Published online Aug 28, 2015. doi: 10.3748/wjg.v21.i32.9554
Peer-review started: February 22, 2015
First decision: April 23, 2015
Revised: May 20, 2015
Accepted: July 8, 2015
Article in press: July 8, 2015
Published online: August 28, 2015
Processing time: 195 Days and 21.2 Hours
AIM: To screen and investigate the effective gRNAs against hepatitis B virus (HBV) of genotypes A-D.
METHODS: A total of 15 gRNAs against HBV of genotypes A-D were designed. Eleven combinations of two above gRNAs (dual-gRNAs) covering the regulatory region of HBV were chosen. The efficiency of each gRNA and 11 dual-gRNAs on the suppression of HBV (genotypes A-D) replication was examined by the measurement of HBV surface antigen (HBsAg) or e antigen (HBeAg) in the culture supernatant. The destruction of HBV-expressing vector was examined in HuH7 cells co-transfected with dual-gRNAs and HBV-expressing vector using polymerase chain reaction (PCR) and sequencing method, and the destruction of cccDNA was examined in HepAD38 cells using KCl precipitation, plasmid-safe ATP-dependent DNase (PSAD) digestion, rolling circle amplification and quantitative PCR combined method. The cytotoxicity of these gRNAs was assessed by a mitochondrial tetrazolium assay.
RESULTS: All of gRNAs could significantly reduce HBsAg or HBeAg production in the culture supernatant, which was dependent on the region in which gRNA against. All of dual gRNAs could efficiently suppress HBsAg and/or HBeAg production for HBV of genotypes A-D, and the efficacy of dual gRNAs in suppressing HBsAg and/or HBeAg production was significantly increased when compared to the single gRNA used alone. Furthermore, by PCR direct sequencing we confirmed that these dual gRNAs could specifically destroy HBV expressing template by removing the fragment between the cleavage sites of the two used gRNAs. Most importantly, gRNA-5 and gRNA-12 combination not only could efficiently suppressing HBsAg and/or HBeAg production, but also destroy the cccDNA reservoirs in HepAD38 cells.
CONCLUSION: These results suggested that CRISPR/Cas9 system could efficiently destroy HBV expressing templates (genotypes A-D) without apparent cytotoxicity. It may be a potential approach for eradication of persistent HBV cccDNA in chronic HBV infection patients.
Core tip: In this manuscript, 15 hepatitis B virus (HBV)-specific gRNAs were designed according to the HBV genome sequences of genotypes A-D. We confirmed that the CRISPR/Cas9 system with these HBV-specific gRNAs could efficiently suppress the replication of multiple HBV genotypes. Further, we demonstrated that dual gRNAs could guide the CRISPR/Cas9 system to efficiently destroy HBV cccDNA and reduce its level in HepAD38 cells. Since cccDNA, the template of HBV replication, accounts for the persistence of HBV infection, our data suggested that CRISPR/Cas9 technique may be a useful tool to eradicate HBV of multiple genotypes.
- Citation: Wang J, Xu ZW, Liu S, Zhang RY, Ding SL, Xie XM, Long L, Chen XM, Zhuang H, Lu FM. Dual gRNAs guided CRISPR/Cas9 system inhibits hepatitis B virus replication. World J Gastroenterol 2015; 21(32): 9554-9565
- URL: https://www.wjgnet.com/1007-9327/full/v21/i32/9554.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i32.9554
Despite prophylactic vaccines available for many years, hepatitis B virus (HBV) infection remains an important public health problem worldwide. Current antiviral agents, including nucleos(t)ide analogues (NAs) and interferon (IFN), can control HBV production but not eliminate HBV, due to the persistence of HBV covalently closed circular DNA (cccDNA) reservoir in the nucleus of hepatocytes. NAs show no effect on cccDNA, thus relapse of hepatitis B occurs frequently in patients who discontinued antiviral treatment[1,2]. Moreover, because the stability of HBV cccDNA was so high that it declines slowly, so life-long treatment of chronic hepatitis B (CHB) is required[1,3,4]. On the other hand, although IFN-α can degrade cccDNA followed by cytidine deamination and apurinic/apyrimidinic site formation, and can further result in virus clearance in a few patients, its efficacy is unsatisfactorily limited[5-7]. Eradication of cccDNA, the only way to reach the clinical cure of CHB, is still an unresolved problem in the treatment of CHB.
Clustered regularly interspaced short palindromic repeats/Cas9 nuclease (CRISPR/Cas9) system is a novel genome editing tool derived from the adaptive immune system of bacteria and archaea[8-10]. CRISPR/Cas9 system promotes genome editing by inducing a double-strand break (DSB) at the target genomic locus. In the absence of a repair template, DSBs are re-ligated through the non-homologous end joining (NHEJ) process, which leads to the insertion/deletion (indel) mutations[11]. CRISPR/Cas9 system has been successfully applied not only for genome editing in cells, but also for disrupting the genome of virus, including adenovirus, herpes simplex virus (HSV) and human immunodeficiency virus (HIV)[12-14]. For HBV, CRISPR/Cas9 system has been proved to efficiently cleave the expressing templates of HBV genotypes A and D[15-17]. Whereas, genotypes A, B, C, D are the predominant genotypes of HBV in East Asia and other part of the world[18-20], so it is necessary to design guide RNAs (gRNAs) specific for HBV genotypes A-D. Here, we evaluated the potential use of CRISPR/Cas9 system to clear the HBV genome of genotypes A-D.
The 1.2xHBV construct (pBB4.5-HBV1.2, genotype C) was constructed using a 1.2-fold length genome of genotype C HBV DNA sequence, and was inserted into the pBB4.5-HBV1.3 (genotype D, G1896A mutation) plasmid digested with PstI and NheI enzymes. The pBB4.5-HBV1.3 (genotype D, G1896A mutation) plasmid was kindly provided by Professor Locarnini SA from the Victorian Infectious Diseases Reference Laboratory, Australia[21]. The 1.2xHBV construct (pBB4.5-HBV1.2, genotype C) has been proved to efficiently produce HBV[22]. The HBV-expression vectors pGEM-HBV1.3A (genotype A) and pGEM-HBV1.3B (genotype B) were kindly provided by Professor Ningshao Xia from School of Public Health, Xiamen University, China.
Cas9 promotes genome editing by inducing a double-strand break (DSB) and re-ligating through the NHEJ process in the absence of a repair template. The gRNA/Cas9 dual expression vector pSpCas9(BB)-2A-GFP (PX458) was obtained from Addgene (Cambridge, MA). PX458 plasmid which expresses a nonsense gRNA (GGGTCTTCGAGAAGACCT) was used as a vector control in each experiment. HBV-specific gRNA/Cas9 dual expression vectors were constructed in our laboratory. The oligonucleotide sequences for the construction of HBV-specific gRNA/Cas9 dual expression vectors are listed in Supplementary Table S1.
Human liver cancer cell lines HuH-7[23] and HepAD38[24] (stable expression of HBV) were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (Gibco, Carlsbad, Calif). HuH-7 and HepAD38 cells were seeded in a 12-well plate at 1.5 × 105 cells/well. HuH-7 cells were co-transfected with HBV expression vectors (HBV of genotype A, B or C) and HBV-specific gRNA/Cas9 dual expression vector [pSpCas9(BB)-2A-GFP] with Lipofectamain 2000 (11668019; Life Technologies). HepAD38 cells were transfected with HBV specific gRNA/Cas9 dual expression vector with Lipofectamain 3000 (L3000015; Life Technologies).
Seventy-two hours after transfection, cell culture supernatants were collected for detection of HBsAg and HBeAg by a time-resolved fluoroimmunoassay (TRFIA) according to manufacturer’s instructions (SY60108A and SY60105A; PerkinElmer). In brief, culture supernatant (100 μL) was added into a microtiter plate coated with anti-HBsAg or anti-HBeAg and shaked for 40 min at room temperature, then washed for 4 times. Europium-labeled anti-HBsAg or anti-HBeAg was diluted 1:50 with HBsAg or HBeAg dilution buffer and added at 100 μL per well, shaked for 40 min in room temperature, then washed 6 times. At last, after incubation with enhancement solution (100 μL) for 5 min, the plates were read using Anytest reader (SYM-BIO), and the concentrations of HBsAg and HBeAg were calculated according to the standard curve. The relative HBsAg or HBeAg level was calculated as the ratio of HBsAg or HBeAg concentration in the cell culture supernatant of gRNA treated and vector control cells.
DNA was extracted from cells using QIAGEN DNA mini kit (51304; QIAGEN), according to the manufacturer’s instruction. Specific primers listed below were used to PCR amplify the cleaved HBV DNA fragments: primer1F (nucleotide position: 1856-1877), 5’-CCTACTGTTCAAGCCTCCAAGC-3’; primer2F (321-342), 5’-CAACCTCCAATCACTCACCAAC-3’; primer1R (434-415), 5’-AGAAGATGAGGCATAGCAGC-3’; primer2R (2006-1986), 5’-CAGAGGCGGTGTCAAGGAGAT-3’; primer3R (1702-1682), 5’-GACTCAAGGTCGGTCGTTGAC-3’; primer4R (1285-1264), 5’-CTAGGAGTTCCGCAGTATGGAT-3’. Primer1F and 1R were used for detection of the fragment cleaved by dual gRNAs of gRNA1 + 13 or gRNA2 + 14, and primer 2F and 2R, 2F and 3R, 2F and 4R were used for detection of the fragment cleaved by dual gRNAs of gRNA3 + 5, gRNA4 + 5 and gRNA5 + 12, respectively.
PCR reaction mixture (20 μL) contained 10 μL 2 × Taq mix (Transgene), 1 μL forward primer (10 μmol/L), 1 μL reverse primer (10 μmol/L), 1 μL DNA template and 7 μL double distilled water (ddH2O). The reaction mixture was denatured at 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min, and at last 72 °C for 5 min. Agarose gel (1.5%) was used for separation of DNA fragments with different length. DNA markers were DL2000 and DL2000 Plus (GenStar). The DNA fragment of expected length was sequenced.
Two gRNA/Cas9 dual expression vectors were co-transfected into HepAD38 cells at least 7 d after tetracycline removed. Cells were collected for DNA extraction at 72 h after transfection using QIAGEN DNA mini kit (51304; QIAGEN), according to the manufacturer’s instruction. To obtain cccDNA, KCl precipitation and plasmid-safeTM ATP-dependent DNase (PSAD) (Epicentre, Madison, WI, United States) were used to remove HBV DNA integrated into cell genome, HBV rcDNA, replicative dsDNA and ssDNA. Afterwards, rolling circle amplification (RCA) was conducted to selectively amplify cccDNA. Finally, PCR was performed using RCA products as template, and using cccDNA specific primers which target the gap region of HBV genome[25,26].
Methyl thiazolyl tetrazolium (MTT) assay is used to monitor cell viability. Cells (1500 cells per well) were plated in 96-well plates and were maintained in DMEM supplemented with 10% fetal bovine serum. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide solution was added into the cell culture at a final concentration of 5 μg/mL and was allowed to remain in culture for 4 h before measurement. Cell viability was monitored every 24 h by measuring the absorbance in a microplate reader (Bio-Rad, Hercules, CA).
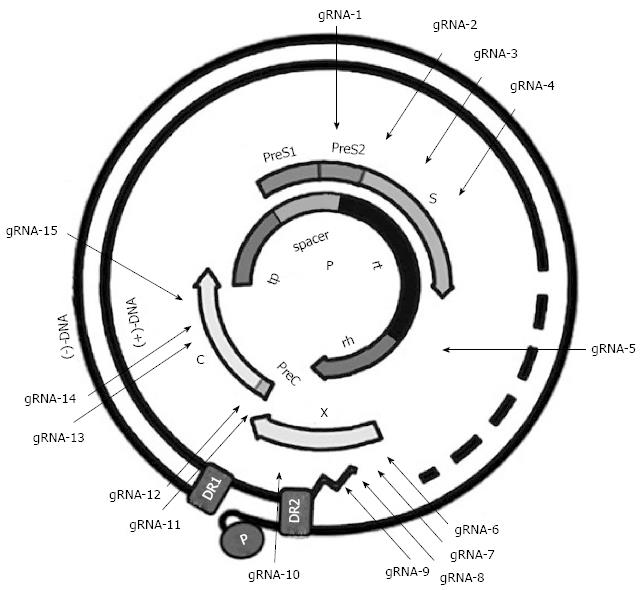
Since SpCas9 in our CRISPR/Cas9 system requires a 5’-NGG protospacer adjacent motif (PAM) sequence, HBV genome of genotypes A-D was searched for potential 18-20 base target sequences called the protospacer with 3’-downstream of PAM, which was shown as GN18-20-NGG. Finally a panel of 15 HBV-specific gRNAs targeting different regions of HBV genome were designed (Figure 1). To avoid the off-target effect, the gRNA sequences were blasted by Nucleotide Blast search (NCBI), and the difference of more than 3 nucleotides to other sequences in human genome was assured. Finally, the gRNA expression vectors were identified by sequencing method. The sequences and locations of these HBV-specific gRNAs are listed in Table 1.
| gRNA No. | Nucleotide position | Sequence (GN18-20NGG, 5’-3’) | Genotype |
| 1 | 56-75 | CCTGCTGGTGGCTCCAGTTC | A/B/C/D |
| 2 | 182-200 | GGACCCCTGCTCGTGTTAC | A/B/C/D |
| 3 | 415-433 | GCTGCTATGCCTCATCTTC | A/B/C/D |
| 4 | 640-658 | ATGGGAGTGGGCCTCAGTC | A/B/C |
| 5 | 1179-1197 | AGTGTTTGCTGACGCAACC | A/B/C/D |
| 6 | 1393-1410 | GCCAACTGGATCCTGCGC | B/C/D |
| 7 | 1521-1540 | GGGGCGCACCTCTCTTTACG | A/B/C/D |
| 8 | 1578-1597 | GAGGTGAAGCGAAGTGCACA | A/B/C/D |
| 9 | 1589-1608 | CTTCACCTCTGCACGTCGCA | B/C/D |
| 10 | 1775-1794 | AGGAGGCTGTAGGCATAAAT | A/B/C/D |
| 11 | 1859-1878 | AGCTTGGAGGCTTGAACAGT | A/B/C/D |
| 12 | 1865-1884 | CAAGCCTCCAAGCTGTGCCT | A/B/C/D |
| 13 | 2336-2355 | ACTACTGTTGTTAGACGACG | C/D |
| 14 | 2367-2386 | CGAGGGAGTTCTTCTTCTAG | A/B/C/D |
| 15 | 2390-2409 | GATTGAGACCTTCGTCTGCG | B/C/D |
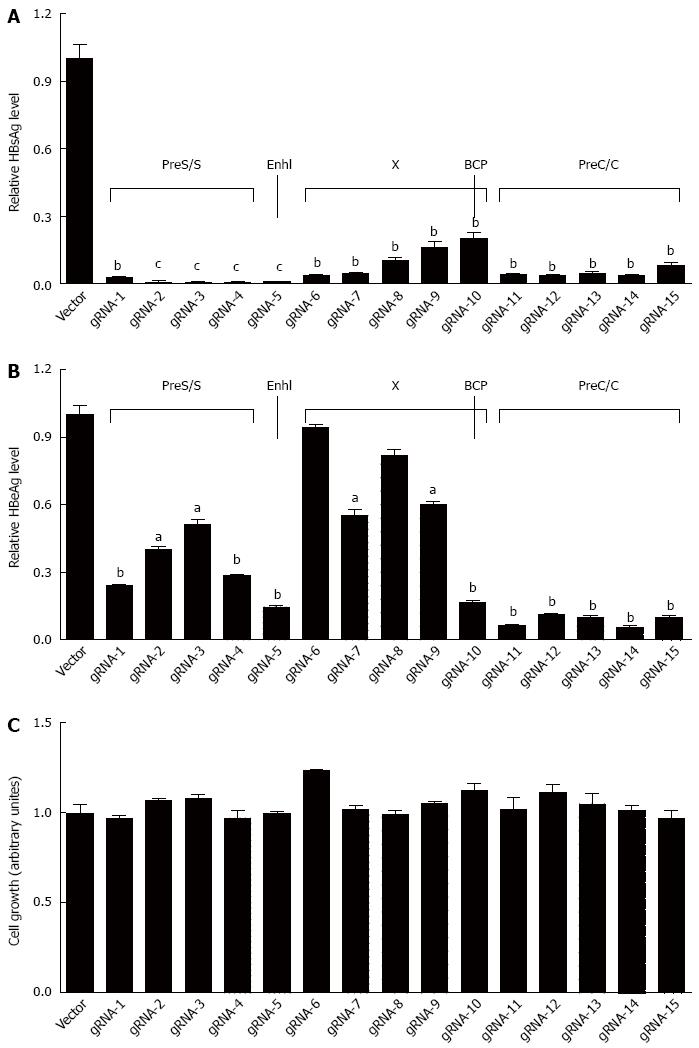
To examine the efficiency of each gRNA in the suppression of HBV replication, the 1.2xHBV construct (pBB4.5-HBV1.2, genotype C) was co-transfected with each gRNA/Cas9 dual expression vector in HuH-7 cells, respectively. As shown in Figure 2A, all designed gRNAs could significantly suppress HBsAg levels in culture supernatants, but with noticeable variation of efficacy, and gRNAs targeting the S region (gRNAs-1, 2, 3, and 4) and enhancer I region (gRNA-5) of HBV genome exhibited higher HBsAg suppressing efficiency than that of the gRNAs targeting either X region (gRNAs-6, 7, 8, 9, and 10) or preC/C (gRNAs-11, 12, 13, 14, and 15) region. For HBeAg, all of gRNAs could significantly reduce HBeAg levels in culture supernatants, with the exception of gRNA-6 and gRNA-8. It is noteworthy that gRNAs targeting preC/C region exhibited higher HBeAg suppressive efficiency than that of those gRNAs targeting either S or X regions (Figure 2B). Intriguingly, gRNA-5 against enhancer I region could efficiently suppress both HBsAg and HBeAg levels (Figure 2A and B). The off-target effect of gRNA may induce cytotoxicity, and then disturb the antiviral effect of HBV-specific gRNAs. The methods for assessing cell viability can test cytotoxicity. To exclude the possibility that the HBsAg or HBeAg suppression observed above was the result of non-specific cytotoxicity, MTT assay, which is a colorimetric assay for assessing cell viability, was conducted. The results revealed that no noticeable cytotoxicity of the gRNAs was observed in HuH-7 cells (Figure 2C).
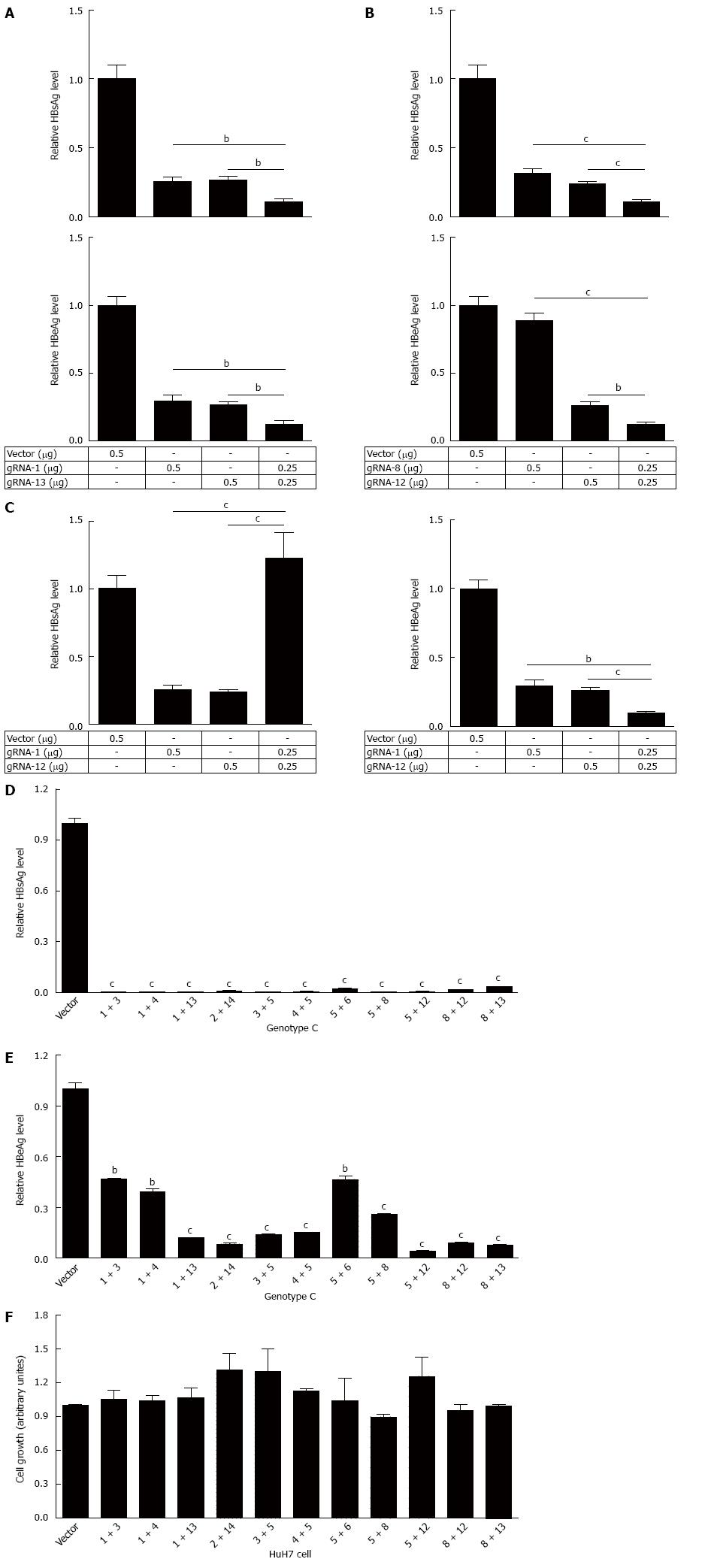
Since CRISPR/Cas9 system can be efficiently used for multiple genome cleavage[11], we wondered if combination of two HBV-specific gRNAs (dual gRNAs) should be more efficient than single one in suppressing HBV replication. To confirm this, pBB4.5-HBV1.2 and combinations of two different gRNA expression vectors were co-transfected into HuH-7 cells. Firstly, we chose the region modulating the expression of HBsAg and HBeAg between two gRNAs in HBV genome, including the combinations of gRNA-1 + 13 covering the S gene promoter and pre S1 region, gRNA-1 + 12 covering the S gene promoter pre S1 region and Core coding DNA sequence (CDS) region, and gRNA-8 + 12 covering Core promoter and Enhancer II region were tested. As expected, the synergistic effect of dual gRNAs in the suppression of HBV replication was observed. Two of the tested dual gRNAs gRNA-1 + 13 (gRNA1 vs gRNA-1 + 13: 0.255 vs 0.110, P = 0.0031; gRNA13 vs gRNA-1 + 13: 0.265 vs 0.110, P = 0.0017) and gRNA-8 + 12 (gRNA8 vs gRNA-8 + 12: 0.319 vs 0.110, P = 0.0004; gRNA12 vs gRNA-8 + 12: 0.240 vs 0.110, P = 0.0007) demonstrated significantly higher suppressive efficiency in HBsAg production, when compared to that of each gRNA used alone (Figure 3A). Consistent with HBsAg, there was synergistic effect of gRNA-1 + 13 (gRNA1 vs gRNA-1 + 13: 0.296 vs 0.124, P = 0.0034; gRNA13 vs gRNA-1 + 13: 0.264 vs 0.124, P = 0.0018) and gRNA-8 + 12 (gRNA8 vs gRNA-8 + 12: 0.889 vs 0.121, P < 0.0001; gRNA12 vs gRNA-8 + 12: 0.260 vs 0.121, P = 0.0018) in the suppression of HBeAg production (Figure 3B). While for the gRNA-1 + 12, such synergistic effect could only be observed in HBeAg production (gRNA1 vs gRNA-1 + 12: 0.296 vs 0.096, P = 0.0014; gRNA12 vs gRNA-1 + 12: 0.260 vs 0.096, P = 0.0006), and an antagonistic effect for HBsAg production (gRNA1 vs gRNA-1 + 12: 0.265 vs 1.125, P = 0.0001; gRNA12 vs gRNA-1 + 12: 0.240 vs 1.125, P = 0.0001) was exhibited (Figure 3C).
Following the above observation, 11 dual gRNAs (two different gRNAs at a ratio of 1:1) were used for further study. The choice of gRNAs for combination was made according to the efficiency of each gRNA and its targeting regions (in promoter, enhancer or reverse transcriptional region of polymerase). Next, the efficiency of those 11 dual gRNAs in the suppression of HBsAg and HBeAg was examined, with different HBV genotypes taken into consideration. The HBV-expression vectors (genotype A, B or C constructs) and two different gRNAs expression vectors were co-transfected into HuH-7 cells. For HBV of genotype C, all of dual gRNAs could significantly suppress HBsAg production (Figure 3D), just as expected. However, although all dual gRNAs could also significantly suppress HBeAg production, the efficacy was lower than that of HBsAg, since higher HBeAg suppressive efficiency (≥ 80%) was detected in only 7 dual gRNAs (Figure 3E). Similar results were observed when HBV of genotypes A and B were tested (Supplementary Figure S1A and B). To exclude the possibility that excess concentration of gRNA may interfere HBV replication, we tested the concentration effect of gRNAs in HBV replication. The result revealed that the suppression efficiency of gRNA on the HBsAg level was gRNA concentration dependent (Figure S2). Similarly, in order to exclude the interference of non-specific cytotoxicity, MTT assay was conducted. The result affirmed that there was no noticeable cytotoxicity of dual gRNAs (Figure 3F).
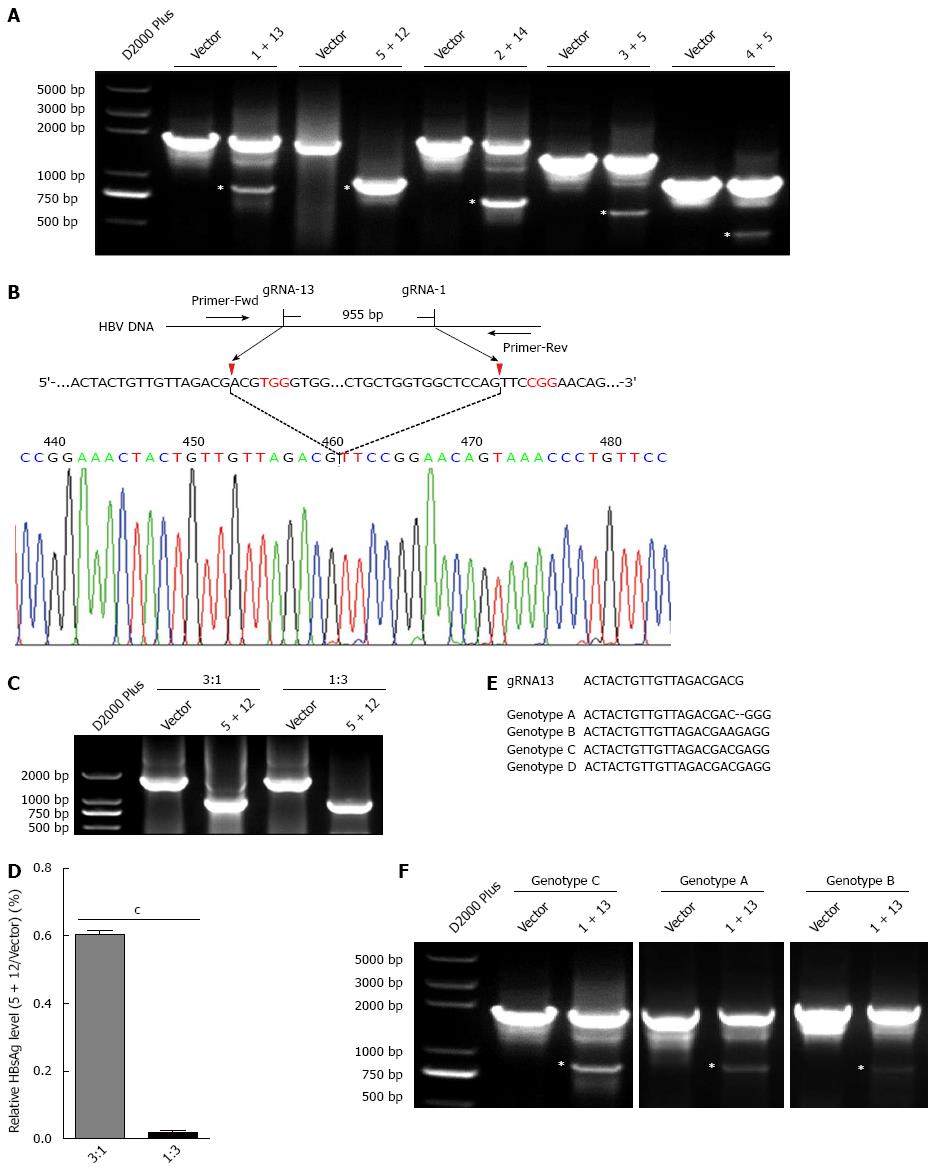
To confirm that the dual gRNAs mediated suppression in HBV replication was at the genome level, the HBV genome region covering the cleavage sites of dual gRNAs was amplified using PCR. If dual gRNAs worked, the fragment between the two cleavage sites would be removed, consequently a relative smaller PCR product would be detected. According to the above suppressive efficiency of dual gRNAs combinations, five dual gRNAs (1 + 13, 5 + 12, 2 + 14, 3 + 5 and 4 + 5) were chosen for further investigation. As expected, all of dual gRNAs could destroy HBV genome, especially the combination of gRNA-5 + 12, in which only the smaller fragment was detected. An up to 100% cleavage efficiency implicated the optimum combination of the two gRNAs (Figure 4A). Furthermore, direct sequencing of the PCR product showed that the smaller fragment was indeed formed by the re-ligation between the ends of two cleavage sites (Figure 4B). Then the plasmid pBB4.5-HBV1.2 was co-transfected with dual gRNA-5 + 12 expression vectors at different ratios to HuH-7 cells and the cleavage efficiency was assessed by PCR amplification. The result revealed that at the ratio of 1:3, almost all of HBV genome DNAs were cleft by gRNA-5 + 12. Surprisingly, even at the ratio of 3:1, still more than 90% of HBV genome DNAs were cleft (Figure 4C). In line with this, the efficiency in suppressing the production of HBsAg in culture supernatants was also gradually but significantly decreased from the ratio of 1:3 (reduced to 0.019% ± 0.007%) to 3:1 (reduced to 0.60% ± 0.014%), as compared to the vector control (Figure 4D). Other four dual gRNAs also showed the capability to destroy HBV genome at different extents (Figure 4A), and all were confirmed by sequencing analysis (data not shown).
As shown in Figure 4E, the target genome sequence of gRNA-13 in genotype B HBV harbors a one base difference as compared to genotypes A/C/D. However, dual gRNAs composed by gRNA-1 and gRNA-13 could also destroy HBV genome of genotype B (Figure 4F).
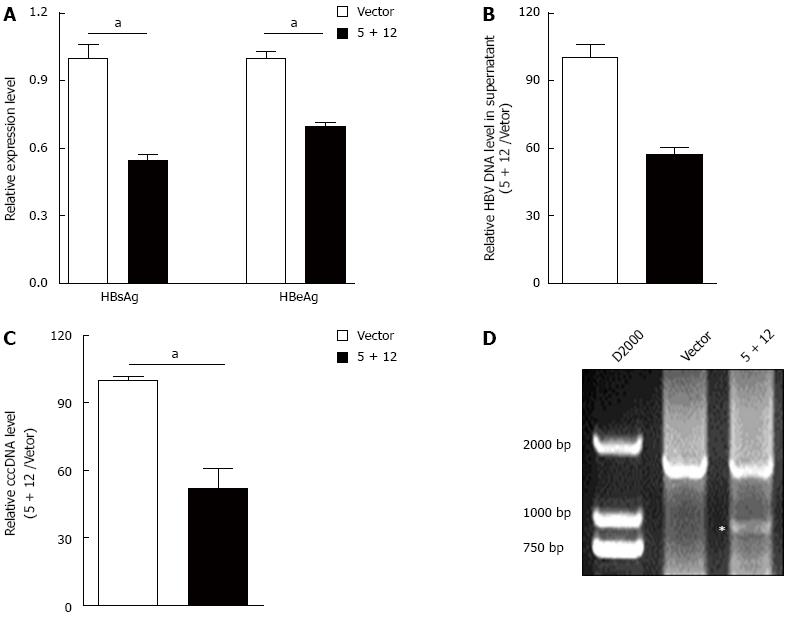
Above data demonstrated that HBV-specific gRNAs could destroy HBV expressing template. Theoretically, such gRNAs could also destroy HBV cccDNA. To confirm this, the most powerful dual gRNAs (gRNA-5 and gRNA-12 expression vectors) were co-transfected into HepAD38 cells which stably express genotype D HBV and produce HBV cccDNA[24]. Firstly, the HBsAg and HBeAg levels in the culture supernatant were measured. The results showed that even with low transfection efficiency, gRNA-5 and gRNA-12 together could significantly suppress both HBsAg and HBeAg levels, particularly for HBsAg which almost dropped 50% off (Figure 5A). Besides, gRNA-5 and gRNA-12 could also significantly reduce the HBV DNA level in the HepAD38 cell culture supernatant (Figure 5B). Since cccDNA reservoirs are the cause of chronic HBV infection, next, the HBV cccDNA level in HepAD38 cells was measured. To make sure that the quantitative measurement of the HBV cccDNA was reliable, KCl precipitation, plasmid-safe ATP-dependent DNase (PSAD) digestion and RCA (Isothermal PCR amplification for the circular HBV cccDNA rather than the linear rcDNA) were employed, followed by quantitative PCR with the cccDNA specific primers (sense primer covers the cleavage site of gRNA-12) that target the gap region of HBV genome. As expected, cccDNA level in HepAD38 cells was significantly suppressed by gRNA-5 and gRNA-12 (Figure 5C). PCR amplification confirmed that the fragment between two cleavage sites of dual gRNAs in HBV expressing template was removed (Figure 4A and C). Similarly, HBV-specific gRNA could also guide Cas9 to destroy HBV cccDNA (Figure 5D). Above results suggested that the downregulation of HBsAg and HBeAg levels was at least partially due to the destruction of HBV cccDNA.
Hepatitis B is an important occupational hazard for health workers. An estimated 240 million people are chronically infected with HBV[27]. Multiple studies demonstrated that without therapy, 15%-40% of HBV infected patients would eventually develop cirrhosis, liver failure, or hepatocellular carcinoma (HCC)[28]. Because of the high HBV-related morbidity and mortality, HBV infection has been a serious public health problem worldwide. Unfortunately, none of the current therapy regimes could eradicate cccDNA in the nucleus of infected hepatocytes, which is the main cause of HBV persistent infection. In recent years, new strategies focused on targeting cccDNA are under intensive study. Three recent studies have suggested the potential use of CRISPR/Cas9 genome editing technique to cut HBV DNA and cccDNA in vitro and in vivo, as well as for chronic or de novo HBV infection[15-17]. However, those studies just focused on single genotype HBV, genotype A or D. Since genotypes B and C are also the major genotypes in the world, particularly in China, in this study, 15 gRNAs were designed according to the HBV genome sequences of genotypes A, B, C and D. These gRNAs covered different regions of HBV genome, including S, P, X, C, enhancer I and basic core promoter (BCP) regions. Our results here indicated that most of the gRNAs could significantly suppress the production of HBsAg and HBeAg. Interestingly, the suppressive efficiency of different gRNAs varied depending on the region in which they against.
As multiple gRNAs could be efficiently used to edit genome, here different combinations of two gRNAs were used to suppress HBV replication in vitro. And high efficiency against genotypes A-D HBV replication was found in different dual gRNAs combinations. However, the combination of gRNA-1 and 12 exhibited an antagonistic effect in the suppression of HBsAg production. Currently we are unable to explain this phenomenon. A potential possibility is that the combination of gRNAs-1 and -12 would cut off and remove the fragment between the two cleavage sites, where C, S promoter (Sp) and preS1 regions located. As a result, the enhancer II, the core promoter (Cp) and the basic core promoter (BCP) regions were re-ligated to the immediate upstream of S gene, and the expression of HBsAg was transcriptionally enhanced. In line with this, we found that dual gRNAs could indeed remove the fragment specifically between the two cleavage sites of gRNAs, which was confirmed by PCR sequencing. Such antagonistic effect reminded us that when multiple gRNAs were used in combination, the side effect should be taken into consideration.
In addition to genotype, due to absence of proofreading function of the viral reverse transcriptase and a high replication rate, the HBV population in an individual was a composition of genetically distinct but closely related variants known as quasispecies[29,30]. It is likely that such variants will cause the imperfect match between the designed gRNAs and their target viral genome sequence. The off-target effect of CRISPR/Cas system has been mentioned, and such shortage could become an advantage when gRNAs were used to guide the cleavage of the high variety HBV genome in an infected individual. Just for this, although the target sequence in HBV genotype B harbors a one base mismatch with gRNA-13, it still could be cleft (Figure 4F). Besides, we and others have proved the oncogenicity of HBV integration[31,32]. Potentially, multiple gRNAs can be used to remove the integrated HBV DNA to cure the HBV-related HCC. Most importantly, we found that wild-type HBV cccDNA level in HepAD38 cells was significantly downregulated by HBV-specific gRNA, which indicated that HBV cccDNA could be destroyed by CRISPR/Cas9 system. It is difficult to confirm the gRNA-induced destruction of cccDNA by Southern blot for the low transfection efficiency of the PX458 plasmid. We will demonstrate the gRNA-induced destruction of cccDNA by Southern blot when the CRISPR/Cas9 system was combined with the recombinant adeno-associated virus system. Above all, the indel mutation and destruction of cccDNA should be the main reason of gRNA-induced suppression of cccDNA replication.
Above all, we confirmed that CRISPR/Cas9 system had the potential to destroy cccDNA which is the template of HBV replication. Our data further demonstrated that dual gRNAs guided CRISPR/Cas9 system may be a useful tool to eradicate CHB and HBV-related disease with infection by multiple HBV genotypes. However, there is still a long way for the treatment of HBV infected patients by CRISPR/Cas9 system. Next, we will validate the effect of HBV-specific gRNA on HBV replication in HBV transgenic mice by hydrodynamic injection method or using the adenovirus or adeno-associated virus system. In the future, dual gRNAs guided CRISPR/Cas9 system will be developed by adeno-associated virus system which is a potential vector for gene therapy, and be used in combination with the current NAs and/or IFN-α based antiviral therapy for the possible clinical cure of CHB.
The CRISPR/Cas9 system is a novel genome editing tool which leads to genome indel mutation by inducing a double-strand break (DSB) at the target genomic locus. The CRISPR/Cas9 system has been successfully applied not only for genome editing in cells, but also for disrupting the genome of virus, including adenovirus, herpes simplex virus, human immunodeficiency virus and hepatitis B virus (HBV). It has become recognized that CRISPR/Cas9 system may be a potential tool to cure viral disease.
Several recent studies have confirmed the potential use of CRISPR/Cas9 system to cleave HBV DNA and cccDNA in vitro and in vivo, as well as for chronic or de novo HBV infection.
Eradication of cccDNA, the only way to reach the clinical cure of chronic hepatitis B (CHB), is still an unresolved problem in the treatment of CHB. Previous studies have reported that the CRISPR/Cas9 system can efficiently cleave the expressing template of HBV. However, those studies just focus on HBV of genotype A or D. Whereas, genotypes A, B, C, D are the predominant genotypes of HBV in East Asia and other part of the world, so it is necessary to design guide RNAs (gRNAs) specific for HBV genotypes A-D. Besides, dual gRNAs can get the synergistic effect in genome editing. In this paper, we evaluated the potential use of dual gRNAs guided CRISPR/Cas9 system to clear the HBV genome of genotypes A-D.
This study provides a potential tool, the dual gRNAs guided CRISPR/Cas9 system, to eradicate CHB and HBV-related disease with infection by multiple HBV genotypes. In the future, the dual gRNAs guided CRISPR/Cas9 system will be developed by adeno-associated virus system which is a potential vector for gene therapy, and be used in combination with the current NAs and/or IFN-α based antiviral therapy for the possible clinical cure of CHB.
The clustered regularly interspaced short palindromic repeats/Cas9 nuclease (CRISPR/Cas9) system is a novel genome editing tool derived from the adaptive immune system of bacteria and archaea. The CRISPR/Cas9 system promotes genome editing by inducing a double-strand break (DSB) at the target genomic locus. In the absence of a repair template, DSBs are re-ligated through the non-homologous end joining process, which leads to the insertion/deletion (indel) mutations.
The authors present a highly interesting functional study in which they showed that dual gRNA guided CRISPR/Cas9 system can suppress replication of multiple HBV genotypes as well as promote clearance of HBV cccDNA in the cell culture. This paper confirms that the presented dual gRNA/CRISPR/Cas9 system might be a potential approach for eradication of HBV cccDNA and thus considered a new and additional antiviral treatment option for CHB.
Biostatistics statement: The statistical methods of this study were reviewed by Xiaomeng Xie from the Department of Epidemiology and Biostatistics, College of Public Health, Zhengzhou University, Zhengzhou, China.
P- Reviewer: Bock CT, Netter HJ, Picardi A, Rodriguez-Frias F, Said ZNA, Wang K S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Zhang DN
| 1. | Zoulim F. Hepatitis B virus resistance to antiviral drugs: where are we going? Liver Int. 2011;31 Suppl 1:111-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 102] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 2. | Dandri M, Burda MR, Will H, Petersen J. Increased hepatocyte turnover and inhibition of woodchuck hepatitis B virus replication by adefovir in vitro do not lead to reduction of the closed circular DNA. Hepatology. 2000;32:139-146. [PubMed] |
| 3. | Caruntu FA, Molagic V. CccDNA persistence during natural evolution of chronic VHB infection. Rom J Gastroenterol. 2005;14:373-377. [PubMed] |
| 4. | Sung JJ, Wong ML, Bowden S, Liew CT, Hui AY, Wong VW, Leung NW, Locarnini S, Chan HL. Intrahepatic hepatitis B virus covalently closed circular DNA can be a predictor of sustained response to therapy. Gastroenterology. 2005;128:1890-1897. [PubMed] |
| 5. | Wursthorn K, Lutgehetmann M, Dandri M, Volz T, Buggisch P, Zollner B, Longerich T, Schirmacher P, Metzler F, Zankel M. Peginterferon alpha-2b plus adefovir induce strong cccDNA decline and HBsAg reduction in patients with chronic hepatitis B. Hepatology. 2006;44:675-684. [PubMed] |
| 6. | Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T, Pollicino T, Petersen J, Raimondo G, Dandri M, Levrero M. IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Invest. 2012;122:529-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 484] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 7. | Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343:1221-1228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 640] [Cited by in RCA: 738] [Article Influence: 67.1] [Reference Citation Analysis (0)] |
| 8. | Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 409] [Cited by in RCA: 430] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 9. | Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1581] [Cited by in RCA: 1655] [Article Influence: 110.3] [Reference Citation Analysis (0)] |
| 10. | Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467-477. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2083] [Cited by in RCA: 1745] [Article Influence: 124.6] [Reference Citation Analysis (0)] |
| 11. | Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281-2308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6962] [Cited by in RCA: 8291] [Article Influence: 690.9] [Reference Citation Analysis (0)] |
| 12. | Bi Y, Sun L, Gao D, Ding C, Li Z, Li Y, Cun W, Li Q. High-efficiency targeted editing of large viral genomes by RNA-guided nucleases. PLoS Pathog. 2014;10:e1004090. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 124] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 13. | Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 386] [Cited by in RCA: 414] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 14. | Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, Li F, Luo B, Alvarez-Carbonell D, Garcia-Mesa Y, Karn J. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci USA. 2014;111:11461-11466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 407] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 15. | Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, Lin YY, Wang HY, Wang CC, Shen YC. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol Ther Nucleic Acids. 2014;3:e186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 270] [Cited by in RCA: 299] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 16. | Seeger C, Sohn JA. Targeting Hepatitis B Virus With CRISPR/Cas9. Mol Ther Nucleic Acids. 2014;3:e216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 220] [Cited by in RCA: 227] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 17. | Kennedy EM, Bassit LC, Mueller H, Kornepati AV, Bogerd HP, Nie T, Chatterjee P, Javanbakht H, Schinazi RF, Cullen BR. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology. 2015;476:196-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 185] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 18. | Schaefer S. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J Gastroenterol. 2007;13:14-21. [PubMed] |
| 19. | Wang Z, Huang Y, Wen S, Zhou B, Hou J. Hepatitis B virus genotypes and subgenotypes in China. Hepatol Res. 2007;37:S36-S41. [PubMed] |
| 20. | Nie JJ, Sun KX, Li J, Wang J, Jin H, Wang L, Lu FM, Li T, Yan L, Yang JX. A type-specific nested PCR assay established and applied for investigation of HBV genotype and subgenotype in Chinese patients with chronic HBV infection. Virol J. 2012;9:121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 21. | Chen RY, Edwards R, Shaw T, Colledge D, Delaney WE, Isom H, Bowden S, Desmond P, Locarnini SA. Effect of the G1896A precore mutation on drug sensitivity and replication yield of lamivudine-resistant HBV in vitro. Hepatology. 2003;37:27-35. [PubMed] |
| 22. | Li WP, Li T, Yan L, Liu BM, Zhuang H. Construction of a 1.2-genome length genotype C HBV construct and study on its expression and replication in HepG2 cells. Ganzang. 2008;13:211-215. [DOI] [Full Text] |
| 23. | Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858-3863. [PubMed] |
| 24. | Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, Seeger C, King RW. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother. 1997;41:1715-1720. [PubMed] |
| 25. | Zhong Y, Han J, Zou Z, Liu S, Tang B, Ren X, Li X, Zhao Y, Liu Y, Zhou D. Quantitation of HBV covalently closed circular DNA in micro formalin fixed paraffin-embedded liver tissue using rolling circle amplification in combination with real-time PCR. Clin Chim Acta. 2011;412:1905-1911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 26. | Li W, Zhao J, Zou Z, Liu Y, Li B, Sun Y, Li X, Liu S, Cai S, Yao W. Analysis of hepatitis B virus intrahepatic covalently closed circular DNA and serum viral markers in treatment-naive patients with acute and chronic HBV infection. PLoS One. 2014;9:e89046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 27. | Hepatitis B. Fact sheet N 204 2015. Available from: http://www.who.int/mediacentre/factsheets/fs204/en/. |
| 28. | Hou J, Liu Z, Gu F. Epidemiology and Prevention of Hepatitis B Virus Infection. Int J Med Sci. 2005;2:50-57. [PubMed] |
| 29. | Kojima N, Horiike N, Michitaka K, Onji M. In situ detection of mutated hepatitis B virus in microdissected, formalin-fixed liver tissues from patients with chronic hepatitis B. J Hepatol. 1999;30:359-365. [PubMed] |
| 30. | Chen L, Zhang Q, Yu DM, Wan MB, Zhang XX. Early changes of hepatitis B virus quasispecies during lamivudine treatment and the correlation with antiviral efficacy. J Hepatol. 2009;50:895-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 54] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 31. | Li X, Zhang J, Yang Z, Kang J, Jiang S, Zhang T, Chen T, Li M, Lv Q, Chen X. The function of targeted host genes determines the oncogenicity of HBV integration in hepatocellular carcinoma. J Hepatol. 2014;60:975-984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 93] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 32. | Tsai WL, Chung RT. Viral hepatocarcinogenesis. Oncogene. 2010;29:2309-2324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 199] [Article Influence: 13.3] [Reference Citation Analysis (0)] |