very-low-density hepatitis C virus particles. Gastroenterology. 2010;139:1774-1783, 1783.e1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 59] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 33. | Fujino T, Nakamuta M, Yada R, Aoyagi Y, Yasutake K, Kohjima M, Fukuizumi K, Yoshimoto T, Harada N, Yada M. Expression profile of lipid metabolism-associated genes in hepatitis C virus-infected human liver. Hepatol Res. 2010;40:923-929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 34. | McPherson S, Jonsson JR, Barrie HD, O’Rourke P, Clouston AD, Powell EE. Investigation of the role of SREBP-1c in the pathogenesis of HCV-related steatosis. J Hepatol. 2008;49:1046-1054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 35. | Cun W, Jiang J, Luo G. The C-terminal alpha-helix domain of apolipoprotein E is required for interaction with nonstructural protein 5A and assembly of hepatitis C virus. J Virol. 2010;84:11532-11541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 105] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 36. | Rubbia-Brandt L, Leandro G, Spahr L, Giostra E, Quadri R, Malé PJ, Negro F. Liver steatosis in chronic hepatitis C: a morphological sign suggesting infection with HCV genotype 3. Histopathology. 2001;39:119-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 401] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 37. | Abid K, Pazienza V, de Gottardi A, Rubbia-Brandt L, Conne B, Pugnale P, Rossi C, Mangia A, Negro F. An in vitro model of hepatitis C virus genotype 3a-associated triglycerides accumulation. J Hepatol. 2005;42:744-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 129] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 38. | Goossens N, Negro F. Is genotype 3 of the hepatitis C virus the new villain? Hepatology. 2014;59:2403-2412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 39. | Waris G, Felmlee DJ, Negro F, Siddiqui A. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J Virol. 2007;81:8122-8130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 212] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 40. | Nakamuta M, Yada R, Fujino T, Yada M, Higuchi N, Tanaka M, Miyazaki M, Kohjima M, Kato M, Yoshimoto T. Changes in the expression of cholesterol metabolism-associated genes in HCV-infected liver: a novel target for therapy? Int J Mol Med. 2009;24:825-828. [PubMed] |
| 41. | Coller KE, Heaton NS, Berger KL, Cooper JD, Saunders JL, Randall G. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog. 2012;8:e1002466. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 140] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 42. | Horner SM, Gale M. Intracellular innate immune cascades and interferon defenses that control hepatitis C virus. J Interferon Cytokine Res. 2009;29:489-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 77] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 43. | Morikawa K, Lange CM, Gouttenoire J, Meylan E, Brass V, Penin F, Moradpour D. Nonstructural protein 3-4A: the Swiss army knife of hepatitis C virus. J Viral Hepat. 2011;18:305-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 44. | Baril M, Racine ME, Penin F, Lamarre D. MAVS dimer is a crucial signaling component of innate immunity and the target of hepatitis C virus NS3/4A protease. J Virol. 2009;83:1299-1311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 124] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 45. | Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA. 2005;102:17717-17722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 630] [Cited by in RCA: 651] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 46. | Bellecave P, Sarasin-Filipowicz M, Donzé O, Kennel A, Gouttenoire J, Meylan E, Terracciano L, Tschopp J, Sarrazin C, Berg T. Cleavage of mitochondrial antiviral signaling protein in the liver of patients with chronic hepatitis C correlates with a reduced activation of the endogenous interferon system. Hepatology. 2010;51:1127-1136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 96] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 47. | Horner SM, Gale M. Regulation of hepatic innate immunity by hepatitis C virus. Nat Med. 2013;19:879-888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 220] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 48. | Romero V, Azocar J, Zúñiga J, Clavijo OP, Terreros D, Gu X, Husain Z, Chung RT, Amos C, Yunis EJ. Interaction of NK inhibitory receptor genes with HLA-C and MHC class II alleles in Hepatitis C virus infection outcome. Mol Immunol. 2008;45:2429-2436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 91] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 49. | Montes-Cano MA, Caro-Oleas JL, Romero-Gómez M, Diago M, Andrade R, Carmona I, Aguilar Reina J, Núñez-Roldán A, González-Escribano MF. HLA-C and KIR genes in hepatitis C virus infection. Hum Immunol. 2005;66:1106-1109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 50. | Cubero M, Gregori J, Esteban JI, García-Cehic D, Bes M, Perales C, Domingo E, Rodríguez-Frías F, Sauleda S, Casillas R. Identification of host and viral factors involved in a dissimilar resolution of a hepatitis C virus infection. Liver Int. 2014;34:896-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 51. | Rojas L, Ampuero J, Del Campo JA, Garcia-Lozano RJ, Solá R, Forns X, Romero-Gómez M. Fine mapping of the butyrophilin genomics region: Role in hepatitis C virus infection (HCV). J Hepatol. 2014;60 Suppl:S139. [DOI] [Full Text] |
| 52. | Ampuero J, Del Campo JA, Rojas L, García-Lozano RJ, Buti M, Solá R, Forns X, Moreno-Otero R, Andrade R, Diago M. Fine-mapping butyrophilin family genes revealed several polymorphisms influencing viral genotype selection in hepatitis C infection. Genes Immun. 2015;16:297-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 53. | Meissner EG, Lee YJ, Osinusi A, Sims Z, Qin J, Sturdevant D, McHutchison J, Subramanian M, Sampson M, Naggie S. Effect of sofosbuvir and ribavirin treatment on peripheral and hepatic lipid metabolism in chronic hepatitis C virus, genotype 1-infected patients. Hepatology. 2015;61:790-801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 121] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 54. | Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci USA. 2007;104:5848-5853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 429] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 55. | Sun HY, Lin CC, Lee JC, Wang SW, Cheng PN, Wu IC, Chang TT, Lai MD, Shieh DB, Young KC. Very low-density lipoprotein/lipo-viro particles reverse lipoprotein lipase-mediated inhibition of hepatitis C virus infection via apolipoprotein C-III. Gut. 2013;62:1193-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 56. | Ikeda M, Abe K, Yamada M, Dansako H, Naka K, Kato N. Different anti-HCV profiles of statins and their potential for combination therapy with interferon. Hepatology. 2006;44:117-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 242] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 57. | Kapadia SB, Chisari FV. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci USA. 2005;102:2561-2566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 386] [Cited by in RCA: 406] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 58. | Rao GA, Pandya PK. Statin therapy improves sustained virologic response among diabetic patients with chronic hepatitis C. Gastroenterology. 2011;140:144-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |