Published online Jan 21, 2015. doi: 10.3748/wjg.v21.i3.878
Peer-review started: April 24, 2014
First decision: May 29, 2014
Revised: June 18, 2014
Accepted: July 15, 2014
Article in press: July 16, 2014
Published online: January 21, 2015
Processing time: 272 Days and 0 Hours
AIM: To determine the role of Notch1 and Hes1 in regulating the activation of hepatic stellate cells (HSCs) and whether Hes1 is regulated by transforming growth factor (TGF)/bone morphogenetic protein (BMP) signaling.
METHODS: Immunofluorescence staining was used to detect the expression of desmin, glial fibrillary acidic protein and the myofibroblastic marker α-smooth muscle actin (α-SMA) after freshly isolated, normal rat HSCs had been activated in culture for different numbers of days (0, 1, 3, 7 and 10 d). The expression of α-SMA, collagen1α2 (COL1α2), Notch receptors (Notch1-4), and the Notch target genes Hes1 and Hey1 were analyzed by reverse transcriptase-polymerase chain reaction. Luciferase reporter assays and Western blot were used to study the regulation of α-SMA, COL1α1, COL1α2 and Hes1 by NICD1, Hes1, CA-ALK3, and CA-ALK5 in HSC-T6 cells. Moreover, the effects of inhibiting Hes1 function in HSC-T6 cells using a Hes1 decoy were also investigated.
RESULTS: The expression of Notch1 and Hes1 mRNAs was significantly down-regulated during the culture of freshly isolated HSCs. In HSC-T6 cells, Notch1 inhibited the promoter activities of α-SMA, COL1α1 and COL1α2. On the other hand, Hes1 enhanced the promoter activities of α-SMA and COL1α2, and this effect could be blocked by inhibiting Hes1 function with a Hes1 decoy. Furthermore, co-transfection of pcDNA3-CA-ALK3 (BMP signaling activin receptor-like kinase 3) and pcDNA3.1-NICD1 further increased the expression of Hes1 compared with transfection of either vector alone in HSC-T6 cells, while pcDNA3-CA-ALK5 (TGF-β signaling activin receptor-like kinase 5) reduced the effect of NICD1 on Hes1 expression.
CONCLUSION: Selective interruption of Hes1 or maintenance of Hes1 at a reasonable level decreases the promoter activities of α-SMA and COL1α2, and these conditions may provide an anti-fibrotic strategy against hepatic fibrosis.
Core tip: In our paper, we found that Notch signaling appears to crosstalk with TGF-β/BMP signaling and co-regulates the expression of Hes1 in hepatic stellate cells. Notch1 may keep Hes1 protein in a reasonable range due to Hes1 auto-negative feedback, allowing Notch1 to repress the expression of α-SMA and collagen1α2 (COL1α2). Over-expression of Hes1 via transfection, thus lacking the negative feedback, enhances the promoter activities of α-smooth muscle actin (α-SMA) and COL1α2, consistent with the finding when Hes1 is captured by a Hes1-decoy oligodeoxynucleotide. This may provide an anti-fibrotic strategy against hepatic fibrosis. Further investigations in the future on the molecular mechanism by which Hes1 regulates the expression of α-SMA and COL1α2 should provide additional support for such an approach.
-
Citation: Zhang K, Zhang YQ, Ai WB, Hu QT, Zhang QJ, Wan LY, Wang XL, Liu CB, Wu JF.
Hes1 , an important gene for activation of hepatic stellate cells, is regulated by Notch1 and TGF-β/BMP signaling. World J Gastroenterol 2015; 21(3): 878-887 - URL: https://www.wjgnet.com/1007-9327/full/v21/i3/878.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i3.878
The activation of hepatic stellate cells (HSCs), a key step in liver fibrosis, can lead to the development of myofibroblast-like cells and contribute to liver fibrosis[1]. The main characteristics of the formation of myofibroblast-like cells are the expression of α-smooth muscle actin (α-SMA) and extracellular matrix (ECM) proteins, particularly collagen I[2]. Because of the importance of myofibroblasts in liver fibrosis, the majority of antifibrotic therapies are designed to inhibit the formation of myofibroblastic cells[3].
Previous studies have revealed that the transforming growth factor-β (TGF-β)/bone morphogenetic protein (BMP) signaling pathway is a pivotal player in the activation of HSCs[4,5]. TGF-β is a key mediator in the activation of HSCs[6]. BMPs are also members of the TGF-β superfamily and are antagonistic against TGF-β[7].
Notch signaling is an evolutionarily conserved pathway controlling diverse aspects of development and disease[8]. In mammals, the Notch family contains five Notch ligands (Jagged1, Jagged2, Delta-like1, Delta-like3, and Delta-like4) and four Notch receptors (Notch1-4). Among the Notch target genes are the basic helix-loop-helix (bHLH) transcription factors of the Hes (Hairy-Enhancer of Split) and HERP (Hes-related repressor protein) families, which function as transcriptional repressors[9,10].
During hematopoiesis and immune development, Notch is critical for T/B lineage specification and for the generation of splenic marginal zone B cells[11,12]. Earlier research has mainly focused on the interconnections between Notch and hematopoiesis or immune system-related molecules, aiming to find a preferable method to modulate hematopoiesis or treat immune disorders.
Recently, several studies have implicated Notch signaling in some human fibrosis diseases, such as renal, pulmonary and even liver fibrosis[13,14]. Evidence suggests that TGF-β/BMP signaling, as a key mediator in the process of HSC activation, has a synergistic effect with Notch in many cell types, enhancing the expression of target genes[15,16]. While the Jagged/Notch pathway may selectively mediate the fibrogenic properties of TGF-β1, which are essential to promote the production and deposition of the ECM[17,18], whether Notch signaling regulates TGF-β/BMP in HSCs remains largely unclear.
We hypothesized that Notch signaling can interact with TGF-β/BMP signaling and play a role in HSC activation. To test this hypothesis, we investigated the relationship between the activation of HSCs and Notch signaling pathways. Our findings demonstrate a pivotal role for Notch signaling in this process.
The use of animals for this study was approved by the laboratory animal center of China, Three Gorges University. Primary rat HSCs were isolated using a previously described method with minor modifications[19]. Freshly isolated, normal rat HSCs were cultured on 6- or 24-well plates in low glucose DMEM (Invitrogen) supplemented with 10% FBS and antibiotics for 0, 1, 3, 7, or 10 d to examine the quiescent, activating, or fully activated cells. HSC-T6 cells, an immortalized rat HSC line provided by the institute of liver disease at Shanghai University of Traditional Chinese Medicine, were cultured in low glucose DMEM supplemented with 10% FBS.
Cells were lysed in lysis buffer (25 mmol/L Tris-HCl pH 7.5, 2.5 mmol/L EDTA, 137 mmol/L NaCl, 2.7 mmol/L KCl, 1% sodium deoxycholic acid, 0.1% SDS, 1% TritonX-100, and 2 mmol/L PMSF) and protease inhibitor cocktail for 30 min at 4 °C. The cell lysates were clarified by centrifugation at 12000 rpm for 20 min at 4 °C, and the supernatants were collected. The protein concentrations were measured using a BCA Protein Assay kit (Thermo). An equal amount of protein from each sample was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane. The membrane was incubated with a mouse monoclonal anti-α-SMA antibody (1:1000 dilution) (Sigma), a goat polyclonal anti-Hes1 antibody (1:1000 dilution) (SANTA CRUZ), a rabbit polyclonal anti-TGF-β(R) antibody (1:1000 dilution) (SANTA CRUZ), a mouse monoclonal anti-HA antibody (1:3000 dilution) (Abmart), a mouse monoclonal anti-c-myc antibody (1:3000 dilution) (Abmart) or a mouse monoclonal anti-β-actin antibody (1:3000 dilution) (Sigma) for 24 h at 4 °C. This primary antibody incubation was followed by incubation with HRP-conjugated anti-mouse (1:3000 dilution), anti-rabbit (1:3000 dilution) or anti-goat (1:12000 dilution) antibody as the secondary antibody for 1 h at RT. These membranes were developed using Immobilon Western Detection Reagents (Millipore) according to the manufacturer’s instructions. The chemiluminescence on the membrane was detected using the VersaDoc system (Bio-Rad). Densitometric analyses of the band intensities were performed using ImageJ software (version 1.38 ×; National Institutes of Health).
Total RNA from freshly isolated HSCs or HSC-T6 cells was extracted using TRIzol reagent (Axygen). The RNA (2 mg) was reverse transcribed using a Transcriptor First Strand cDNA Synthesis Kit according to the manufacturer’s instructions (Fermentas). The primers used in the polymerase chain reaction (PCR) reactions to detect the mRNA levels were synthesized by Sangon Biotech (Shanghai, China) and are described in Table 1.
| Gene | Forward primer | Reverse primer | Bp | Accession No. |
| Notch1 | AGAGCTTTTCCTGTGTCTGTCC | CGGTACAGTCAGGTGTGTTGTT | 414 | NM_001105721 |
| Notch2 | GACTGCCAATACTCGACCTC | TTCAGAAGTGAAGTCTCCAG | 438 | NM_024358.1 |
| Notch3 | CCTCTTTCACCTGTACCTGTCC | ACACAGTAGTGGGAGTGGTCCT | 496 | NM_020087 |
| Notch4 | CAACTCTGCGAGAACGGTGG | TGGAAGGAGCCCAAGGTGTT | 443 | NM_001002827.1 |
| hes1 | GCCAGCTGATATAATGGA | TAGGTCATGGCGTTGATC | 462 | NM_024360.3 |
| hey1 | CTACAGCTCCTCTGATAGTG | GAGGCATCGAGTCCTTCAAT | 420 | NM_001191845 |
| TGF-β1 | GGTGAAACGGAAGCGCATCG | CTTGAATCTCTGCAGGCGCA | 364 | NM_021578.2 |
| E-cadherin | CTCGTGGCTTTGTCAGCA | GACCCAGTCTCGTTTCTG | 432 | NM_031334.1 |
| α-SMA | GTGTGAAGAGGAAGACAG | TTGGCCTTAGGGTTCAGC | 348 | NM_031004.2 |
| β-catenin | CTGACAGAGTTGCTCCACTC | CAGCCCATCAACTGGATAGT | 342 | NM_053357.2 |
| Bmp7 | CAGAGCATCAACCCCAAGTT | GATGAAGTGAACCAGTGTCT | 396 | NM_001191856.1 |
| COL1α2 | ACCTCAGGGTGTTCAAGGTG | CGGATTCCAATAGGACCAGA | 222 | NM_053356 |
| GAPDH | ACCACAGTCCATGCCATCAC | TCCACCACCCTGTTGCTGTA | 480 | NM_017008.4 |
Freshly isolated, normal rat HSCs cultured for varying lengths of time (1, 3, 7, 10 d) were fixed in phosphate buffered saline containing 4% paraformaldehyde at room temperature for 30 min and were then treated with blocking solution containing 0.1% Triton X-100. The fixed and blocked cells were incubated for 1 h with either a mouse monoclonal anti-α-SMA (1:100) (Sigma), a mouse monoclonal anti-GFAP (1:250) (Sigma) or a mouse monoclonal anti-desmin (1:600) (Sigma) antibody in 3% NCBS. The cells were then incubated with a Cy3-conjugated anti-mouse secondary antibody (1:1000) (NovoGene) for 1 h at 37 °C. After incubation, the cells were stained with the nuclear stain 4’,6-diamidino-2-phenyl-indole (DAPI). The cells were then examined under a fluorescence microscope (Nikon, Japan).
The pcDNA3-CA-ALK3 (BMP signaling activin receptor-like kinase 3)[20] and pcDNA3-CA-ALK5 (TGF-β signaling activin receptor-like kinase 5)[21] plasmids were gifts from Professor Miyazono at Tokyo University. The rat Notch1 intracellular domain (NICD1) and Hes1 cDNA (Hes1) were cloned into the pcDNA3.1(+) vector. The promoters of α-SMA (pSMA), COL1α1 (P-COL1α1), COL1α2 (P-COL1α2) and Hes1 (P-Hes1) were cloned into the pGLuc-Basic vector (N8082S, NEB, United States) for luciferase assays. The HSC-T6 cells were seeded in a 24- or 6-well plate at 60% confluence. After 24 h, the cells were transfected with different plasmids using the FuGENE HD Transfection Reagent (Roche) according to the manufacturer’s instructions. The cells were lysed after 36 h in culture for Western blot analyses. For luciferase assays, the supernatants were collected 24 h after transfection, and the assays were performed using the Gaussia Luciferase Assay Kit (BioLux) according to the manufacturer’s instructions. The reactions were examined using a Fluorescence Detector (Brethold).
The Hes1 oligodeoxynucleotide (ODN) decoy (5’-TTT CAC GAG TTT TCA CGA GTT T-3’, 5’-AAA CTC GTG AAA ACT CGT GAA A-3’) and scrambled (Scr) ODN decoy (5’-TTT ACA GAG TTT TAC AGA GTT T-3’, 5’- AAA CTC TGT AAA ACT CTG TAA A-3’) were synthesized by Sangon Biotech (Shanghai, China). These ODN decoys were annealed overnight while the temperature decreased from 80 °C to 25 °C. After GLuc reporter plasmids were transfected into HSC-T6 cells for 12 h, the Hes1 ODN decoy was delivered by Mirus (Mirus Bio Corporation). The supernatant was collected for luciferase reporter analysis after 24 h of culture.
Data are presented as mean ± SE of several experiments. The statistical significance was assessed using a two-tailed Student’s t-test. P < 0.05 was considered statistically significant.
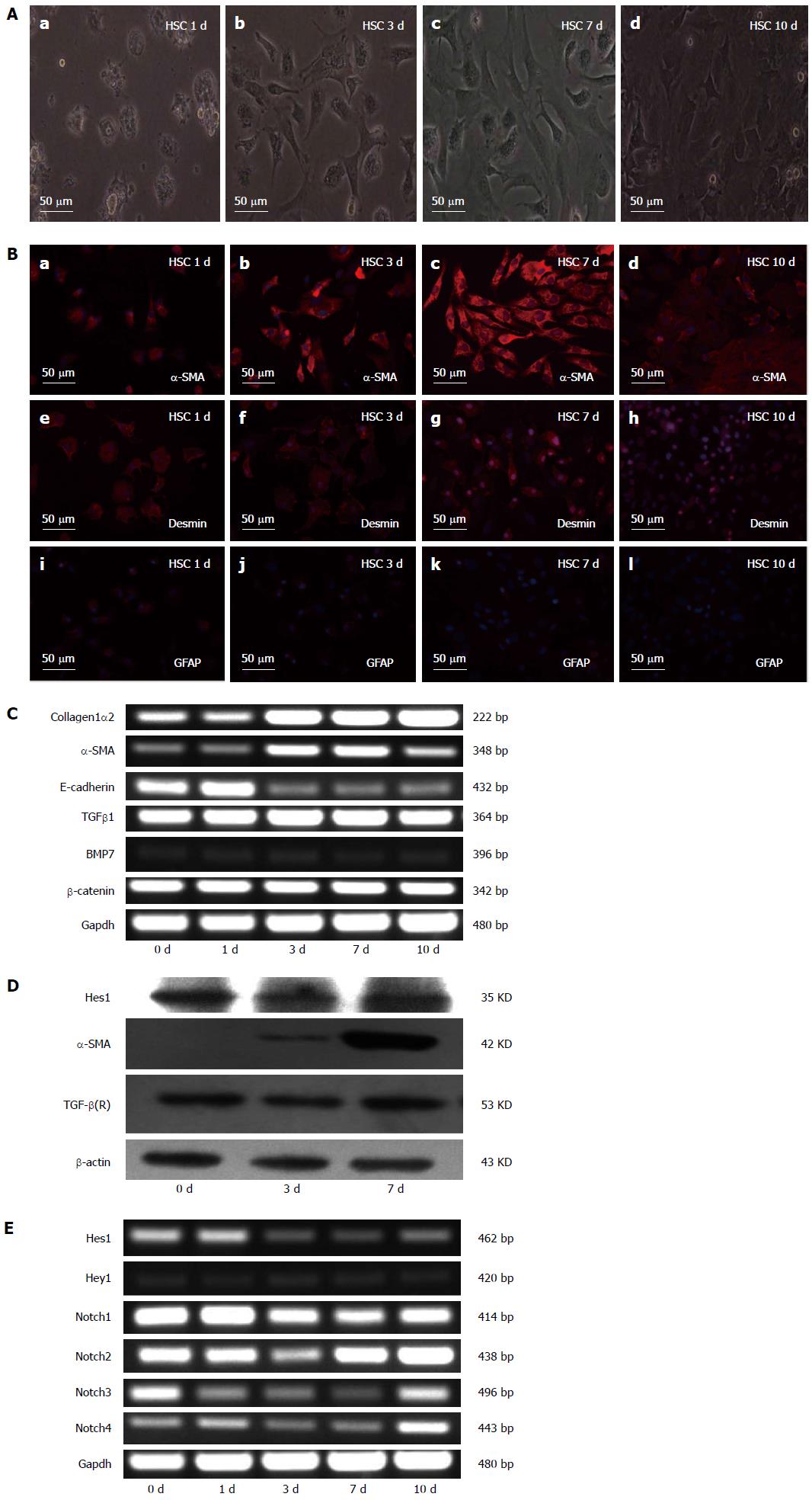
The activation of HSCs can be modeled in vitro by culturing freshly isolated (quiescent) rodent or human HSCs on plastic in serum-containing media. Freshly isolated HSCs from normal rats were cultured for various lengths of time (1, 3, 7 and 10 d), and the results showed that the HSCs lost their typical lipid droplets and developed into myofibroblast-like cells during culture (Figure 1A). When various HSC markers were analyzed by immunofluorescence, the expression of the HSC marker GFAP had decreased, desmin synthesis had increased, and the myofibroblast marker α-SMA had also increased (Figure 1B). To further characterize the quiescent and activated HSCs, we analyzed the expression of α-SMA, COL1α2, E-cadherin, TGF-β1, BMP7, and β-catenin on different days during the culture period (0, 1, 3, 7, and 10 d) by RT-PCR. The results showed that the mRNA levels of TGF-β1, BMP7 and β-catenin did not change during the activation of HSCs; the mRNA levels of E-cadherin, a marker of epithelial-mesenchymal transition (EMT), decreased, while the mRNA levels of the hepatic fibrosis-related genes α-SMA and COL1α2 increased (Figure 1C). We also used RT-PCR to analyze the expression of the Notch receptors (Notch1-4) and their target genes Hes1 and Hey1 on different days during the culture period. The mRNA levels of the Notch1 receptor decreased during culture, while the mRNA levels of the other Notch receptors (Notch2-4) changed inconsistently (Figure 1E). The mRNA levels of the Notch target gene Hes1 decreased, but the protein did not significantly change (Figure 1D).
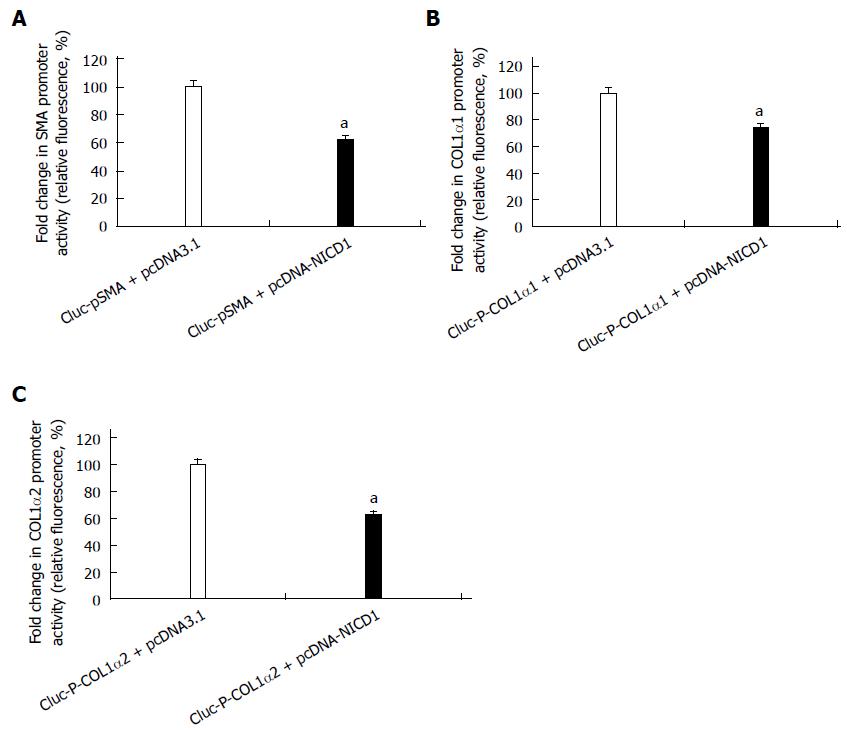
To investigate the role of Notch1 on the activation of HSCs, we examined the effect of over-expression of Notch1 in the HSC-T6 cell line. After the pcDNA3.1-NICD1 was co-transfected with reporter plasmids of GLuc-pSMA, GLuc-P-COL1α1 or GLuc-P-COL1α2, respectively, into HSC-T6 for 24 h, the supernatant was collected for a luciferase reporter assay. The results revealed that the promoter activities of α-SMA, COL1α1 and COL1α2 were reduced (Figure 2).
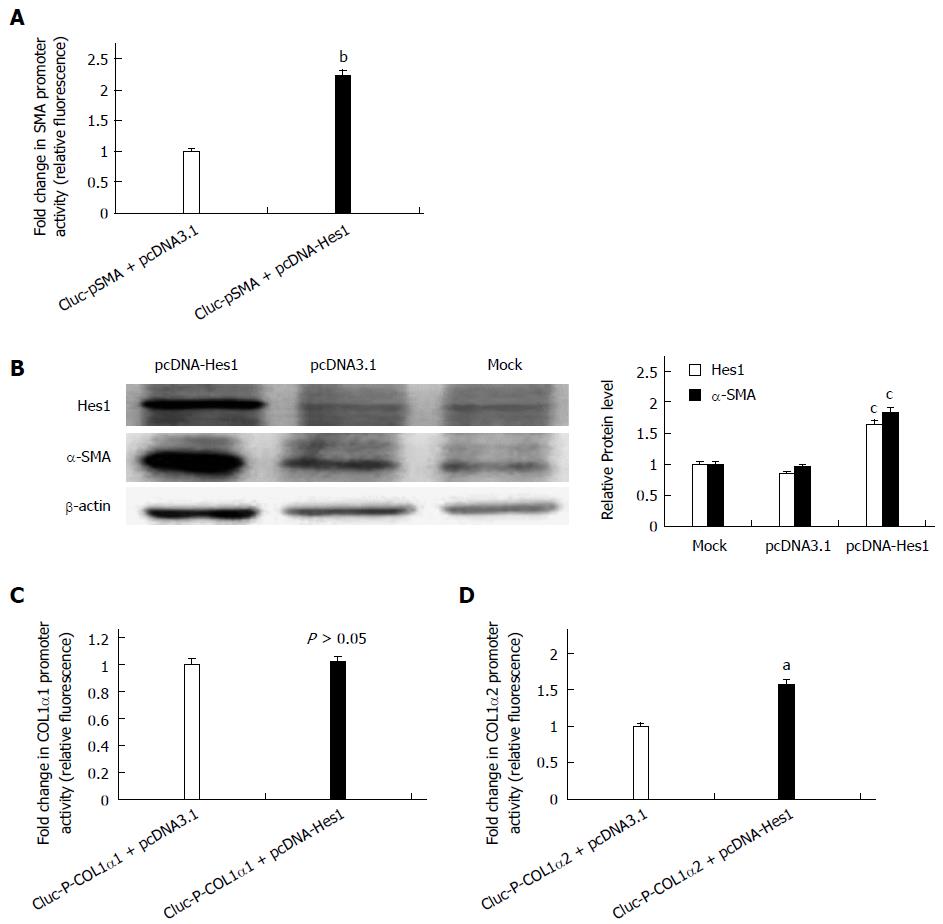
To determine whether the Notch1 target gene Hes1 is involved in the activation of HSCs, the pcDNA-Hes1 plasmid was co-transfected with the reporter plasmids GLuc-pSMA, GLuc-P-COL1α1 or GLuc-P-COL1α2 into HSC-T6 cells for 24 h. The supernatant was collected for luciferase assays. The activities of GLuc-pSMA and GLuc-P-COL1α2 were higher in the pcDNA-Hes1 transfected cells than in the empty vector control cells (Figure 3A, C), while over-expression of Hes1 did not alter the activity of the COL1α1 promoter (Figure 3B). Over-expression of Hes1 also increased the expression of endogenous α-SMA, as revealed by Western blot analysis (Figure 3D). Thus, Hes1, a Notch signaling target gene, has an opposite effect on the activation of HSCs to that of the Notch1 receptor.
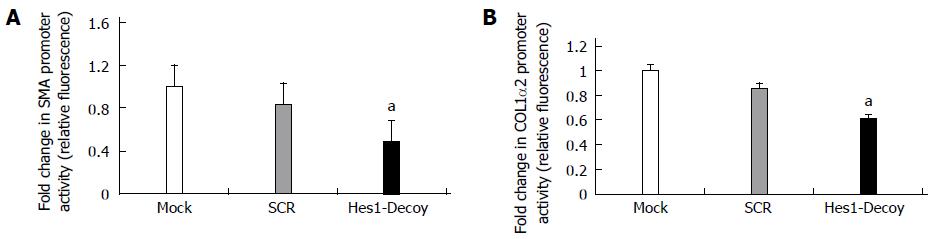
To inhibit Hes1, an ODN decoy was transfected into the HSCs. The Hes1 ODN decoy is a double-stranded DNA containing a Hes1 binding site. After the Hes1 ODN decoy was delivered into the HSC-T6 cells, the luciferase reporter activities for the pSMA and P-COL1α2 reporter constructs decreased compared with those in the control group (Scr) (Figure 4A, B). Thus, inhibiting the function of the Hes1 transcription factor in HSC-T6 cells could down-regulate the expression of α-SMA and COL1α2.
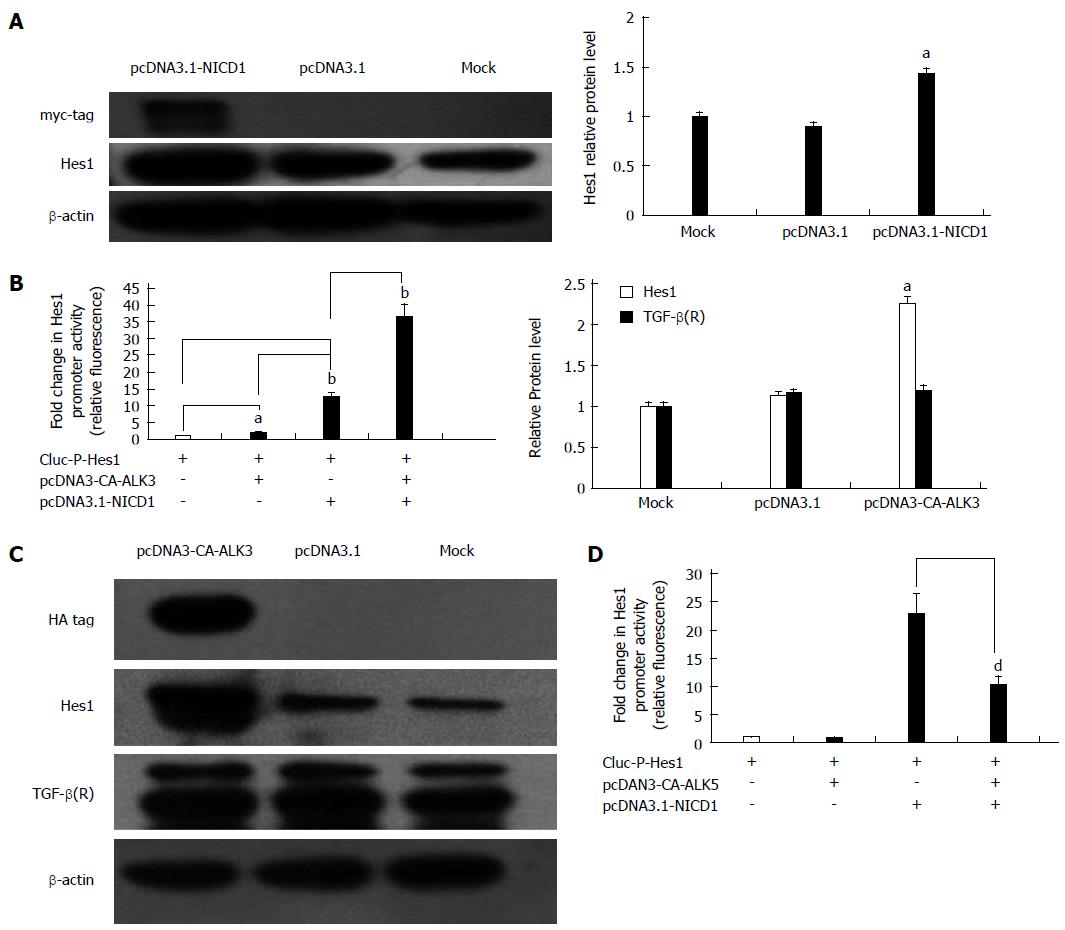
It is well known that Notch1 can induce the expression of Hes1 directly in many mammalian cell types. To determine whether Notch1 up-regulates the expression of Hes1 in HSC-T6 cells, the GLuc-P-Hes1 and pcDNA3.1-NICD1 plasmids were co-transfected into HSC-T6 cells, and GLuc reporter analysis showed that the promoter activity of Hes1 was enhanced approximately 5.5-fold compared with the activity in cells transfected with the empty plasmid pcDNA3.1 (Figure 5B). At the protein level, pcDNA3.1-NICD1 also increased the expression of Hes1 based on Western blot analysis (Figure 5A). Next, we studied the relationship between the TGF-β/BMP signaling pathway and Hes1. To investigate whether the Notch pathway interacts with the TGF-β/BMP signaling pathway in HSC-T6 cells, a pcDNA3-CA-ALK3 (BMP signaling activin receptor-like kinase 3) expression vector was transfected into HSC-T6 cells, and the Notch target gene Hes1 and the TGF-β receptor were analyzed by Western blot. We found that over-expression of CA-ALK3 increased the expression of Hes1 approximately 2.5-fold (Figure 5C), while the expression of the TGF-β receptor was not altered (Figure 5C). We also obtained the same results using a luciferase reporter assay. When pcDNA3-CA-ALK3 and GLuc-P-Hes1 were co-transfected into HSC-T6 cells, the reporter activity increased approximately 2-fold compared with the control (empty vector pcDNA3.1). In addition, co-transfection with pcDNA3-CA-ALK3 and pcDNA3.1-NICD1 cooperatively enhanced the expression of Hes1 compared to transfection with pcDNA3-CA-ALK3 or pcDNA3.1-NICD1 alone in HSC-T6 cells (Figure 5B). Thus, CA-ALK3 enhances NICD1-mediated up-regulation of Hes1 in HSC-T6 cells. We also analyzed the effect of CA-ALK5, an antagonist of CA-ALK3, and found that it could not affect Hes1 expression by itself in HSC-T6 cells but reduced the effects of NICD1 on Hes1 expression (Figure 5D).
It is well accepted that HSCs play a major role in the progression of liver fibrosis because the cells can develop into myofibroblast cells that synthesize extracellular matrix proteins[22]. Thus, a better understanding of the molecular mechanisms underlying HSC activation is an important prerequisite for developing new therapeutic modalities for liver fibrosis. TGF-β signaling has been shown to be the key event in promoting fibrogenesis in vivo and in vitro; however, the development of anti-fibrotic strategies targeting the TGF-β axis is uncertain because of the pleiotropic nature of TGF-β action[23].
Notch signaling is critical for the regulation of cell differentiation, and its aberrant activation has been implicated in human fibrosis diseases. It has ben confirmed that Notch3 may regulate the activation of HSCs, but it is unknown whether other members of the Notch family, such as Notch1, Notch2 and Notch4, play a part in activating HSCs and if so, what the underlying mechanism may be[24]. In studies of the HSC niche in the rat liver, it has been found that Notch1 is involved in the regulation of β-catenin-dependent Wnt signaling and may be a marker for activated somatic stem/progenitor cells[25,26]. Some studies have also shown that, in response to inflammatory zone 1, Notch1 signaling may play a significant role in myofibroblast differentiation during lung fibrosis[14]. Thus, we hypothesized that Notch1 signaling may play an important role in liver fibrosis.
Our present study suggests that Notch1 and its target gene, Hes1, play crucial roles in liver fibrosis. Based on the evidence described above, we focused on Notch1 and its target gene Hes1 in our study. Our findings showed that Notch1 and Hes1 mRNA expression decreased upon the activation of freshly isolated HSCs, in agreement with an earlier report[27]. To investigate the effects of the Notch1 receptor and Hes1 in HSC-T6 cells, we over-expressed Notch1 and Hes1 in HSC-T6 cells and found that Notch1 inhibited α-SMA, COL1α1 and COL1α2 promoter activities, while Hes1 enhanced the α-SMA and COL1α2 promoter activities. These findings suggest that the Notch1 receptor, as an upstream gene in the Notch signaling pathway, has pleiotropic effects not displayed by its downstream target, the Hes1 transcription factor. Hes1 is a bHLH transcription factor, and Hes1 homodimers auto-inhibit their own transcription by directly binding to the N-box in the Hes1 promoter. This negative feedback leads to the rapid disappearance of the extremely unstable Hes1 protein and allows a new round of expression[16]. This negative feedback may keep expression of the Hes1 protein in a reasonable range when pcDNA3.1-NICD1 is transfected into HSCs. On the other hand, Hes1 protein expression will be much higher when pcDNA3.1-Hes1 is transfected into HSCs. Because of the pleiotropic effects of Notch1 and the negative feedback of Hes1, Notch1 may keep the Hes1 protein at a reasonable level and restrain the promoter activities of α-SMA, COL1α1 and COL1α2 in HSCs, while over-expression of Hes1 may enhance the promoter activities of α-SMA and COL1α2.
We have shown that Hes1 participated in the activation of HSCs. Thus, another important question is which factors affect Hes1 expression in HSCs. Previous studies have suggested that Notch signaling may interact with TGF-β/BMP signaling and regulate the expression of Hes1 in many cell types[16]. To determine whether a similar mechanism exists in HSC-T6 cells, we analyzed the effect of CA-ALK3 and CA-ALK5. CA-ALK3 alone increased the expression of Hes1, and more importantly, CA-ALK3 strengthened the NICD1-mediated effects on Hes1 expression in HSC-T6 cells. CA-ALK5, on the other hand, could not alone affect Hes1 expression in HSC-T6 cells, but it did reduce the effects of NICD1 on the expression of Hes1. Earlier studies suggested a mechanism in which NICD1 binds phosphorylated SMAD1/5/8 (P-SMAD1/5/8) and forms a NICD1-P-SMAD1/5/8 complex, which then migrates into the nucleus, causing the activation of the Hes1 promoter in HSC-T6 cells[28,29]. CA-ALK5 can increase SMAD2/3 phosphorylation (P-SMAD2/3) and attenuate the levels of P-SMAD1/5/8 through SMAD6, ultimately leading to a significant reduction in the NICD1-P-SMAD1/5/8 complex and inhibition of Hes1 promoter activity[16]. Thus, our results demonstrate that TGF/BMP signaling interacts with Notch1 to regulate Hes1 expression in HSC-T6 cells. Clearly, the role of Notch1 and TGF-β1/BMP signaling in the regulation of the Hes1 promoter should be further investigated.
In conclusion, Notch signaling appears to interact with TGF-β/BMP signaling and co-regulates Hes1 expression in HSCs. Notch1 may keep Hes1 protein expression at a reasonable level through Hes1 auto-negative feedback, allowing Notch1 to repress the expression of α-SMA and COL1α2. This repression is consistent with the effect of capturing Hes1 using a Hes1 ODN decoy, which could, nonetheless, reduce the effects of NICD1 on Hes1 expression. Over-expression of Hes1 via transfection, which bypasses the negative feedback, enhances the promoter activities of α-SMA and COL1α2. This may provide an anti-fibrotic strategy against hepatic fibrosis. Further investigations of the molecular mechanisms of Hes1-mediated regulation of α-SMA and COL1α2 should provide additional support for such an approach.
We are grateful to the Institute of Molecular Biology of China, Three Gorges University, for providing us with an experimental platform, and thank Li-Li Zou for technical assistance. We thank Dr. Yun-Bo Shi of the American NIH for his helpful suggestions on the manuscript.
It is well known that a better understanding of the molecular mechanisms underlying hepatic stellate cell (HSC) activation is an important prerequisite for developing new therapeutic modalities for liver fibrosis. The Notch signaling pathway is critical for the regulation of cell differentiation. Recently, Notch has been implicated in human liver fibrosis diseases. It has been confirmed that Notch3 may regulate the activation of HSCs, but it is unknown whether other members of the Notch family, such as Notch1, Notch2 and Notch4, play a part in HSC activation and what the underlying mechanisms might be.
During hematopoiesis and immune development, Notch is critical for T/B lineage specification and for the generation of splenic marginal zone B cells. Earlier research mainly focused on the interconnections between Notch and hematopoiesis or immune system-related molecules, aiming to find a preferable method to modulate hematopoiesis or treat immune disorders. In our paper, we explored whether Notch signaling has a function in regulating HSC activation.
This study illuminates the role of Notch1 and its target gene Hes1 in HSC activation. Furthermore, we found that Notch signaling appears to interact with transforming growth factor-β (TGF-β)/bone morphogenetic protein (BMP) signaling and co-regulates Hes1 expression in HSCs. This finding indicates that Notch1 and its target gene Hes1 may contribute to liver fibrosis by regulating HSC activation.
This study demonstrated that over-expression of Notch1 or selective interruption of Hes1 decreases the expression of myofibroblastic markers in rat hepatic stellate cells, which provides a potential novel therapeutic target for liver fibrosis.
The submitted manuscript entitled, “Hes1, an important gene for activation of hepatic stellate cells, is regulated by Notch1 and TGF-β/BMP signaling” presents a very interesting study. The finding highlights the crosstalk between Notch and TGF-β/BMP signaling modules in the regulation of Hes1 in hepatic stellate cells towards the understanding of anti-fibrotic mechanism. Overall, the study is very well conceived, designed and executed as well as well written.
P- Reviewer: Higuera-de la Tijera MF, Kovacs SJ, Parvez MK S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Liu XM
| 1. | Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125-172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2244] [Cited by in RCA: 2200] [Article Influence: 129.4] [Reference Citation Analysis (0)] |
| 2. | Wells RG. The role of matrix stiffness in hepatic stellate cell activation and liver fibrosis. J Clin Gastroenterol. 2005;39:S158-S161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 176] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 3. | Bataller R, Brenner DA. Hepatic stellate cells as a target for the treatment of liver fibrosis. Semin Liver Dis. 2001;21:437-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 397] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 4. | Purps O, Lahme B, Gressner AM, Meindl-Beinker NM, Dooley S. Loss of TGF-beta dependent growth control during HSC transdifferentiation. Biochem Biophys Res Commun. 2007;353:841-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 5. | Choi SS, Diehl AM. Epithelial-to-mesenchymal transitions in the liver. Hepatology. 2009;50:2007-2013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 257] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 6. | Sugiyama D, Kulkeaw K, Mizuochi C. TGF-beta-1 up-regulates extra-cellular matrix production in mouse hepatoblasts. Mech Dev. 2013;130:195-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 7. | Weiskirchen R, Meurer SK, Gressner OA, Herrmann J, Borkham-Kamphorst E, Gressner AM. BMP-7 as antagonist of organ fibrosis. Front Biosci (Landmark Ed). 2009;14:4992-5012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 72] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 8. | Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4480] [Cited by in RCA: 4554] [Article Influence: 175.2] [Reference Citation Analysis (0)] |
| 9. | Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 942] [Cited by in RCA: 971] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 10. | Kageyama R, Ohtsuka T, Tomita K. The bHLH gene Hes1 regulates differentiation of multiple cell types. Mol Cells. 2000;10:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 125] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 11. | Harman BC, Jenkinson EJ, Anderson G. Entry into the thymic microenvironment triggers Notch activation in the earliest migrant T cell progenitors. J Immunol. 2003;170:1299-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 44] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 12. | Maillard I, Adler SH, Pear WS. Notch and the immune system. Immunity. 2003;19:781-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 165] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 13. | Bielesz B, Sirin Y, Si H, Niranjan T, Gruenwald A, Ahn S, Kato H, Pullman J, Gessler M, Haase VH. Epithelial Notch signaling regulates interstitial fibrosis development in the kidneys of mice and humans. J Clin Invest. 2010;120:4040-4054. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 296] [Cited by in RCA: 302] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 14. | Liu T, Hu B, Choi YY, Chung M, Ullenbruch M, Yu H, Lowe JB, Phan SH. Notch1 signaling in FIZZ1 induction of myofibroblast differentiation. Am J Pathol. 2009;174:1745-1755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 103] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 15. | Fu Y, Chang A, Chang L, Niessen K, Eapen S, Setiadi A, Karsan A. Differential regulation of transforming growth factor beta signaling pathways by Notch in human endothelial cells. J Biol Chem. 2009;284:19452-19462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 94] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 16. | Beets K, Huylebroeck D, Moya IM, Umans L, Zwijsen A. Robustness in angiogenesis: notch and BMP shaping waves. Trends Genet. 2013;29:140-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 17. | Nyhan KC, Faherty N, Murray G, Cooey LB, Godson C, Crean JK, Brazil DP. Jagged/Notch signalling is required for a subset of TGFβ1 responses in human kidney epithelial cells. Biochim Biophys Acta. 2010;1803:1386-1395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Dees C, Zerr P, Tomcik M, Beyer C, Horn A, Akhmetshina A, Palumbo K, Reich N, Zwerina J, Sticherling M. Inhibition of Notch signaling prevents experimental fibrosis and induces regression of established fibrosis. Arthritis Rheum. 2011;63:1396-1404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 110] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 19. | Eda H, Kulig KM, Steiner TA, Shimada H, Patel K, Park E, Kim ES, Borenstein JT, Neville CM, Keller BT. A nanofiber membrane maintains the quiescent phenotype of hepatic stellate cells. Dig Dis Sci. 2012;57:1152-1162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 20. | Fujii M, Takeda K, Imamura T, Aoki H, Sampath TK, Enomoto S, Kawabata M, Kato M, Ichijo H, Miyazono K. Roles of bone morphogenetic protein type I receptors and Smad proteins in osteoblast and chondroblast differentiation. Mol Biol Cell. 1999;10:3801-3813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 347] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 21. | Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin CH, Miyazono K. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16:5353-5362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 806] [Cited by in RCA: 855] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 22. | Tsukada S, Parsons CJ, Rippe RA. Mechanisms of liver fibrosis. Clin Chim Acta. 2006;364:33-60. [PubMed] |
| 23. | Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1096] [Cited by in RCA: 1380] [Article Influence: 98.6] [Reference Citation Analysis (0)] |
| 24. | Chen YX, Weng ZH, Zhang SL. Notch3 regulates the activation of hepatic stellate cells. World J Gastroenterol. 2012;18:1397-1403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (2)] |
| 25. | Sawitza I, Kordes C, Reister S, Häussinger D. The niche of stellate cells within rat liver. Hepatology. 2009;50:1617-1624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 117] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 26. | Reister S, Kordes C, Sawitza I, Häussinger D. The epigenetic regulation of stem cell factors in hepatic stellate cells. Stem Cells Dev. 2011;20:1687-1699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 27. | Xie G, Karaca G, Swiderska-Syn M, Michelotti GA, Krüger L, Chen Y, Premont RT, Choi SS, Diehl AM. Cross-talk between Notch and Hedgehog regulates hepatic stellate cell fate in mice. Hepatology. 2013;58:1801-1813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 105] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 28. | Larrivée B, Prahst C, Gordon E, del Toro R, Mathivet T, Duarte A, Simons M, Eichmann A. ALK1 signaling inhibits angiogenesis by cooperating with the Notch pathway. Dev Cell. 2012;22:489-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 301] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 29. | Moya IM, Umans L, Maas E, Pereira PN, Beets K, Francis A, Sents W, Robertson EJ, Mummery CL, Huylebroeck D. Stalk cell phenotype depends on integration of Notch and Smad1/5 signaling cascades. Dev Cell. 2012;22:501-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 186] [Article Influence: 14.3] [Reference Citation Analysis (0)] |