Published online Jan 21, 2015. doi: 10.3748/wjg.v21.i3.803
Peer-review started: July 16, 2014
First decision: September 15, 2014
Revised: September 30, 2014
Accepted: November 7, 2014
Article in press: November 11, 2014
Published online: January 21, 2015
Processing time: 188 Days and 21 Hours
There are more than 1000 microbial species living in the complex human intestine. The gut microbial community plays an important role in protecting the host against pathogenic microbes, modulating immunity, regulating metabolic processes, and is even regarded as an endocrine organ. However, traditional culture methods are very limited for identifying microbes. With the application of molecular biologic technology in the field of the intestinal microbiome, especially metagenomic sequencing of the next-generation sequencing technology, progress has been made in the study of the human intestinal microbiome. Metagenomics can be used to study intestinal microbiome diversity and dysbiosis, as well as its relationship to health and disease. Moreover, functional metagenomics can identify novel functional genes, microbial pathways, antibiotic resistance genes, functional dysbiosis of the intestinal microbiome, and determine interactions and co-evolution between microbiota and host, though there are still some limitations. Metatranscriptomics, metaproteomics and metabolomics represent enormous complements to the understanding of the human gut microbiome. This review aims to demonstrate that metagenomics can be a powerful tool in studying the human gut microbiome with encouraging prospects. The limitations of metagenomics to be overcome are also discussed. Metatranscriptomics, metaproteomics and metabolomics in relation to the study of the human gut microbiome are also briefly discussed.
Core tip: Metagenomics plays a role in understanding the human gut microbiome, including the diversity of the gut microbiome, identifying novel genes, and determining the etiology of functional dysbiosis. A combination of metagenomics, metatranscriptomics, metaproteomics and metabolomics can also promote an understanding of the functional activity of the human gut microbiome and possibly provide a new strategy for disease diagnosis and treatment.
- Citation: Wang WL, Xu SY, Ren ZG, Tao L, Jiang JW, Zheng SS. Application of metagenomics in the human gut microbiome. World J Gastroenterol 2015; 21(3): 803-814
- URL: https://www.wjgnet.com/1007-9327/full/v21/i3/803.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i3.803
The human gastrointestinal tract harbors an extremely complex and dynamic microbial community, including archaea, bacteria, viruses and eukaryota[1]. Most of the microorganisms residing in the gastrointestinal tract are bacteria, with a density of approximately 1013-1014 cells/g fecal matter, in which 70% of the total microbes colonize the colon[2]. The gut microbial community plays an important role in protecting the host against pathogenic microbes[3-5], modulating immunity[6,7], regulating metabolic processes[8,9], and is regarded as a neglected endocrine organ[10]. Recently, the role of the human gut microbiome has been well reviewed[11-13].
Classical studies of the gut microbiome have been largely dependent on cultivation techniques. However, traditional culture methods only cultivate 10%-30% of gut microbiota[14-16]. With the rapid development of advanced molecular technologies such as PCR-denaturing gradient gel electrophoresis, it has been shown that the gut microbial ecosystem is far more complex than previously thought[17]. In recent years, several next-generation sequencing technologies have been developed[18,19], which further facilitate the analysis of a large number of microorganisms in different environments[20-22] and human body sites[23], including the human gut[24-26]. 16S rDNA sequence analysis and metagenomics are two effective DNA sequencing approaches, and both have been used to study uncultivated gut microbial communities. The former focuses on the sequencing of the conserved 16S rDNA gene present in all microbes[27,28], and has established a series of novel connections between intestinal microbiota composition and disease[29-31]. The research based on 16S rDNA sequence attempts to reveal “who’s there?” in a given microbial community, while shotgun metagenomic sequencing can be used to answer the complementary question of “what can they do?”[32,33].
Metagenomics was first described in 1998 by Handelsman and Rodon[34,35], and became another DNA sequencing approach to study the complex gut microbial community. It aims to catalog all the genes from a community by the random sequencing of all DNA extracted from the sample[36-38]. Firstly, the total DNA of all microorganisms is extracted from fecal samples. Before being sequenced, total DNA samples are randomly sheared by a “shotgun” approach. The comprehensive sequences are then analyzed to obtain either species profiles based on phylogenetic markers (16S rDNA)[39] or genomic profiles based on whole genomes[22]. The shotgun sequence reads are filtered to obtain the high-quality sequences for the whole genomic profile by metagenomics. Based on sequence overlaps, the filtered sequences are then assembled to form longer genomic sequence contigs. Computational methods are needed to code sequences in the contigs. Data mining and database searches applying different powerful algorithms are then used to annotate genes[40]. The information obtained from the sequence-based and functional metagenomics enables a more comprehensive understanding of the structure and function of microbial communities than ever before.
The European project, MetaHIT[37], and the American Human Microbiome Project[25,41] have contributed to the availability of the reference gene catalog. These projects have facilitated the study of the human intestinal microbiome using metagenomics. By applying metagenomics to investigate fecal samples from 124 European individuals, the MetaHIT consortium found 3.3 million non-redundant genes in the human gut microbiome for the first time. Surprisingly, the gene set was 150 times larger than the human gene complement. Moreover, over 99% of the genes in the human gut microbial communities were bacterial, which indicated that the entire cohort harbored more than 1000 bacterial species[37]. However, the number of genes in the human gut microbiome was expanded more than threefold by subsequent analyses[42,43]. These observations further completed the catalog of reference genes in the human gut microbiome.
A core human gut microbiome has also been explored[44]. The average human intestinal microbiome is now better defined and comprises approximately 160 bacterial species in each individual[37]. Moreover, on average, individual microbiota could have long-term stability[45,46]. By applying low-error 16S ribosomal RNA amplicon sequencing and whole-genome sequencing methods to characterize bacterial strain composition in the fecal microbiota of 37 patients in the United States, Jeremiah and colleagues[45] found that on average, their individual microbiota was remarkably stable and 60% of strains remained over the course of five years. From the in-depth profiling of metagenomic datasets derived from fecal metagenomes of healthy individuals, the human gut has been postulated to consist of three enterotypes, typified by the relative dominance of particular groups of organisms: Prevotella, Ruminococcus and Bactericides spp[47].
In addition, studies using different methods including metagenomics have found a series of factors that can influence the composition and diversity of the gut microbiome, such as diet[48-50], age[51,52], geography[51,53], drugs[54,55], and environmental substances[56]. For example, a study found that due to the difference in long-term dietary habits, human gut microbiome abundance and proportions varied between United States individuals. Furthermore, species composition, but not enterotype, in these subjects was affected by short-term changes in diet[57]. In the MARS-500 study, Mardanov et al[58] found dynamic changes in the gut microbiome in participants using metagenomic analysis.
In recent years, due to the rapidly developing computational methods critical for the analysis of metagenomic data and earlier surveys performed on marine and other environmental microbiome, more and more studies have focused on functional metagenomics of the human gut microbiome[37,38,51,59]. Escherichia coli (E. coli) is the most commonly used host for functional metagenomics, and genes from a large diversity of bacteria can be expressed within E. coli[33]. Furthermore, other species such as Streptomyces, Bacillus subtilis and Lactococcus lactis can also be used to promote the heterologous expression of Gram-positive bacterial DNA[60].
Hehemann and colleagues[61] found genes encoding the enzymes porphyranase and agarase from the gut microbiome in Japanese individuals, but not in North Americans. Interestingly, the marine Bacteroidetes are widely found in seaweed, and many Japanese eat seaweed regularly[61], thus it is likely that these functions were acquired from these organisms by lateral gene transfer.
The catabolism of dietary fibers is important for human health[32]. Carbohydrate active enzymes (CAZymes), produced by human intestinal microorganisms, can degrade components of dietary fiber into metabolizable monosaccharides and disaccharides. However, before metagenomics were used to study the human gut microbiome, the study of CAZymes was restricted to cultivated bacterial species. Several metagenomics studies focusing on the gut microbiome determined the diversity of CAZymes, revealing the human gut microbiome to be a surprisingly rich source of carbohydrate active enzymes[36,38,62,63]. In addition, Tasse et al[64] detected novel CAZymes using functional metagenomics on human gut microbial genes. The function-based screening of intestinal metagenome libraries also identified a large genetic repertoire of genes encoding bile-salt hydrolase enzymes[65]. These functions were selectively enriched in intestinal microorganisms. Furthermore, novel β-glucuronidase activity, dominant in healthy adults and children, from Firmicutes was revealed by the functional screening of large insert metagenomic libraries of E. coli clones independent of culturability[66].
Recently Vital and colleagues[67] performed an extensive analysis of butyrate-producing pathways and individual genes by screening thousands of sequenced bacterial genomes from the Integrated Microbial Genome database. They found that the genomes of 225 bacteria had the potential to produce butyrate, including many previously unknown candidates. Most candidates belonged to distinct families within Firmicutes[67]. These authors also used the established gene catalog to screen for butyrate synthesis pathways in 15 metagenomes obtained from the fecal samples of healthy individuals. The results revealed that a high percentage of total genomes exhibited a butyrate-producing pathway, and the most prevalent was the acetyl-coenzyme A pathway, followed by the lysine pathway[67].
Many bacterial infections are becoming more and more difficult to treat, partly due to the increasing antibiotic resistance of human pathogens[68-70]. Recent studies have revealed that some human commensal microbiota harbor numerous antibiotic-resistance genes (ARGs), which have led to the human gut-associated resistome[71,72]. Thus, it is important to understand the contribution of the whole microbiota of particular systems to antibiotic resistance[73].
The first population-level analysis of resistance gene prevalence in the human gut was performed by Seville et al[74]. Interestingly, they found that several of the tested genes were common in human microbiomes using microarray probes to identify 14 tetracycline and macrolide-resistance genes in the fecal and saliva samples of 20 healthy volunteers from England, Finland, France, Italy, Norway, and Scotland. However, the fecal samples from France and Italy showed significantly higher levels of some tetracycline and erythromycin genes than those of the samples from Scandinavia or the United Kingdom.
Compared with previous methods, the metagenomic approach is a powerful tool that can help us to gain a more comprehensive understanding of the ARGs in human gut microbes. By analyzing the genomic content of a microorganism, it is possible to predict the resistance phenotype and adapt a specific treatment. Functional metagenomics can be used to isolate completely novel ARGs from the non-cultivable fraction of the microbiota and reveal the complex background context in which antibiotic resistance evolves in both microbial and host communities.
By employing fecal metagenomic data from different countries, studies have confirmed that resistance gene prevalence exists in the human gut microbiome and the distribution of ARGs are different between countries[75,76]. Moreover, Forslund et al[75] found associations between transposable elements in the tested genes, which is consistent with the finding that ARGs can be exchanged among gastrointestinal microbes[77], particularly during host stress[78].
Sommer et al[72] screened the gene inserts that caused resistance in E. coli to 13 different antibiotics by performing functional metagenomics screening of fecal and oral samples from two human donors. They then compared these genes with previous homologs in pathogens and found a considerable diversity of ARGs in the microbes. Cheng et al[79] first applied the strategy of screening a relatively large-insert fosmid library generated from the gut microbiota of four healthy candidates. The library was used to screen for ARGs against seven antibiotics. The authors identified a variety of previously unknown resistance determinants and found that only the N-terminus conferred kanamycin resistance following a functional study on the new kanamycin resistance gene.
Recently, Moore et al[80] used functional metagenomics in fecal microbiota from 22 healthy infants and children to identify ARGs. They not only identified three novel resistance genes, but also reported their results on resistance to folate-synthesis inhibitors conferred by a predicted Nudix hydrolase, which was an important part of the folate-synthesis pathway. In addition, their functional metagenomic investigations demonstrated that fecal resistomes of healthy children had a higher diversity than previously suspected.
As reported, dysbiosis of the intestinal microbial community is associated with some diseases, including inflammatory bowel disease[81-83], obesity[38,84-86], diabetes[87-89], allergy[90,91], irritable bowel syndrome (IBS)[92], colorectal cancer[93-96], liver cirrhosis[97-99], nonalcoholic steatohepatitis[100,101], neurodevelopmental disorders[102,103], cardiovascular disorders[104], cholesterol gallstones[105], diarrhea[106], malnutrition[107], kidney disease[108], and colon polyps[109]. Recently, in a metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation, Holler et al[110] found a relative shift from a predominance of commensal bacteria toward Enterococci, which was particularly marked in patients who subsequently developed or suffered from active gastrointestinal graft-vs-host disease.
In addition to identifying dysbiosis of the human gut microbiome in some diseases, metagenomics can determine novel changes in microbial functions. A study using the metagenomic approach in patients with type 2 diabetes performed by Qin et al[87] found a moderate variation in the microbial composition of the gut between cases and controls. Microbial functions conferring sulfate reduction and oxidative stress resistance were also more abundant in patients with type 2 diabetes than in healthy controls. Similarly, Wei et al[98] analyzed the fecal microbiota of 20 hepatitis B liver cirrhosis patients and 20 healthy controls using metagenomic methods, and found an obvious change in fecal microbiota between the two groups. Importantly, compared with the controls, functional diversity was significantly reduced in the fecal microbiota in the patients. In addition, the fecal microbiota in the patients showed abundant metabolism of glutathione, gluconeogenesis, branched-chain amino acids, nitrogen and lipids, but a decrease in the level of aromatic amino acids, bile acid and cell cycle related metabolism.
Metagenomics can also be used to determine the interactions between intestinal bacteria and the host. Intestinal bacteria play an integral role in human health. However, the possible mechanisms related to the interactions between intestinal bacteria and the host are not understood. Lakhdari et al[111] used high-throughput screening technology to investigate intestinal microbial pathways, and found that E. coli metagenomic clones could modulate intestinal mucosal proliferation by activating the nuclear factor-κB pathway in epithelial cells[112-114]. Recently, by applying functional metagenomics, Dobrijevic et al[115] found that secreted and surface-exposed proteins from Gram-positive bacteria in the human gut microbiota played a role in immune modulation.
Metagenomics is also helpful in screening plasmid-encoding elements, especially with the improvement of methods for purifying high-quality and high-yield plasmid DNA[116]. The plasmid harbors numerous mobile genetic elements. A profound understanding of the mobile genetic elements associated with the human gut microbiota is meaningful as they can reflect the co-evolution of host and microbe in the human gut[32].
Jones and Marchesi[117] isolated novel plasmids from the human gut microbiota and revealed that some crucial genes were enriched in the human gut compared with other systems using the culture independent transposon-aided capture system. Recently, based on metagenomic analysis, several studies indicated that the horizontal gene could transfer between phylogenetically distant bacterial groups[64,65,118]. However, the triggers that promote this gene exchange are not known. Smillie et al[119] showed that ecology could be the main driver of gene exchange.
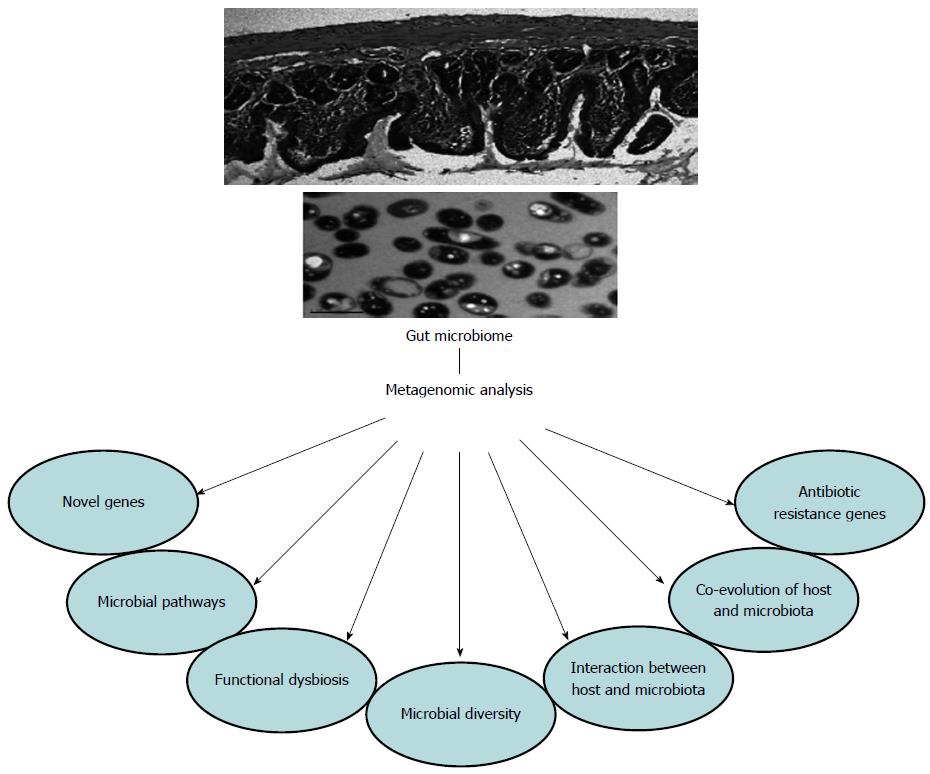
From the above findings (Figure 1), metagenomics have been shown to be an incredibly powerful technology in studying the human gut microbiome. However, there are still some limitations in the use of metagenomics, as shown in Figure 2. Firstly, it is not possible to identify microbial expression. Secondly, as metagenomics require much higher sequence coverage than 16S rDNA sequence analysis[120], the costs and time involved in DNA sequencing projects for gut metagenomics are far greater than those of 16S rDNA sequence analysis. Thirdly, to obtain high coverage required for metagenomics, a sufficient quantity and high quality of DNA samples are essential. Although precautionary steps are performed, human contaminants are found in 50%-90% of sequences[24]. Different DNA extraction kits and laboratories also have an impact on the assessment of human gut microbiota[121]. Comparing data across studies that use different bacterial DNA extraction methods is difficult[122]. Fourthly, to perform a metagenomic study successfully, the quality of the underlying functional annotations of metagenomic sequence fragments is very important. However, a significant proportion of data cannot be assigned a function due to a lack of close matches in reference databases[37]. For viral data, this situation is particularly severe, as 80% or more of sequence reads lack known matches[123].
Millions of sequences in each sample are required for functional gene analysis of a complex microbial community. It is difficult to identify and improve the accuracy of information derived from the relatively short gene fragments generated by next-generation sequencing, due to the many bioinformatics challenges proposed by the huge metagenomic shotgun sequencing. It is also difficult to assign function unambiguously based on sequence similarity alone, which may cause misannotation[124]. Moreover, when there are less abundant members of the microbiome or a community containing many closely related species, it may be difficult to assemble genomes[125]. This can lead to a situation where, even if a function can be ascertained, it may be a challenge to assign it to specific species within the microbial community. In addition, DNA is the material used in metagenomic sequencing, and the expression of each functional gene in a sample in a given environment is very difficult to determine.
Metagenomics is an extremely powerful tool that can be used to describe the genetic potential of the microorganisms present in a given environment. However, it has a very limited function in revealing their activity or gene expression. With the rapid development of metatranscriptomics[126], metaproteomics[127] and metabolomics[128], the functional activity of a microbial community can be identified.
Metatranscriptomic sequencing can be used to determine the activity of genes in a defined environment. Gosalbes et al[129] used metatranscriptomic analysis of fecal microbiomes from ten healthy humans, and found that carbohydrate metabolism, energy production and synthesis of cellular components were the main functional roles of the gut microbiota. In contrast, amino acid and lipid metabolism were reduced in the metatranscriptome.
Metatranscriptomics also has some limitations. Firstly, it is very difficult to obtain high-quality and sufficient amounts of RNA from environmental samples. Secondly, it is a challenge to separate the mRNA of interest from the more abundant types of RNA such as rRNA. Thirdly, the short half-life of mRNA leads to difficulty in the detection of rapid and short-term responses to environmental changes[130]. Fourthly, the reference databases are insufficient.
An analysis of proteins is also important to understand microbial functions. Recently, a study demonstrated that the fecal metaproteome in healthy adults was subject-specific and relatively stable over a one-year period[131]. In addition, some core functions, including carbohydrate metabolism and transport, were also determined. However, similar to metatranscriptomics, the ability to assign functional classifications is limited due to insufficient reference databases. It is a significant challenge to disentangle the complex array of proteins produced by the intestinal microbiota.
Currently, metabolomics is increasingly used to study the gut microbiome[132]. Some human intestinal disorders, such as colorectal cancer, inflammatory bowel disease and IBS, have been studied using metabolomics[133-136]. For instance, a study on patients with ulcerative colitis and IBS revealed that, compared with controls, the patients with ulcerative colitis had increased quantities of taurine and cadaverine. Importantly, a higher bile acid concentration and lower levels of branched-chain fatty acids were found in IBS patients[136]. Moreover, no significant changes were found in short-chain fatty acid and amino acid concentrations. However, some limitations restrict the development and clinical application of metabolomics. On the one hand, the metabolomics databases are incomplete and insufficient, and there are many metabolites that are not included in the databases. On the other hand, the obtained metabolites are mixed, thus it is very difficult to identify the information from the host and the microbial metabolites.
As shown in Figure 2, though there are some limitations with these approaches, they have significant potential clinical applications. The combination of the meta-omics may be sufficiently powerful to elucidate the ecologic roles of the human gut microbiome[137].
In conclusion, metagenomics can not only identify the diversity of the human gut microbiome, but can also reveal new genes and microbial pathways, and uncover functional dysbiosis. The application of metagenomics has huge potential in revealing the mechanisms and correlations between the human intestinal microbiome and diseases. However, metagenomics also has limitations and requires improvement[138].
With the rapid development and application of metagenomics, as well as metatranscriptomics, metaproteomics and metabolomics, it is possible to identify new microbial diagnostic markers that will provide early diagnosis and novel treatments. Maximizing the contribution of microorganisms and identifying more probiotics are also very promising. Based on an increased understanding of the role of the human microbiome in diseases and their interactions, as well as inter-individual differences and physiologic parameters, the exploration of personalized medicine will progress immensely. In addition, it is possible to explore new antibiotics that target antibiotic resistance microbiomes based on a profound understanding of ARGs in the gut microbiome.
Current metagenomic studies of the human gut microbiome have been performed in limited cohorts, thus, it is necessary to enhance our understanding of the human gut microbiome by investigating human populations from different countries, for longer periods, and include multiple age groups[32], and various disease stages. A study of the characteristics of the human gut microbiome in different disease stages would help us understand the relationship between the gut microbiome and disease development, and thus would help to establish the optimal strategies for preventing, improving and even reversing diseases.
As metagenomics still has some limitations, it is necessary to combine other microbiome approaches, including cultivation methods, with a study of metagenomics in the intestinal microbiome. This will ensure that the results are more accurate and convincing. Recently, several studies have successfully used this combination and obtained meaningful findings[56,139-141]. To overcome the limitations of metagenomics, it is also important to create a unified microbial DNA extraction method, improve computational algorithms, and complete the reference databases.
The application of metagenomic technology in the human gut microbiome is in its infancy. However, it has been used in other environments, including soil and the sea, for some time. Thus, the success of applying metagenomic technology in studying these environments can be followed by further study in the human gut microbiome. In addition, the human gut harbors not only bacteria, but also eukaryota and viruses. To date, few studies on eukaryota and viruses using the metagenomics approach have been carried out, thus, the future study of the human gut microbiome using the metagenomics approach is promising and more efforts are urgently needed.
P- Reviewer: Chen CY, Chen YC, Streba LAM S- Editor: Qi Y L- Editor: AmEditor E- Editor: Liu XM
| 1. | Weinstock GM. Genomic approaches to studying the human microbiota. Nature. 2012;489:250-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 360] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 2. | Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7222] [Cited by in RCA: 6409] [Article Influence: 337.3] [Reference Citation Analysis (0)] |
| 3. | Endt K, Stecher B, Chaffron S, Slack E, Tchitchek N, Benecke A, Van Maele L, Sirard JC, Mueller AJ, Heikenwalder M. The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog. 2010;6:e1001097. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 253] [Cited by in RCA: 262] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 4. | Fukuda S, Toh H, Taylor TD, Ohno H, Hattori M. Acetate-producing bifidobacteria protect the host from enteropathogenic infection via carbohydrate transporters. Gut Microbes. 2012;3:449-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 160] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 5. | Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1444] [Cited by in RCA: 1691] [Article Influence: 120.8] [Reference Citation Analysis (0)] |
| 6. | Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 952] [Cited by in RCA: 1108] [Article Influence: 85.2] [Reference Citation Analysis (0)] |
| 7. | Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1147] [Cited by in RCA: 1543] [Article Influence: 128.6] [Reference Citation Analysis (0)] |
| 8. | Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2779] [Cited by in RCA: 3081] [Article Influence: 237.0] [Reference Citation Analysis (0)] |
| 9. | Cani PD. Metabolism in 2013: The gut microbiota manages host metabolism. Nat Rev Endocrinol. 2014;10:74-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 123] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 10. | Clarke G, Stilling RM, Kennedy PJ, Stanton C, Cryan JF, Dinan TG. Minireview: Gut microbiota: the neglected endocrine organ. Mol Endocrinol. 2014;28:1221-1238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 607] [Cited by in RCA: 783] [Article Influence: 71.2] [Reference Citation Analysis (1)] |
| 11. | Sommer F, Bäckhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2011] [Cited by in RCA: 2403] [Article Influence: 200.3] [Reference Citation Analysis (0)] |
| 12. | Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258-1270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2236] [Cited by in RCA: 2559] [Article Influence: 196.8] [Reference Citation Analysis (0)] |
| 13. | Owyang C, Wu GD. The gut microbiome in health and disease. Gastroenterology. 2014;146:1433-1436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 14. | Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Doré J. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799-4807. [PubMed] |
| 15. | Sokol H, Seksik P. The intestinal microbiota in inflammatory bowel diseases: time to connect with the host. Curr Opin Gastroenterol. 2010;26:327-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 116] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 16. | Tannock GW. Molecular assessment of intestinal microflora. Am J Clin Nutr. 2001;73:410S-414S. [PubMed] |
| 17. | Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635-1638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5700] [Cited by in RCA: 5590] [Article Influence: 279.5] [Reference Citation Analysis (2)] |
| 18. | Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135-1145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2905] [Cited by in RCA: 2469] [Article Influence: 145.2] [Reference Citation Analysis (0)] |
| 19. | Fuller CW, Middendorf LR, Benner SA, Church GM, Harris T, Huang X, Jovanovich SB, Nelson JR, Schloss JA, Schwartz DC. The challenges of sequencing by synthesis. Nat Biotechnol. 2009;27:1013-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 164] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 20. | Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3035] [Cited by in RCA: 2469] [Article Influence: 117.6] [Reference Citation Analysis (0)] |
| 21. | Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1639] [Cited by in RCA: 1355] [Article Influence: 64.5] [Reference Citation Analysis (0)] |
| 22. | Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC. Comparative metagenomics of microbial communities. Science. 2005;308:554-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1167] [Cited by in RCA: 980] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 23. | Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 551] [Cited by in RCA: 616] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 24. | Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012;486:215-221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2152] [Cited by in RCA: 1814] [Article Influence: 139.5] [Reference Citation Analysis (0)] |
| 25. | Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207-214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9292] [Cited by in RCA: 8066] [Article Influence: 620.5] [Reference Citation Analysis (2)] |
| 26. | Tyakht AV, Kostryukova ES, Popenko AS, Belenikin MS, Pavlenko AV, Larin AK, Karpova IY, Selezneva OV, Semashko TA, Ospanova EA. Human gut microbiota community structures in urban and rural populations in Russia. Nat Commun. 2013;4:2469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 224] [Cited by in RCA: 185] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 27. | Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169-D172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 874] [Cited by in RCA: 775] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 28. | Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088-5090. [PubMed] |
| 29. | Blaser M, Bork P, Fraser C, Knight R, Wang J. The microbiome explored: recent insights and future challenges. Nat Rev Microbiol. 2013;11:213-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 134] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 30. | Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2636] [Cited by in RCA: 2255] [Article Influence: 173.5] [Reference Citation Analysis (0)] |
| 31. | Ren Z, Cui G, Lu H, Chen X, Jiang J, Liu H, He Y, Ding S, Hu Z, Wang W. Liver ischemic preconditioning (IPC) improves intestinal microbiota following liver transplantation in rats through 16s rDNA-based analysis of microbial structure shift. PLoS One. 2013;8:e75950. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 32. | Lepage P, Leclerc MC, Joossens M, Mondot S, Blottière HM, Raes J, Ehrlich D, Doré J. A metagenomic insight into our gut’s microbiome. Gut. 2013;62:146-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 243] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 33. | Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1529] [Cited by in RCA: 1192] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 34. | Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998;5:R245-R249. [PubMed] |
| 35. | Rondon MR, Raffel SJ, Goodman RM, Handelsman J. Toward functional genomics in bacteria: analysis of gene expression in Escherichia coli from a bacterial artificial chromosome library of Bacillus cereus. Proc Natl Acad Sci USA. 1999;96:6451-6455. [PubMed] |
| 36. | Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355-1359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3709] [Cited by in RCA: 3192] [Article Influence: 168.0] [Reference Citation Analysis (0)] |
| 37. | Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9101] [Cited by in RCA: 7838] [Article Influence: 522.5] [Reference Citation Analysis (4)] |
| 38. | Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP. A core gut microbiome in obese and lean twins. Nature. 2009;457:480-484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6397] [Cited by in RCA: 5666] [Article Influence: 354.1] [Reference Citation Analysis (1)] |
| 39. | Sunagawa S, Mende DR, Zeller G, Izquierdo-Carrasco F, Berger SA, Kultima JR, Coelho LP, Arumugam M, Tap J, Nielsen HB. Metagenomic species profiling using universal phylogenetic marker genes. Nat Methods. 2013;10:1196-1199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 348] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 40. | Thomas T, Gilbert J, Meyer F. Metagenomics - a guide from sampling to data analysis. Microb Inform Exp. 2012;2:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 628] [Cited by in RCA: 474] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 41. | Blottière HM, de Vos WM, Ehrlich SD, Doré J. Human intestinal metagenomics: state of the art and future. Curr Opin Microbiol. 2013;16:232-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 42. | Gevers D, Knight R, Petrosino JF, Huang K, McGuire AL, Birren BW, Nelson KE, White O, Methé BA, Huttenhower C. The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS Biol. 2012;10:e1001377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 284] [Cited by in RCA: 306] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 43. | Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1198] [Cited by in RCA: 1406] [Article Influence: 127.8] [Reference Citation Analysis (0)] |
| 44. | Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153-4158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 707] [Article Influence: 44.2] [Reference Citation Analysis (0)] |
| 45. | Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1329] [Cited by in RCA: 1452] [Article Influence: 121.0] [Reference Citation Analysis (0)] |
| 46. | Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426-429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 381] [Cited by in RCA: 460] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 47. | Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM. Enterotypes of the human gut microbiome. Nature. 2011;473:174-180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5822] [Cited by in RCA: 5036] [Article Influence: 359.7] [Reference Citation Analysis (2)] |
| 48. | Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O’Sullivan O. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1993] [Cited by in RCA: 2303] [Article Influence: 177.2] [Reference Citation Analysis (0)] |
| 49. | Chewapreecha C. Your gut microbiota are what you eat. Nat Rev Microbiol. 2014;12:8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 50. | David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559-563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5625] [Cited by in RCA: 6827] [Article Influence: 568.9] [Reference Citation Analysis (0)] |
| 51. | Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP. Human gut microbiome viewed across age and geography. Nature. 2012;486:222-227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4762] [Cited by in RCA: 5466] [Article Influence: 420.5] [Reference Citation Analysis (0)] |
| 52. | Biagi E, Candela M, Turroni S, Garagnani P, Franceschi C, Brigidi P. Ageing and gut microbes: perspectives for health maintenance and longevity. Pharmacol Res. 2013;69:11-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 180] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 53. | Suzuki TA, Worobey M. Geographical variation of human gut microbial composition. Biol Lett. 2014;10:20131037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 143] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 54. | Xu D, Gao J, Gillilland M, Wu X, Song I, Kao JY, Owyang C. Rifaximin alters intestinal bacteria and prevents stress-induced gut inflammation and visceral hyperalgesia in rats. Gastroenterology. 2014;146:484-496.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 205] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 55. | Vrieze A, Out C, Fuentes S, Jonker L, Reuling I, Kootte RS, van Nood E, Holleman F, Knaapen M, Romijn JA. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol. 2014;60:824-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 428] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 56. | Guo M, Huang K, Chen S, Qi X, He X, Cheng WH, Luo Y, Xia K, Xu W. Combination of metagenomics and culture-based methods to study the interaction between ochratoxin a and gut microbiota. Toxicol Sci. 2014;141:314-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 57. | Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4098] [Cited by in RCA: 4541] [Article Influence: 324.4] [Reference Citation Analysis (1)] |
| 58. | Mardanov AV, Babykin MM, Beletsky AV, Grigoriev AI, Zinchenko VV, Kadnikov VV, Kirpichnikov MP, Mazur AM, Nedoluzhko AV, Novikova ND. Metagenomic Analysis of the Dynamic Changes in the Gut Microbiome of the Participants of the MARS-500 Experiment, Simulating Long Term Space Flight. Acta Naturae. 2013;5:116-125. [PubMed] |
| 59. | Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4578-4585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1666] [Cited by in RCA: 1742] [Article Influence: 116.1] [Reference Citation Analysis (0)] |
| 60. | Gabor EM, Alkema WB, Janssen DB. Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ Microbiol. 2004;6:879-886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 142] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 61. | Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 752] [Cited by in RCA: 737] [Article Influence: 49.1] [Reference Citation Analysis (1)] |
| 62. | Turnbaugh PJ, Henrissat B, Gordon JI. Viewing the human microbiome through three-dimensional glasses: integrating structural and functional studies to better define the properties of myriad carbohydrate-active enzymes. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1261-1264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 63. | Li LL, McCorkle SR, Monchy S, Taghavi S, van der Lelie D. Bioprospecting metagenomes: glycosyl hydrolases for converting biomass. Biotechnol Biofuels. 2009;2:10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 64. | Tasse L, Bercovici J, Pizzut-Serin S, Robe P, Tap J, Klopp C, Cantarel BL, Coutinho PM, Henrissat B, Leclerc M. Functional metagenomics to mine the human gut microbiome for dietary fiber catabolic enzymes. Genome Res. 2010;20:1605-1612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 180] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 65. | Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci USA. 2008;105:13580-13585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 591] [Cited by in RCA: 774] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 66. | Gloux K, Berteau O, El Oumami H, Béguet F, Leclerc M, Doré J. A metagenomic β-glucuronidase uncovers a core adaptive function of the human intestinal microbiome. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4539-4546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 155] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 67. | Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014;5:e00889. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 562] [Cited by in RCA: 829] [Article Influence: 75.4] [Reference Citation Analysis (1)] |
| 68. | Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089-1093. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1477] [Cited by in RCA: 1280] [Article Influence: 80.0] [Reference Citation Analysis (0)] |
| 69. | Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037-1050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 914] [Cited by in RCA: 974] [Article Influence: 54.1] [Reference Citation Analysis (0)] |
| 70. | Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:629-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 637] [Cited by in RCA: 636] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 71. | Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004;12:412-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 547] [Cited by in RCA: 546] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 72. | Sommer MO, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128-1131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 662] [Cited by in RCA: 609] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 73. | D’Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1023] [Cited by in RCA: 984] [Article Influence: 51.8] [Reference Citation Analysis (0)] |
| 74. | Seville LA, Patterson AJ, Scott KP, Mullany P, Quail MA, Parkhill J, Ready D, Wilson M, Spratt D, Roberts AP. Distribution of tetracycline and erythromycin resistance genes among human oral and fecal metagenomic DNA. Microb Drug Resist. 2009;15:159-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 89] [Article Influence: 5.6] [Reference Citation Analysis (1)] |
| 75. | Forslund K, Sunagawa S, Kultima JR, Mende DR, Arumugam M, Typas A, Bork P. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 2013;23:1163-1169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 286] [Cited by in RCA: 289] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 76. | Hu Y, Yang X, Qin J, Lu N, Cheng G, Wu N, Pan Y, Li J, Zhu L, Wang X. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat Commun. 2013;4:2151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 473] [Cited by in RCA: 555] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 77. | Karami N, Martner A, Enne VI, Swerkersson S, Adlerberth I, Wold AE. Transfer of an ampicillin resistance gene between two Escherichia coli strains in the bowel microbiota of an infant treated with antibiotics. J Antimicrob Chemother. 2007;60:1142-1145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 89] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 78. | Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ, Barthel M, Westendorf AM, Krogfelt KA, Walker AW. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc Natl Acad Sci USA. 2012;109:1269-1274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 346] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 79. | Cheng G, Hu Y, Yin Y, Yang X, Xiang C, Wang B, Chen Y, Yang F, Lei F, Wu N. Functional screening of antibiotic resistance genes from human gut microbiota reveals a novel gene fusion. FEMS Microbiol Lett. 2012;336:11-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 80. | Moore AM, Patel S, Forsberg KJ, Wang B, Bentley G, Razia Y, Qin X, Tarr PI, Dantas G. Pediatric fecal microbiota harbor diverse and novel antibiotic resistance genes. PLoS One. 2013;8:e78822. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 81. | Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1633] [Cited by in RCA: 1677] [Article Influence: 88.3] [Reference Citation Analysis (0)] |
| 82. | Ray K. IBD. Understanding gut microbiota in new-onset Crohn’s disease. Nat Rev Gastroenterol Hepatol. 2014;11:268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 83. | Bye W, Ishaq N, Bolin TD, Duncombe VM, Riordan SM. Overgrowth of the indigenous gut microbiome and irritable bowel syndrome. World J Gastroenterol. 2014;20:2449-2455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 12] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 84. | Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2727] [Cited by in RCA: 3213] [Article Influence: 267.8] [Reference Citation Analysis (2)] |
| 85. | Tarantino G. Gut microbiome, obesity-related comorbidities, and low-grade chronic inflammation. J Clin Endocrinol Metab. 2014;99:2343-2346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 86. | Ohtani N, Yoshimoto S, Hara E. Obesity and cancer: a gut microbial connection. Cancer Res. 2014;74:1885-1889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 87. | Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3971] [Cited by in RCA: 4829] [Article Influence: 371.5] [Reference Citation Analysis (1)] |
| 88. | de Goffau MC, Fuentes S, van den Bogert B, Honkanen H, de Vos WM, Welling GW, Hyöty H, Harmsen HJ. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia. 2014;57:1569-1577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 244] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 89. | Tilg H, Moschen AR. Microbiota and diabetes: an evolving relationship. Gut. 2014;63:1513-1521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 519] [Cited by in RCA: 559] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 90. | Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012;129:434-440, 440.e1-e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 518] [Cited by in RCA: 584] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 91. | Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2014;44:842-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 458] [Cited by in RCA: 528] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 92. | Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:521-530, e248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 326] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 93. | Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, Goedert JJ, Hayes RB, Yang L. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105:1907-1911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 566] [Cited by in RCA: 713] [Article Influence: 59.4] [Reference Citation Analysis (1)] |
| 94. | Ou J, Carbonero F, Zoetendal EG, DeLany JP, Wang M, Newton K, Gaskins HR, O’Keefe SJ. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr. 2013;98:111-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 462] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 95. | Candela M, Turroni S, Biagi E, Carbonero F, Rampelli S, Fiorentini C, Brigidi P. Inflammation and colorectal cancer, when microbiota-host mutualism breaks. World J Gastroenterol. 2014;20:908-922. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 153] [Cited by in RCA: 152] [Article Influence: 13.8] [Reference Citation Analysis (1)] |
| 96. | Yang T, Owen JL, Lightfoot YL, Kladde MP, Mohamadzadeh M. Microbiota impact on the epigenetic regulation of colorectal cancer. Trends Mol Med. 2013;19:714-725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 97. | Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, Wang Y, Zhu B, Li L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 674] [Cited by in RCA: 797] [Article Influence: 56.9] [Reference Citation Analysis (3)] |
| 98. | Wei X, Yan X, Zou D, Yang Z, Wang X, Liu W, Wang S, Li X, Han J, Huang L. Abnormal fecal microbiota community and functions in patients with hepatitis B liver cirrhosis as revealed by a metagenomic approach. BMC Gastroenterol. 2013;13:175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 106] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 99. | Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1230] [Cited by in RCA: 1536] [Article Influence: 139.6] [Reference Citation Analysis (40)] |
| 100. | Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, Gill SR. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1280] [Article Influence: 106.7] [Reference Citation Analysis (1)] |
| 101. | Miura K, Ohnishi H. Role of gut microbiota and Toll-like receptors in nonalcoholic fatty liver disease. World J Gastroenterol. 2014;20:7381-7391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 252] [Cited by in RCA: 284] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 102. | Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451-1463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2084] [Cited by in RCA: 2343] [Article Influence: 195.3] [Reference Citation Analysis (0)] |
| 103. | Flight MH. Neurodevelopmental disorders: the gut-microbiome-brain connection. Nat Rev Neurosci. 2014;15:65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 104. | Karlsson FH, Fåk F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, Bäckhed F, Nielsen J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 711] [Cited by in RCA: 933] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 105. | Wu T, Zhang Z, Liu B, Hou D, Liang Y, Zhang J, Shi P. Gut microbiota dysbiosis and bacterial community assembly associated with cholesterol gallstones in large-scale study. BMC Genomics. 2013;14:669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 160] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 106. | Pop M, Walker AW, Paulson J, Lindsay B, Antonio M, Hossain MA, Oundo J, Tamboura B, Mai V, Astrovskaya I. Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol. 2014;15:R76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 186] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 107. | Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417-421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 807] [Cited by in RCA: 897] [Article Influence: 81.5] [Reference Citation Analysis (0)] |
| 108. | Ramezani A, Raj DS. The gut microbiome, kidney disease, and targeted interventions. J Am Soc Nephrol. 2014;25:657-670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 423] [Cited by in RCA: 529] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 109. | Brim H, Yooseph S, Zoetendal EG, Lee E, Torralbo M, Laiyemo AO, Shokrani B, Nelson K, Ashktorab H. Microbiome analysis of stool samples from African Americans with colon polyps. PLoS One. 2013;8:e81352. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 110. | Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, Zhu W, Sporrer D, Hehlgans T, Kreutz M. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant. 2014;20:640-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 415] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 111. | Lakhdari O, Cultrone A, Tap J, Gloux K, Bernard F, Ehrlich SD, Lefèvre F, Doré J, Blottière HM. Functional metagenomics: a high throughput screening method to decipher microbiota-driven NF-κB modulation in the human gut. PLoS One. 2010;5:e13092. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 112. | Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1766] [Cited by in RCA: 2145] [Article Influence: 178.8] [Reference Citation Analysis (0)] |
| 113. | Lakhdari O, Tap J, Béguet-Crespel F, Le Roux K, de Wouters T, Cultrone A, Nepelska M, Lefèvre F, Doré J, Blottière HM. Identification of NF-κB modulation capabilities within human intestinal commensal bacteria. J Biomed Biotechnol. 2011;2011:282356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 114. | Gloux K, Leclerc M, Iliozer H, L’Haridon R, Manichanh C, Corthier G, Nalin R, Blottière HM, Doré J. Development of high-throughput phenotyping of metagenomic clones from the human gut microbiome for modulation of eukaryotic cell growth. Appl Environ Microbiol. 2007;73:3734-3737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 30] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 115. | Dobrijevic D, Di Liberto G, Tanaka K, de Wouters T, Dervyn R, Boudebbouze S, Binesse J, Blottière HM, Jamet A, Maguin E. High-throughput system for the presentation of secreted and surface-exposed proteins from Gram-positive bacteria in functional metagenomics studies. PLoS One. 2013;8:e65956. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 116. | Brown Kav A, Benhar I, Mizrahi I. A method for purifying high quality and high yield plasmid DNA for metagenomic and deep sequencing approaches. J Microbiol Methods. 2013;95:272-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 117. | Jones BV, Marchesi JR. Transposon-aided capture (TRACA) of plasmids resident in the human gut mobile metagenome. Nat Methods. 2007;4:55-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 79] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 118. | Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169-181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 647] [Cited by in RCA: 610] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 119. | Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 605] [Cited by in RCA: 633] [Article Influence: 45.2] [Reference Citation Analysis (1)] |
| 120. | Raes J, Korbel JO, Lercher MJ, von Mering C, Bork P. Prediction of effective genome size in metagenomic samples. Genome Biol. 2007;8:R10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 208] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 121. | Kennedy NA, Walker AW, Berry SH, Duncan SH, Farquarson FM, Louis P, Thomson JM, Satsangi J, Flint HJ, Parkhill J. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS One. 2014;9:e88982. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 204] [Cited by in RCA: 217] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 122. | Wesolowska-Andersen A, Bahl MI, Carvalho V, Kristiansen K, Sicheritz-Pontén T, Gupta R, Licht TR. Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome. 2014;2:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 199] [Cited by in RCA: 199] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 123. | Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334-338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 988] [Cited by in RCA: 850] [Article Influence: 56.7] [Reference Citation Analysis (0)] |
| 124. | Schnoes AM, Brown SD, Dodevski I, Babbitt PC. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol. 2009;5:e1000605. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 536] [Cited by in RCA: 487] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 125. | Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31:533-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 937] [Cited by in RCA: 865] [Article Influence: 72.1] [Reference Citation Analysis (0)] |
| 126. | Poretsky RS, Hewson I, Sun S, Allen AE, Zehr JP, Moran MA. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol. 2009;11:1358-1375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 191] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 127. | Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, Lefsrud MG, Apajalahti J, Tysk C, Hettich RL. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3:179-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 394] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 128. | Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 682] [Cited by in RCA: 655] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 129. | Gosalbes MJ, Durbán A, Pignatelli M, Abellan JJ, Jiménez-Hernández N, Pérez-Cobas AE, Latorre A, Moya A. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One. 2011;6:e17447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 306] [Cited by in RCA: 252] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 130. | Simon C, Daniel R. Metagenomic analyses: past and future trends. Appl Environ Microbiol. 2011;77:1153-1161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 465] [Cited by in RCA: 352] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 131. | Kolmeder CA, de Been M, Nikkilä J, Ritamo I, Mättö J, Valmu L, Salojärvi J, Palva A, Salonen A, de Vos WM. Comparative metaproteomics and diversity analysis of human intestinal microbiota testifies for its temporal stability and expression of core functions. PLoS One. 2012;7:e29913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 143] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 132. | Heinken A, Khan MT, Paglia G, Rodionov DA, Harmsen HJ, Thiele I. Functional metabolic map of Faecalibacterium prausnitzii, a beneficial human gut microbe. J Bacteriol. 2014;196:3289-3302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 170] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 133. | Phua LC, Chue XP, Koh PK, Cheah PY, Ho HK, Chan EC. Non-invasive fecal metabonomic detection of colorectal cancer. Cancer Biol Ther. 2014;15:389-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 134. | Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, Wilson ID, Wang Y. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 455] [Cited by in RCA: 475] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 135. | Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J, Tysk C, Schmitt-Kopplin P. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS One. 2009;4:e6386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 372] [Cited by in RCA: 405] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 136. | Le Gall G, Noor SO, Ridgway K, Scovell L, Jamieson C, Johnson IT, Colquhoun IJ, Kemsley EK, Narbad A. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res. 2011;10:4208-4218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 279] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 137. | Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, Earl AM, Giannoukos G, Boylan MR, Ciulla D, Gevers D. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci USA. 2014;111:E2329-E2338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 429] [Cited by in RCA: 477] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 138. | Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, Sunagawa S, Plichta DR, Gautier L, Pedersen AG, Le Chatelier E. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol. 2014;32:822-828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 753] [Article Influence: 68.5] [Reference Citation Analysis (0)] |
| 139. | Mitra S, Förster-Fromme K, Damms-Machado A, Scheurenbrand T, Biskup S, Huson DH, Bischoff SC. Analysis of the intestinal microbiota using SOLiD 16S rRNA gene sequencing and SOLiD shotgun sequencing. BMC Genomics. 2013;14 Suppl 5:S16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 140. | Culligan EP, Marchesi JR, Hill C, Sleator RD. Combined metagenomic and phenomic approaches identify a novel salt tolerance gene from the human gut microbiome. Front Microbiol. 2014;5:189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 141. | Lee S, Cantarel B, Henrissat B, Gevers D, Birren BW, Huttenhower C, Ko G. Gene-targeted metagenomic analysis of glucan-branching enzyme gene profiles among human and animal fecal microbiota. ISME J. 2014;8:493-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |