Published online Jun 14, 2015. doi: 10.3748/wjg.v21.i22.6884
Peer-review started: December 17, 2014
First decision: January 8, 2015
Revised: February 8, 2015
Accepted: March 30, 2015
Article in press: March 31, 2015
Published online: June 14, 2015
Processing time: 184 Days and 1.8 Hours
AIM: To study the potential prognostic role of microRNA-382 (miR-382) in esophageal squamous cell carcinoma (ESCC).
METHODS: Forty six patients were divided into 2 groups according to postoperative survival time: the poor outcome group (28 patients), who showed early metastasis but no recurrence, and died within 1 year after surgery, 12 patients of the group received postoperative chemotherapy treatment that was given after early metastasis happening; the good outcome group (18 patients), who had no clinical metastasis and recurrence, and survived 5 years or more after surgery, all patients did not receive any postoperative treatment. Total RNA was extracted from the patients’ formalin-fixed and paraffin-embedded esophageal cancer tissues. miR-382 level was evaluated using high-throughput real-time quantitative polymerase chain reaction analysis. The correlation between miR-382 level and clinicopathologic features was analyzed through COX regression model, and Kaplan-Meier analysis was used to analyze the relationship between miR-382 level and patient survival time.
RESULTS: miR-382 was differentially expressed in the two groups. Overall the average miR-382 level in the ESCC patients with good outcome was 9.8 ± 3.8, while miR-382 level in the ESCC patients with poor outcome was 3.0 ± 0.8. The differences of miR-382 levels between two groups were significant (P < 0.05). Kaplan-Meier analysis results showed that miR-382 expression level generally had a significant reverse-correlation with ESCC patient survival time (P < 0.001), in which the patients with higher expressions of miR-382 had a longer survival time either among individuals with the same tumor stage or among the overall patients.
CONCLUSION: miR-382 levels are reverse-correlated with ESCC poor outcomes, suggesting that miR-382 could be a potential predictive biomarker for both prognosis and treatment of ESCC.
Core tip: Esophageal squamous cell carcinoma (ESCC) patients often have significantly different outcomes due to early metastasis happening or not, although the patients are at the same pathological stage and receive the similar surgical therapy. Exploring novel biomarkers related with ESCC metastasis is required for monitoring the progression of the disease, and predicting the outcome of the patient after clinical intervention. Current research addressed a potential prognostic role of microRNA-382 in ESCC.
- Citation: Qi B, Lu JG, Yao WJ, Chang TM, Qin XG, Ji YH, Wang TY, Liu SG, Li HC, Liu YZ, Zhao BS. Downregulation of microRNA-382 is associated with poor outcome of esophageal squamous cell carcinoma. World J Gastroenterol 2015; 21(22): 6884-6891
- URL: https://www.wjgnet.com/1007-9327/full/v21/i22/6884.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i22.6884
Esophageal cancer (EC) is one of the most common gastrointestinal cancers. Up to 300 thousand people worldwide die from this disease each year[1]. More than 50% of the global incidence of EC is in China[2]. Histologically, EC is divided into two main types: squamous cell carcinoma and adenocarcinoma. Esophageal squamous cell carcinoma (ESCC) is the major histological type of EC in China. The global 5-year survival rate of EC is only about 15% with most patients dying withing one year of diagnosis[3]. This is mainly based on EC’s highly invasive characteristic, which often leads to early metastasis and reduces treatment efficacy[4,5]. Although Tumor Node Metastasis (TNM) staging system is still a major criteria for EC prognosis, our clinical observations show that ESCC patients receiving similar therapy (e.g., surgical therapy by the same surgeon) at the same pathological stage had significantly different outcomes. Moreover, metastasis in the early postoperative stage also occurred in early-stage ESCC patients. These observations suggested the hypothesis that metastasis-related molecules may be heterogeneously present in ESCC individuals. Therefore, exploring novel biomarkers related to ESCC metastasis is required for monitoring the progression of the disease, and predicting the prognosis of the patient after clinical intervention.
MicroRNA (miRNA) is a small non-coding RNA with 22-25 nucleotides in length, and controls gene expression via the regulation of translation efficiency and mRNA stability by binding to the complementary site in 3’-untranslated region (UTR) of the mRNA[6,7]. miRNAs are abundantly expressed and play an essential role in the regulation of a large number of biological processes, including cancer[8,9]. The expression patterns and biological functions of miRNAs in ESCC have been investigated in recent years[10-12].
Human miR-382 (has-mir-382, MIMAT0000737, 5’-GAAGUUGUUCGUGGUGGAUUCG-3’) resides in a miRNA cluster in the imprinted DLK1-DIO3 region on the 14q32 locus which hosts one of the largest miRNA clusters in the genome. Many of these miRNAs are differentially expressed in several pathologic processes and various cancers. Recent studies reported that miR-382 was decreased or increased in several types of human malignancy[13,14], suggesting that the role of miR-382 contributing to tumor development and metastasis is tissue specific.
Using high-throughput real-time quantitative polymerase chain reaction, we initially evaluated the expression profiles of 754 miRNAs in paraffin-embedded tumor specimens from two ESCC patients with TNM IIa stage. These patients had different outcomes although they both received same postoperative treatment (chemotherapy twice)[15]. Our results showed that the levels of multiple miRNAs were significantly different between the two patients. One of the findings was that the ESCC patient who had neck lymph node metastasis which occurred four months after surgery and died 1 year after surgery had significantly lower miR-382 level compared to another patient who did not have metastasis and survived five years after surgery.
The present study was to further validate a potential role for miR-382 as an ESCC prognostic biomarker. For this purpose, miR-382 levels were examined from 46 ESCC patients with different outcomes; afterwards the relationship between miR-382 level and clinicopathological characteristics of the patients was assessed. We found that expression of miR-382 was weaker in the specimens from the patients with poor outcome compared to those with good outcome (P < 0.05) and miR-382 level was inversely correlated with ESCC patient survival time (P < 0.001).
Our aim was to examine miR-382 levels in cancer tissues from ESCC patients who were at the same TNM stage but had different outcomes. Forty-six patients with ESCC diagnosed by histopathological examination between 2006 and 2009 were enrolled in this study. All patients’ clinicopathological information had been recorded and the specimens had been collected before this work started. Additionally, all patients had not received any radiotherapy or chemotherapy prior to the surgical procedure performed in the Department of Thoracic Surgery, the First Affiliated Hospital of Xinxiang Medical University. TNM classifications after surgery were made according to International Union Against Cancer (UICC) staging criteria for esophageal cancer, sixth edition (2002). The 46 patients were divided into 2 groups according to postoperative survival time: the poor outcome group, who showed early metastasis but no recurrence, and died within 1 year after surgery; the good outcome group, who had no clinical metastasis and recurrence, and survived 5 years or more after surgery. This study did not include the patients surviving between 1-5 years after surgery. In addition, postoperative survival time of all stage IV patients undergoing surgical procedure in our department was less than 5 years when we started this work. Thus, patients with stage IV disease were excluded from this study. The postoperative follow-up was a standardized process that included routine computed tomography scan and upper gastrointestinal endoscopy. All patients (n = 18) with good outcome did not receive any postoperative treatment, and 12 out of 28 patients with poor outcome received postoperative chemotherapy treatment that was given after early metastasis happening.
The present study was conducted in accordance with the declaration of Helsinki, and approved by the Institutional Review Board for Human Research of the First Affiliated Hospital of Xinxiang Medical University. Written informed consent form was obtained from all patients.
Formalin-fixed and paraffin-embedded esophageal cancer tissues from the patients were used for RNA extraction. Using a microtome, 10-μm slices of tissue from each patient were captured and placed into 1.5 mL microcentrifuge tubes. Paracancerous normal esophageal mucous membranes (8 cm distant to the verge of the tumor tissue) from 4 ESCC patients were taken as controls. Total RNA (including miRNAs) was extracted using TRI Reagent (Applied Biosystems, Foster City, United States) according to manufacturer’s instructions. The RNA yield was determined using a UV spectrophotometer, and then stored at -80 °C for further processing.
Total RNA samples were reverse-transcribed using Taqman MicroRNA reverse transcription kit in combination with Megaplex reverse transcription (RT) primer Human pool set v3.0 (Applied Biosystems, Foster City, United States). Briefly, 3 μL of total RNA was supplemented with Megaplex RT primer mix (× 10), RT buffer (× 10), Multiscribe reverse transcriptase (50 U/μL), dNTPs with dTTP (100 mmol/L), MgCl2 (25 mmol/L), and RNase inhibitor (20 U/μL) in a total reaction volume of 8 μL. RT reaction was performed for 40 cycles of 16 °C for 2 min, 42 °C for 1 min and 50 °C for 1 s, followed by a final reverse transcriptase inactivation at 85 °C for 5 min. cDNA samples were kept at -80 °C until PCR analysis.
2.5 μL of cDNA samples was pre-amplified using Applied Biosystems’Taqman preamp master mix (× 2) and Megaplex preamp primers (× 5) in a 25 μL PCR. Megaplex™ PreAmp Primers (Applied Biosystems, Foster City, United States) that contained forward primers specific to miR-382 were used. The pre-amplification cycling conditions were as follows: 95 °C for 10 min, 55 °C for 2 min and 75 °C for 2 min, followed by 12 cycles of 95 °C for 15 s and 60 °C for 4 min.
qPCR was performed through 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, United States). Pre-amplified cDNA samples were diluted with low-EDTA (0.1 mmol/L) TE buffer (1:50) and qPCR reaction included TaqMan® 2X Universal PCR Master Mix (No AmpErase® UNG) 10 μL, TaqMan® MicroRNA Assays, 20X TaqMan® Assay 1 μL, PreAmp Product 1 μL and nuclease free water 8 μL. qPCR cycling conditions were as follows: 95 °C for 10 min, followed by 40 cycles of 97 °C for 30 s and 59.7 °C for 1 min. The small nRNA U6 (Assay ID: 001093) was used as an endogenous control in RT-qPCR. We run PCR in triplicate for each sample and all samples’ Ct values are less 30, which is in a reasonable range. The relative quantitative method was used. Gene expression was quantitated based on the following formula: F = 2-∆∆Ct where ∆∆Ct = (mean CT of has-mir-382 in the test sample - mean Ct of the housekeeping gene in the test sample) - (mean Ct of has-mir-382 in the control sample - mean Ct of the housekeeping gene in the control sample). A high F-value indicates a relatively high expression of miR-382.
Two-tailed student t test was used to determine the expression levels of miR-382, and results were expressed as mean ± SE. P < 0.05 was considered statistically significant. The χ2 test was used to determine the relationship between miR-382 expression level and clinicopathological features, and Kaplan-Meier was used to analyze the relationship between miRNA expression and survival time. COX regression model was used to analyze the influence of the related factor on the survival time of patients with ESCC. SPSS 17.0 software (SPSS Inc., Chicago, United States) was used for data analysis.
The study included 46 patients with ESCC. The patients were followed up for 3 to 84 mo, and were subsequently divided into two groups - good outcome and poor outcome. The group with good outcome included 18 (39.1%) patients who showed neither clinical metastasis nor recurrence, and survived 5 years or more after surgery; the group with poor outcome included 28 (60.9%) patients who showed early metastasis but no recurrence, and died within 1 year after surgery.
Demographic variables of the patients are listed in Table 1. In this study, there were 23 (50%) males and 23 (50%) females with ages ranging from 45 to 71 years (median, 59 years). The tumor size in 19 (41.3%) cases was more than 5 cm and in 27 (58.7%) cases was less than 5 cm. Regarding the histological differentiation, 11 (23.9%) patients had well differentiated ESCC, 19 (41.3%) patients had moderately differentiated ESCC, and 16 (34.8%) had poorly differentiated ESCC. Lymph node metastasis occurred in 19 (41.3%) patients, and did not occur in 27 (58.7%) cases. 7 (15.2%) cases were at stage 0 and I (1 case at stage 0 and 6 cases at stage I), 21 (45.7%) cases were at stage II (IIa and IIb), and 18 (39.1%) cases were at stage III. This study did not include the patients at stage IV and the patients living between 1-5 years.
| Characteristic | n (%) |
| All patients | 46 (100) |
| Sex | |
| Male | 23 (50.0) |
| Female | 23 (50.0) |
| Age | |
| < 60 | 25 (54.3) |
| ≥ 60 | 21 (45.7) |
| Size of tumor | |
| ≤ 5 cm | 27 (58.7) |
| > 5 cm | 19 (41.3) |
| Site of tumor | |
| Upper thoracic | 4 (8.7) |
| Middle thoracic | 35 (76.1) |
| Lower thoracic | 7 (15.2) |
| Differentiation | |
| Good | 11 (23.9) |
| Moderate | 19 (41.3) |
| Poor | 16 (34.8) |
| Depth of invasion | |
| Tis, T1 | 8 (17.4) |
| T2 | 6 (13.0) |
| T3 | 32 (69.6) |
| Stage (TNM) | |
| 0-I | 7 (15.2) |
| IIa-IIb | 21 (45.7) |
| III | 18 (39.1) |
| Lymph node metastasis | |
| Negative | 27 (58.7) |
| Positive | 19 (41.3) |
| Metastasis | |
| Early metastasis | 28 (60.9) |
| Non metastasis | 18 (39.1) |
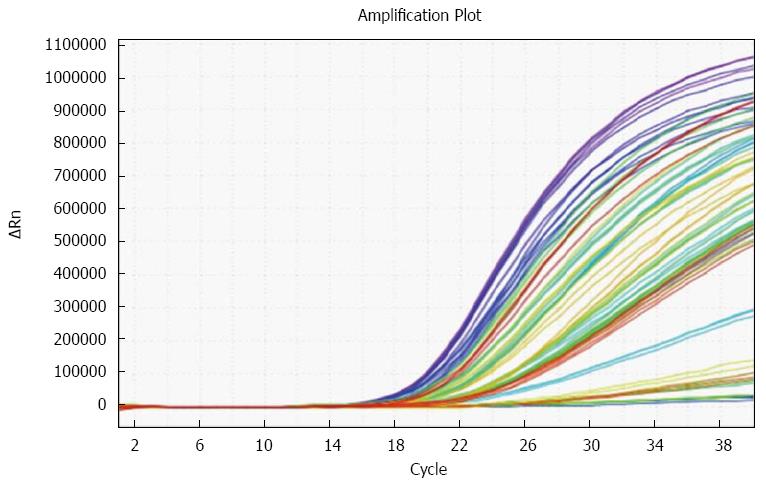
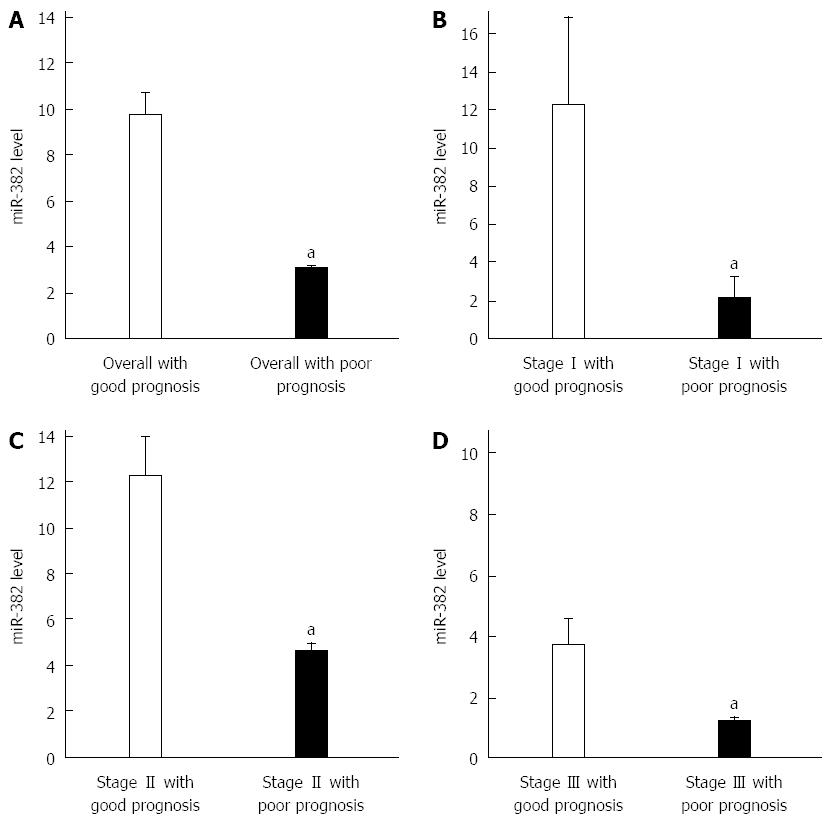
RT-qPCR was performed to examine the expression level of miR-382. qPCR amplification plot with ∆Ct values of miR-382 is shown as Figure 1. All Ct values from each sample were less than 30 (data not shown). miR-382 levels shown in Figure 2 were differentially expressed in ESCC patients. Overall the miR-382 level (Figure 2A) in the patients with good outcome was 9.8 ± 3.8 (mean ± SE), while miR-382 level in the patients with poor outcome was 3.0 ± 0.8 (mean ± SE). Therefore, miR-382 average level from all ESCC patients with poor outcome was lower than that from all ESCC patients with good outcome. The differences of miR-382 levels between two groups were significant (P < 0.05). Furthermore, in each TNM stage shown in Figure 2B-D, miR-382 levels were decreased in the patients with poor outcome when compared with the patients with good outcome.
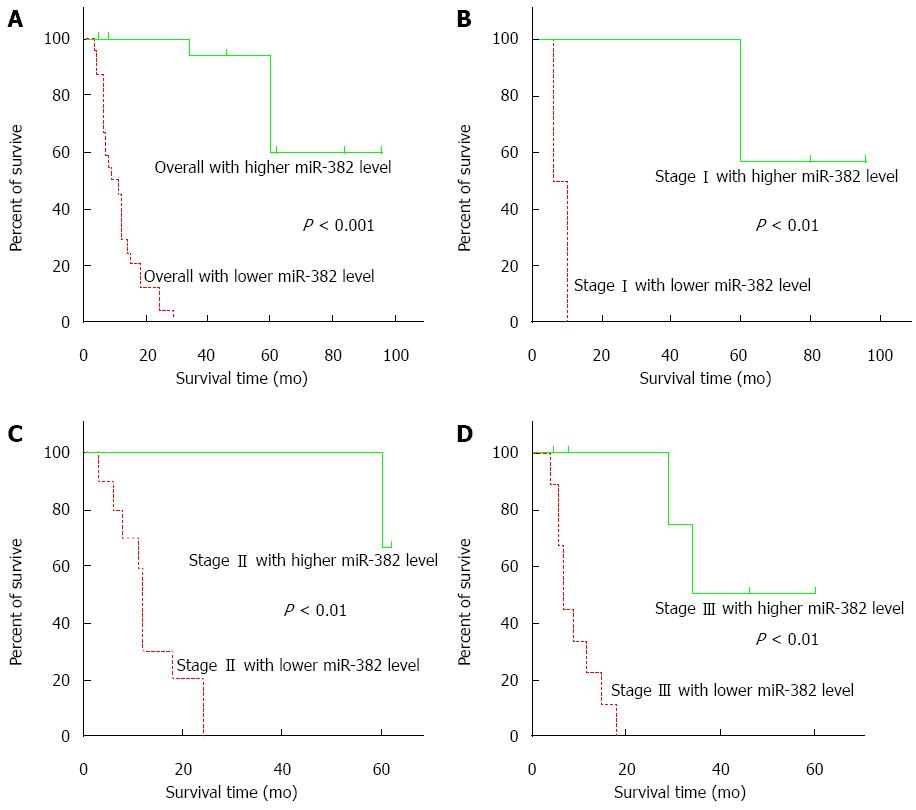
Kaplan-Meier analysis results showed that miR-382 expression level generally had a significant reverse-correlation with ESCC patient survival time (Figure 3A, P < 0.001). Similarly, at individual stage as showed in Figure 3B-D, expression levels of miR-382 were reversely correlated with patient postoperative survival time. Cox single factor related risk analysis results showed that patient TNM stage (P = 0.023), tumor size (P = 0.006), postoperative time (P = 0.010), miR-382 level (P = 0.007), and patient survival time had a significant correlation.
In the current study, we investigated the prognostic impact of miR-382 level in 46 patients with ESCC. We discovered that miR-382 was differentially expressed in cancer specimens from ESCC patients with different outcomes, although the patients were at the same pathological stage and received similar surgical treatment. We showed that miR-382 expression was significantly decreased in specimens from ESCC patients with poor outcomes compared to the patients with good outcomes. Moreover, our clinical results revealed that miR-382 level was inversely correlated with ESCC patient survival time, and significantly linked to the patients’ outcomes. These data suggest a potential role for miR-382 as a biomarker for identifying patients who will experience an unfavorable clinical outcome.
ESCC is a common malignancy[2]. Accurate judgments of the clinical stage and prognosis of ESCC are important bases for clinicians to take a rational approach to the treatment of this disease. Although TNM staging is traditionally considered the single most important factor to guide treatment decisions and prognosis for ESCC, clinical observations showed that same TNM stage ESCC patients who received the same surgical treatment by the same surgeon had different outcomes, indicating that ESCC metastasis susceptibility is somehow individually specific. Other traditional examinations based on medical equipment such as CT, X-ray barium meal fluoroscopy, gastroscopy, and B-ultrasonography, can be utilized to discover cancer metastases, whereas all these medical examinations are unable to predict distinct metastasis susceptibility. New research methods in molecular techniques have facilitated the biomarker discoveries of cancer metastasis susceptibility[16]. The current clinical biomarkers for ESCC prognosis are not ideal, as there remains a lack of reliable biomarkers that can specifically distinguish between ESCC patients who are susceptible to metastasis and those who are not.
Numerous studies have established miRNAs as broad and powerful regulators of protein expression in physiology and diseases[17]. Correlation analyses between miRNAs and cancers have shown miRNA as potential prognostic biomarkers in variant cancers[18-20]. miRNAs that are upregulated in cancers are proposed to be oncogenes, whereas those that are downregulated are considered tumor suppressors[21]. Some miRNAs, such as has-miR-335, has-miR-181d, has-miR-25, has-miR-7, and has-miR-495, have been reported to directly participate in the initiation and development of ESCC[22-24].
The relationship between miR-382 levels and prognoses has been examined in several types of human malignancy. Decreased miR-382 levels were associated with poor survival in osteosarcoma patients[14], indicating that the role of miR-382 in osteosarcoma as a tumor suppressor. Conversely increased miR-382 levels were found in acute myeloid leukemia tumor tissue[15], indicating that miR-382 plays the role of an oncogene in this disease. Consequently the role of miR-382 in tumor development and metastasis is a heterogeneous one for which the physical, cellular and molecular determinants adapt and react throughout the progression of the disease in a cell-driven and tissue-driven manner.
Our study revealed that the average level of miR-382 was significantly lower in specimens from ESCC patients who showed a poor outcome than those in the patients who showed a good outcome, and miR-382 level was reverse-correlated with patient’s survival time and linked to a poor outcome. Metastasis had occurred in all ESCC patients with poor outcome but not in the patients with good outcome in our study. Thus, our results indicate that miR-382 is involved in the ESCC metastasis process and is a potential biomarker for ESCC patients who are individual susceptible to metastasis. This could at least partially explain why the ESCC patients at similar clinicopathological stages and receiving similar surgical treatment had completely different outcomes.
The mechanism by which miR-382 affects ESCC behavior is not yet clear. One study reported that miR-382 as a tumor suppresser in osteosarcoma negatively regulated the expression of KLF12 and HIPK3 by directly targeting their 3’ UTR sequences to inhibit tumor cell growth in vivo and in vitro[25]. The downstream signals of miRNA contributing to tumor development and metastasis are tissue heterogeneous. Thus our future work will be aimed at determining the specific downstream molecules by which miR-382 affects ESCC behavior[26].
The specimens we used in this study were formalin-fixed and paraffin-embedded esophageal cancer tissues. The expression level of miR-382 in the cancer tissue is dependent on the individual ESCC patient situation rather than the specimen type used for examination. Hence, our results should be applicable to other specimen types such as fresh surgical specimen and forceps biopsies obtained during upper gastrointestinal endoscopy.
In this study, the level of miR-382 we mentioned just means the relative quantification when comparing the results from two groups. The total number of patients assessed in our study was relatively small, especially for the number of individual pathological stage patients. Accordingly, it is difficult to determine the cutoff level of miR-382 as a biomarker for clinical utility from our current results. A large size cohort study must therefore be an objective of future projects to determine the miR-382 cutoff level, which can be used not only for predicting ESCC outcomes but also for being a supplemental criterion to TNM staging or postoperative treatment decision.
In conclusion, it is the first study to show that miR-382 was downregulated in ESCC patients with early metastasis, and that miR-382 levels were significantly reverse-correlated with ESCC patient outcomes. Therefore, miR-382 could be a potential predictive biomarker for both outcome prognosis and treatment of ESCC. Further studies are needed to define the detailed mechanisms and determine the cutoff level of miR-382.
Different outcomes often happen to esophageal squamous cell carcinoma (ESCC) patients with the same pathological stage and receiving similar therapy due to early metastasis happening or not. It is important to explore reliable biomarkers that can specifically distinguish between ESCC patients who are susceptible to metastasis and those who are not. microRNAs (miRNAs) as broad and powerful regulators of protein expression are potential prognostic biomarkers of cancers. microRNA-382 (miR-382) has been shown to relate with tumor development and metastasis in variant cancers.
It was reported that miR-382 was reduced and associated with poor survival in osteosarcoma patients. Conversely increased miR-382 levels were found in acute myeloid leukemia tumor tissue. Consequently the role of miR-382 in tumor development and metastasis is a heterogeneous one in a cell-driven and tissue-driven manner.
This study revealed that miR-382 was downregulated in ESCC patients with early metastasis comparing with the patients without metastasis. This is the first study to show that miR-382 levels were significantly reverse-correlated with ESCC patient outcomes. Therefore, the results indicate that miR-382 is involved in ESCC metastasis process and is a potential biomarker for ESCC patients who are individual susceptible to metastasis.
By understanding the differential expression of miR-382 in the specimen of ESCC patient with or without early metastasis, and its relationship with clinicopathological characteristics and patient outcomes, this study may provide a future strategy in the development of a novel biomarker for both metastasis predictor and treatment of ESCC.
Metastasis is a complex process that involves the spread of a tumor or cancer from its original site to other places in the body. miRNA is a small non-coding RNA with about 22 nucleotides in length, and controls gene expression via the regulation of translation efficiency and mRNA stability. miRNAs have been shown as potential prognostic biomarkers in variant cancers. miR-382 is one of these, the role of miR-382 to tumor development and metastasis is tissue specific.
This is an interested topic aiming to advance management for this difficult disease. The authors first provide the evidence of the divergent expression of microRNA-382 in tumor tissues from ESCC patients with different outcomes, which indicate that it is associated with prognosis and may develop a novel biomarker for both diagnosis and treatment of ESCC.
P- Reviewer: Carrara S, Le Page PA, Liu DL S- Editor: Qi Y L- Editor: O’Neill M E- Editor: Ma S
| 1. | Sun X, Chen W, Chen Z, Wen D, Zhao D, He Y. Population-based case-control study on risk factors for esophageal cancer in five high-risk areas in China. Asian Pac J Cancer Prev. 2010;11:1631-1636. [PubMed] |
| 2. | Mao WM, Zheng WH, Ling ZQ. Epidemiologic risk factors for esophageal cancer development. Asian Pac J Cancer Prev. 2011;12:2461-2466. [PubMed] |
| 3. | Siegel R, Naishadham D, Jemal A. Cancer statistics for Hispanics/Latinos, 2012. CA Cancer J Clin. 2012;62:283-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 389] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 4. | Zhu ZJ, Hu Y, Zhao YF, Chen XZ, Chen LQ, Chen YT. Early recurrence and death after esophagectomy in patients with esophageal squamous cell carcinoma. Ann Thorac Surg. 2011;91:1502-1508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 5. | Griffin SM, Burt AD, Jennings NA. Lymph node metastasis in early esophageal adenocarcinoma. Ann Surg. 2011;254:731-76; discussion 731-76;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 6. | Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519-1524. [PubMed] |
| 7. | Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13:e249-e258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 623] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 8. | Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175-205. [PubMed] |
| 9. | Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524-15529. [PubMed] |
| 10. | Zhou ZQ, Cao WH, Xie JJ, Lin J, Shen ZY, Zhang QY, Shen JH, Xu LY, Li EM. Expression and prognostic significance of THBS1, Cyr61 and CTGF in esophageal squamous cell carcinoma. BMC Cancer. 2009;9:291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 11. | He B, Pan Y, Cho WC, Xu Y, Gu L, Nie Z, Chen L, Song G, Gao T, Li R. The association between four genetic variants in microRNAs (rs11614913, rs2910164, rs3746444, rs2292832) and cancer risk: evidence from published studies. PLoS One. 2012;7:e49032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 12. | Yang M, Liu R, Sheng J, Liao J, Wang Y, Pan E, Guo W, Pu Y, Yin L. Differential expression profiles of microRNAs as potential biomarkers for the early diagnosis of esophageal squamous cell carcinoma. Oncol Rep. 2013;29:169-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 13. | Sarver AL, Thayanithy V, Scott MC, Cleton-Jansen AM, Hogendoorn PC, Modiano JF, Subramanian S. MicroRNAs at the human 14q32 locus have prognostic significance in osteosarcoma. Orphanet J Rare Dis. 2013;8:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 85] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 14. | Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, Chen P, Wang Y, Yan M, Qian Z. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA. 2008;105:15535-15540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 359] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 15. | Zhao BS, Liu SG, Wang TY, Ji YH, Qi B, Tao YP, Li HC, Wu XN. Screening of microRNA in patients with esophageal cancer at same tumor node metastasis stage with different prognoses. Asian Pac J Cancer Prev. 2013;14:139-143. [PubMed] |
| 16. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] |
| 17. | Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11:537-561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1008] [Cited by in RCA: 1332] [Article Influence: 95.1] [Reference Citation Analysis (0)] |
| 18. | Gomes AQ, Nolasco S, Soares H. Non-coding RNAs: multi-tasking molecules in the cell. Int J Mol Sci. 2013;14:16010-16039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 196] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 19. | Liang M. MicroRNA: a new entrance to the broad paradigm of systems molecular medicine. Physiol Genomics. 2009;38:113-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 20. | Lin RJ, Xiao DW, Liao LD, Chen T, Xie ZF, Huang WZ, Wang WS, Jiang TF, Wu BL, Li EM. MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J Surg Oncol. 2012;105:175-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 93] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 21. | Guo Y, Chen Z, Zhang L, Zhou F, Shi S, Feng X, Li B, Meng X, Ma X, Luo M. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 268] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 22. | Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1-12. [PubMed] |
| 23. | Mathé EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192-6200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 452] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 24. | Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, Swanson SJ, Godfrey TE, Litle VR. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255-60; discussion 260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 298] [Cited by in RCA: 304] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 25. | Kong KL, Kwong DL, Chan TH, Law SY, Chen L, Li Y, Qin YR, Guan XY. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61:33-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 194] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 26. | Xu M, Jin H, Xu CX, Sun B, Mao Z, Bi WZ, Wang Y. miR-382 inhibits tumor growth and enhance chemosensitivity in osteosarcoma. Oncotarget. 2014;5:9472-9483. [PubMed] |