Published online Jun 7, 2015. doi: 10.3748/wjg.v21.i21.6621
Peer-review started: November 29, 2014
First decision: December 26, 2014
Revised: January 24, 2015
Accepted: February 13, 2015
Article in press: February 13, 2015
Published online: June 7, 2015
Processing time: 193 Days and 22.7 Hours
AIM: To analyze RASSF6 expression in pancreatic ductal adenocarcinoma (PDAC) and to determine whether RASSF6 has an independent prognostic value in PDAC.
METHODS: We studied RASSF6 expression in 96 histologically confirmed PDAC samples and 20 chronic pancreatitis specimens using immunohistochemistry and real-time quantitative reverse transcription-PCR. PDAC issues were then classified as RASSF6 strongly positive, weakly positive or negative. RASSF6 mRNA and protein expression in PDAC samples with strong positive staining was further evaluated using real-time PCR and Western blot analysis. Lastly, correlations between RASSF6 staining and patients’ clinicopathological variables and outcomes were assessed.
RESULTS: RASSF6 was negatively expressed in 51 (53.1%) PDAC samples, weakly positively expressed in 29 (30.2%) and strongly positively expressed in 16 (16.7%), while its expression was much higher in para-tumor tissues and chronic pancreatitis tissues. Positive relationships between RASSF6 expression and T-stage (P = 0.047) and perineural invasion (P = 0.026) were observed. The median survival time of strongly and weakly positive and negative RASSF6 staining groups was 33 mo, 15 mo and 11 mo, respectively. Cox multivariate analysis indicated that RASSF6 was an independent prognostic indicator of overall survival in patients with PDAC. A survival curve analysis revealed that increased RASSF6 expression was correlated with better overall survival (P = 0.009).
CONCLUSION: RASSF6 expression is an independent biomarker of an unfavorable prognosis in patients with PDAC.
Core tip: This is the first study that provides evidence for the prognostic value and potential tumor suppressor activity of RASSF6 in pancreatic ductal adenocarcinoma. The prominent findings in this study are that RASSF6 is an independent prognostic marker for predicting the survival of pancreatic ductal adenocarcinoma patients after curative operations and that the expression of RASSF6 might decrease with the development of cancer.
- Citation: Ye HL, Li DD, Lin Q, Zhou Y, Zhou QB, Zeng B, Fu ZQ, Gao WC, Liu YM, Chen RW, Li ZH, Chen RF. Low RASSF6 expression in pancreatic ductal adenocarcinoma is associated with poor survival. World J Gastroenterol 2015; 21(21): 6621-6630
- URL: https://www.wjgnet.com/1007-9327/full/v21/i21/6621.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i21.6621
Pancreatic cancer is a highly malignant neoplasm and the fourth-leading cause of cancer death. Pancreatic ductal adenocarcinoma (PDAC) accounts for the majority (> 90%) of pancreatic malignancies[1]. The 5-year survival of PDAC is only approximately 5%, and this figure has remained nearly unchanged over the past two decades, but the incidence of PDAC has been rising worldwide[2,3]. Unlike in breast and other carcinomas, no molecular markers have been established to date for estimating prognosis or providing information for treatment decision-making in patients with PDAC. The identification of biomarkers that accurately predict disease recurrence or response to chemotherapy would be of substantial aid in individual risk assessment and treatment selection and may even lead to novel therapies by becoming targets for molecular intervention in specific subsets of patients.
Common genetic changes have been validated in PDAC pathogenesis in several whole-genome sequencing studies. The genetic landscape of the PDAC genome is notable for 4 frequently mutated genes, including KRAS, CDKN2A/p16, TP53, and SMAD4/DPC4[4,5]. Among all of the mutated genes, KRAS is among the earliest and most pervasive alteration in pancreatic carcinogenesis, and the KRAS gene is mutated in virtually all PDAC patients[6-8]. In addition, a number of genetic studies have shown that such activating K-ras gene mutations are necessary for the onset of pancreatic cancer[9]. An inducible pancreas-specific expression system was recently used to show that K-RASG12D expression is also required for tumor maintenance[10]. These findings indicate the importance of K-ras gene in PDAC. However, K-ras gene showed low prognostic and predictive value because this gene was mutated in almost all of the PDAC patients.
The inappropriate activation of Ras proteins promotes a variety of malignant phenotypes, including enhanced growth, loss of contact inhibition, reduced requirement for growth factors, enhanced motility and invasion and resistance to apoptosis[11]. Interestingly, activated Ras can also induce various aspects of growth inhibition and death[12-14]. RASSF family proteins have now been identified as potential mediators of some of the growth inhibitory effects of Ras, and RASSF family proteins are often down-regulated during tumorigenesis[15]. RASSF6 was identified as a new negative effector of the RAS protein, which demonstrates a tumor suppressor function, including the inhibition of growth, promotion of cell cycle arrest, and induction of apoptosis[16]. In 239-T cells, a dramatic synergistic activation of cell death was observed when the cells were transfected with activated KRAS and RASSF6 together, and siRNA-mediated knockdown of RASSF6 expression in human lung tumor cells increases tumorigenicity[15]. In addition, studies showed that the inactivation of RASSF6 is also an extremely frequent event in the pathogenesis of childhood leukemia[17].
Collectively, these findings above indicate that RASSF6 might play an important role in regulating PDAC progression. However, to our knowledge, no data concerning the role of RASSF6 in PDAC is available. The objective of the current study was therefore to clarify the clinical implications of the status of RASSF6 in PDAC. We investigated the status of RASSF6 expression in pancreatitis tissues, PDAC primary lesions, and lymph node metastases, and evaluated the impact of RASSF6 expression on outcomes in patients undergoing resection for PDAC.
This study involved patients with PDAC undergoing radical operations in a retrospective study cohort between January 2000 and June 2012 at Sun Yat-sen Memorial Hospital of Sun Yat-sen University and a prospective cohort from June 2012 to May 2013 (registration number: ChiCTR-TRC-12002548). The study was approved by the Ethics Committee of Sun Yat-sen Memorial Hospital, and all study participants provided informed written consent prior to study enrollment. Data were collected on age, sex, histological and pathological findings, lymph node metastasis, distant metastases, tumor stage (UICC 6th edition), and clinical follow-up. Patients who died of postoperative complications and patients lost during follow-up were excluded. Overall survival (OS) was calculated from the date of operation to the date of death or last follow-up. Through searching the electronic medical record system including the clinical and pathological records of inpatients, 96 surgically treated patients diagnosed with histologically confirmed stage I or II PDAC were included.
Paraffin-embedded samples of the primary carcinomas from 96 patients were stained for RASSF6. The pathological sections were deparaffinized in xylene and rehydrated in a graded ethanol series followed by heat-induced epitope retrieval in citrate buffer (pH 6.0). Antigen retrieval was carried out in 10 mmol/L citrate buffer (pH 6.0) in a microwave oven for 15 min. The activity of endogenous peroxidase was exhausted with 3% hydrogen peroxide for 10 min at room temperature. A rabbit RASSF6 polyclonal antibody (Proteintech Group, Chicago, IL) was applied overnight at 4 °C at an optimal working concentration of 1:200. After sufficient phosphate buffered saline rinses, the sections were immunostained with goat anti-rabbit polymers. The staining was scored by two independent investigators, without knowledge of the patient outcomes, clinical features or pathological characteristics, according to the staining intensity and extent as described previously[18]. The sum of staining intensity and extent was designated as follows: 0-2, negative expression; 3-4, weak expression; and 5-6, strong expression. If a disagreement occurred, the slides were re-examined to obtain a final consensus.
The total RNA of 96 frozen primary pancreatic cancer tissues was prepared with Trizol (TRIzol, Invitrogen, United States) according to the manufacturer’s instructions and reverse transcribed into cDNA. The mRNA levels of RASSF6 and GAPDH were analyzed using the SYBR Green Realtime PCR Master Mix with gene-specific primers on the ABI 7500 Fast Real-Time PCR System, according to the manufacturer’s instructions. The primers for qRT-PCR were: RASSF6, sense 5’-AGCTGCCAGTTCTTGGAATG-3’ and antisense 5’-AGGCCAGACAGCTCTGATGT-3’; GAPDH, sense 5’-GTCCACCACCCTGTTGCTGTA-3’ and antisense 5’-CTTCAACAGCGACACCCACTC-3’. The cycle number at which the reaction crossed an arbitrarily placed threshold (Ct) was determined for each gene. The RASSF6 mRNA level for each tumor was calculated using the △Ct method: △Ct = CtRASSF6 - CtGAPDH.
The total protein content was extracted from frozen PDAC and para-tumor tissues stored in liquid nitrogen using a Whole Cell Extraction Kit (Pierce, Rockford, IL). For equal protein loading, a bicinchoninic acid protein assay kit (Pierce) was used to calculate the protein concentration in each sample. Equivalent amounts of proteins were separated by SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes for immunoblotting. The membranes were blocked in 5% fat-free milk for 2 h at room temperature and washed 3 times, and then the membranes were incubated with the following primary antibodies: a rabbit anti-human RASSF6 polyclonal antibody (1:1000, Proteintech Group, Chicago, IL) and a polyclonal rabbit anti-human β-actin polyclonal antibody (1:1000, Abcam, Cambridge, MA). β-actin was used as a loading control. Horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology) and an ECL chemiluminescence kit (Pierce) were used to detect the bound antibodies.
All of the statistical analyses were performed with the SPSS 16.0 statistical software package (SPSS Inc., Chicago, IL). Frequency distributions were compared by χ2 test. Continuous variables were compared using the Student’s t-test. The principal outcome measure was the length of survival as measured from the time of the original surgery. The patients alive at the time of the follow-up point were censored. The last follow-up period for patients still alive was May 2013. A Kaplan- Meier survival analysis was used to assess the significance of RASSF6 protein alone or together with other clinical factors. The significant differences between the survival curves were determined using the log-rank test. The 1-, 3- and 5-year survival rates were estimated using life tables. Variables that were significant after univariate analysis at P < 0.1 were included in multivariate analysis. Cox proportional hazards models were generated for multivariate analysis. A P value < 0.05 was considered statistically significant in a two-tailed test. The statistical methods of this study were reviewed by Professor Jing Gu from the School of Public Health of Sun Yat-sen University.
Demographic data and tumor characteristics are summarized in Table 1. The cohort of 96 patients consisted of 45 women and 51 men. The mean age at operation was 60.7 years, with a median age of 61 years and a range of 31-77. At the time of the last clinical follow-up (May 2013), 23 (24.0%) patients were alive. None of the patients died during the perioperative period because of post-operative complications. The median OS was 15.5 mo, with 1-, 3-, and 5-year survival rates of 57.5%, 23.0%, and 14.0%, respectively, in all 96 patients. The majority of tumors were moderately differentiated (38.5%), followed by well differentiated (36.5%), and 25.0% of tumors were poorly differentiated. Most of the tumors (n = 81) were located in the head of the pancreas (84.4%), and 80 (83.3%) were more than 2.0 cm in maximal diameter. Eighty-one (84.4%) patients had T3 tumors. Lymph node metastases were present in 61 (63.5%) patients, and portal vein invasion was present in 12 (12.5%) of the 96 patients. All of the patients presented with the Union for International Cancer Control (UICC) stage I or II disease, and none of the patients presented with UICC stage III or IV disease. All of the patients had undergone primary curative resection, and 17 (17.7%) patients had a positive surgical margin. If indicated, patients received either gemcitabine-based or 5-FU-based chemotherapy as first-line chemotherapy according to their clinical disease stage and performance status.
| Parameter | Total | RASSF6 | RASSF6 | RASSF6 | P value |
| n = 96 | negative | weak | positive | ||
| n = 51 | n = 29 | n = 16 | |||
| Age (yr) | 0.115 | ||||
| mean (range) | 60.7 (31-77) | 59.3 (77-45) | 61.3 (31-77) | 64.3 (54-70) | |
| Gender | 0.563 | ||||
| Male | 51 | 29 | 13 | 9 | |
| Female | 45 | 22 | 16 | 7 | |
| Location of tumor | 0.777 | ||||
| Head | 81 | 42 | 22 | 13 | |
| Body/tail | 15 | 9 | 7 | 3 | |
| Differentiation | 0.138 | ||||
| Well | 35 | 14 | 11 | 10 | |
| Moderate | 37 | 22 | 12 | 3 | |
| Poor | 24 | 15 | 6 | 3 | |
| Tumor diameter (cm) | 0.613 | ||||
| mean (range) | 3.7 (1.5-8.0) | 3.8 (1.5-8.0) | 3.5 (1.5-6.0) | 3.9 (7.5-5.5) | |
| UICC/TNM stage | 0.200 | ||||
| pI | 6 | 4 | 0 | 2 | |
| pII | 90 | 47 | 29 | 14 | |
| T-stage | 0.0471, 0.0762 | ||||
| pT1 | 5 | 3 | 2 | 0 | |
| pT2 | 10 | 3 | 2 | 5 | |
| pT3 | 81 | 45 | 25 | 11 | |
| N-stage | 0.124 | ||||
| pN0 | 35 | 23 | 9 | 3 | |
| pN1 | 61 | 28 | 20 | 13 | |
| Portal vein invasion | 0.698 | ||||
| Negative | 84 | 45 | 26 | 13 | |
| Positive | 12 | 6 | 3 | 3 | |
| Perineural invasion | 0.026 | ||||
| Negative | 31 | 11 | 11 | 9 | |
| Positive | 65 | 40 | 18 | 7 | |
| Surgical margins | 0.694 | ||||
| Negative | 79 | 43 | 24 | 12 | |
| Positive | 17 | 8 | 5 | 4 |
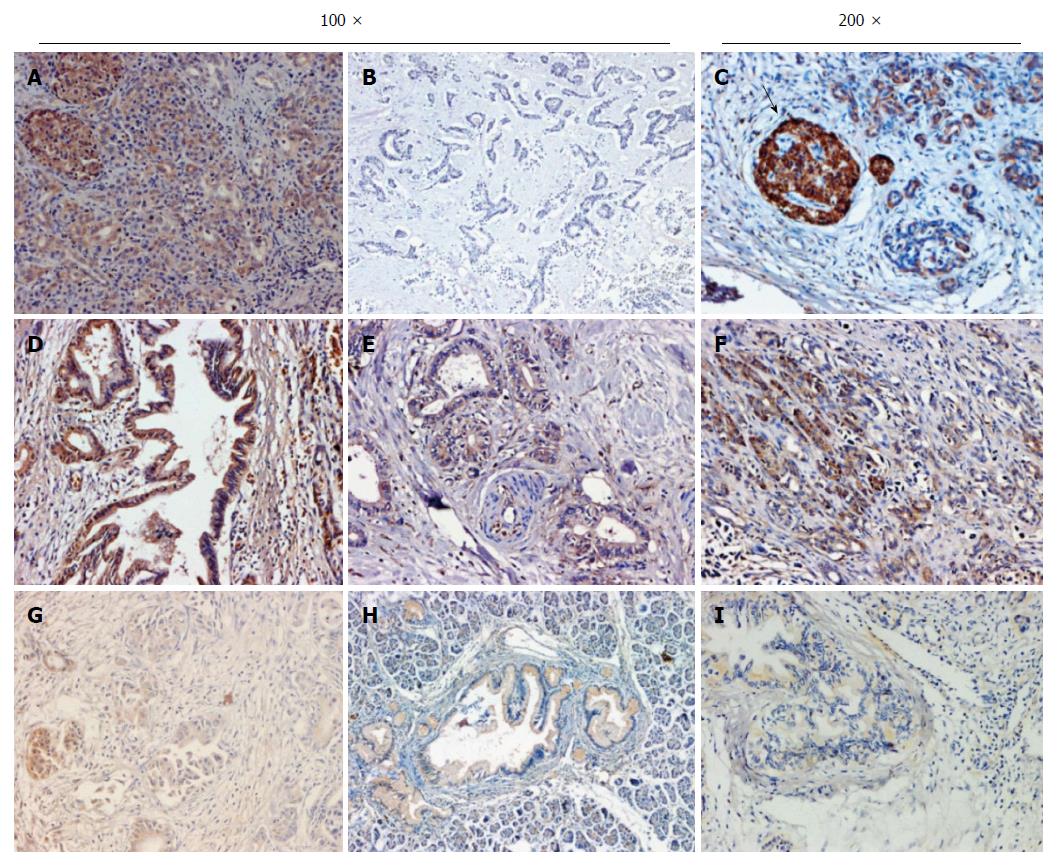
The immunohistochemical analysis of RASSF6 was performed on the 96 primary lesions of pancreatic ductal adenocarcinoma and 20 resected lesions of chronic pancreatitis. Figure 1 represents the immunohistochemical results of RASSF6 expression in patients with pancreatic ductal adenocarcinoma and chronic pancreatitis. RASSF6 was detected in carcinoma cells in the tumor tissues and was localized predominantly in the cytoplasm, without nuclear staining. In addition, islet cells showed an extremely strong staining of RASSF6 expression. Tumor cytoplasmic expression of RASSF6 was classified as negative (Figure 1A), strongly positive (Figure 1B) and weakly positive (Figure 1C). Of the 96 patients with pancreatic cancer, 51 (53.1%) showed negative RASSF6 expression, 29 (30.2%) had weak RASSF6 staining, and 16 (16.7%) displayed strongly positive staining. Among the 20 patients with chronic pancreatitis, strong RASSF6 expression and weak RASSF6 expression were observed in 80% (16/20) and 20% (4/20) of cases, respectively.
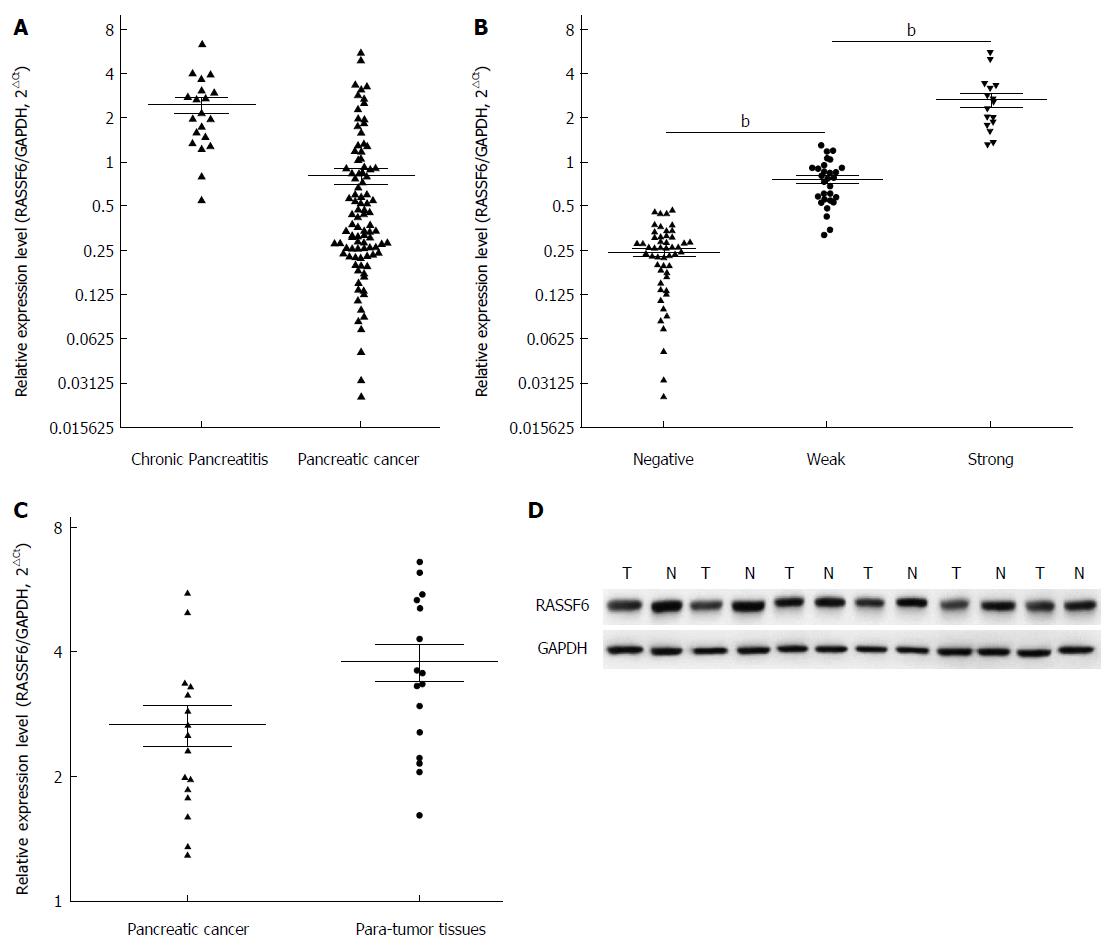
Immunohistochemical analysis demonstrated a significantly higher proportion of positive RASSF6 staining in chronic pancreatitis compared with pancreatic cancer (Figure 1) (P = 0.015, Fisher’s exact test). In addition, qRT-PCR results revealed that the mRNA levels of RASSF6 were also down-regulated in PDAC compared with chronic pancreatitis (Figure 2A, P < 0.001). The relationship between the immunohistochemical stage of RASSF6 and its mRNA level was further evaluated (Figure 2B), and we found a clear difference in measured mRNA levels between negative, weak and strong RASSF6 staining groups. This finding indicates that the immunohistochemical scores are consistent with qRT-PCR results. Subsequently, RASSF6 mRNA expression and protein expression in 16 cases of PDAC samples with strong positive RASSF6 staining were evaluated using real-time PCR and Western blot analysis. The results confirmed that RASSF6 protein and mRNA levels were significantly down-regulated in cancer tissues (Figure 2C and D).
Tumors with perineural invasion demonstrated less frequent RASSF6 expression, and a significant association between RASSF6 expression level and the presence of perineural invasion was observed (P = 0.026; Table 1). In addition, RASSF6 expression seemed to be associated with the UICC T stage. We found that higher-stage tumors demonstrated RASSF6 expression less frequently with a significant difference at the boundary (Table 1). No significant association was observed between age, sex, location of tumor, differentiation, lymph node status, portal vein invasion, or surgical margins.
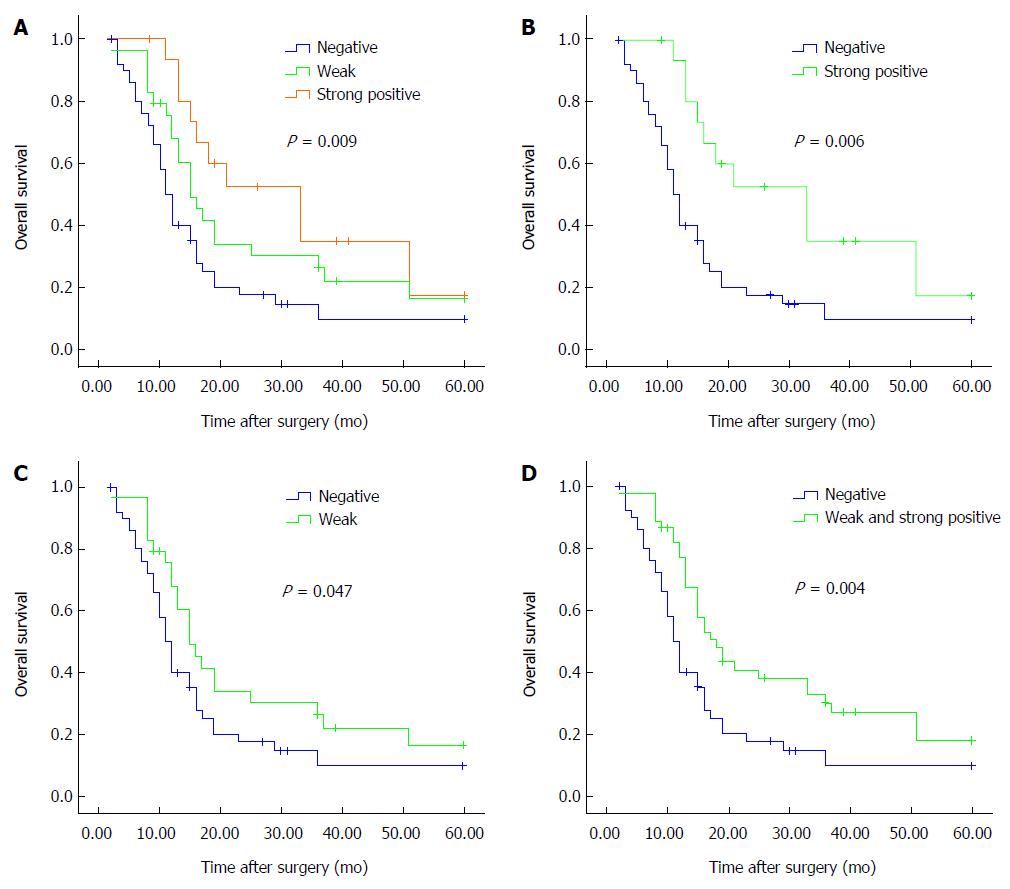
Based on the immunohistochemical staining of RASSF6, 96 patients were then stratified into 3 groups (strongly positive vs weak vs negative). Kaplan-Meier curves were constructed to determine whether RASSF6 expression was correlated with survival in these groups. The median survival time of the strongly positive group, weak group and negative group was 33 mo, 15 mo and 11 mo, respectively. In this cohort, there was a significant correlation between RASSF6 expression levels and OS (P = 0.009; Figure 3A). Compared with patients with negative RASSF6A expression, patients with both strongly positive RASSF6 staining and weak RASSF6 staining showed improved OS time (strongly positive vs negative, P = 0.006; Figure 3B; weak vs negative, P = 0.047; Figure 3C). Furthermore, positive (weakly and strongly) RASSF6 expression was also strongly associated with an increased OS time compared with negative RASSF6 expression (P = 0.004) (Figure 3D).
The variables most likely to impact survival by univariate analysis (P≤ 0.1) were entered into the multivariate analysis model (Table 2). Cox regression analysis demonstrated that negative RASSF6 expression remained an independent prognostic factor of poor prognosis HR = 4.620, 95%CI: 2.027-10.527, P = 0.006).
| Variable | Univariate | Multivariate | ||
| P value | HR | 95%CI | P value | |
| Tumor diameter | 0.010 | 1.172 | 0.992-1.385 | 0.062 |
| T stage | 0.012 | 2.355 | 1.413-3.925 | 0.031 |
| Differentiation | 0.016 | 2.397 | 1.665-3.451 | < 0.001 |
| Perineural invasion | 0.076 | 2.236 | 1.163-4.300 | 0.016 |
| Rassf6 expression | 0.021 | 4.620 | 2.027-10.527 | 0.006 |
For the first time, we examined the mRNA and protein expression of RASSF6 in PDAC and evaluated its prognostic value. We confirmed that both the protein and mRNA expression levels of RASSF6 were down-regulated in PDAC. In addition, down-regulation of RASSF6 is a predictor of poor survival outcome. Our findings suggested that RASSF6 was a valuable prognostic indicator of PDAC patients undergoing radical operations, and a decrease in RASSF6 expression might be associated with the progression of PDAC.
The Ras proteins perform their diverse functions by interacting with RASS effectors, which have conserved Ras interacting domains. The RA-domain, a type of Ras interacting domain, is a main characteristic of the Ras-association domain family (RASSF) proteins[19]. RASSF family proteins have now been identified as potential mediators of some of the growth inhibitory effects of Ras[20-23]. Interestingly, RASSF family proteins are often down-regulated during the process of oncogenesis[22,23], and genes of the RASSF family have been reported to be epigenetically silenced in certain types of human cancers[23]. RASSF6 is the last reported member of the RASSF family proteins. Of the 96 patients, we found that more than half of the tumors showed negative RASSF6 expression (53.1%), and only 16 (16.7%) patients’ tumor tissues stained strongly for RASSF6. Our findings agree with earlier studies demonstrating that RASSF6 is often down-regulated in primary human tumors. We did not find any significant differences in the distribution of age, gender and tumor location between groups based upon RASSF6 expression. The PDAC in the present study tended to be larger (mean diameter = 3.7 cm, with 86% being T3); this fact can be explained by difficult early diagnoses, especially in developing regions, such as where our hospital located.
Until now, only one study investigated the clinical value of RASSF6 expression in cancer: Wen et al[18] reported that RASSF6 expression was an independent prognostic factor for gastric cancer patients and strong RASSF6 expression was associated with better OS compared with patients whose tumors are RASSF6 negative. Using immunohistochemistry, we found that decreased RASSF6 protein expression seemed to be associated with higher T stage tumors, and RASSF6 was moderately or strongly expressed to a greater degree in tumors without perineural invasion. Multivariate analysis confirmed that the RASSF6 expression level was an independent prognostic factor for OS (P < 0.001). In the present study, survival analysis also showed that increased RASSF6 expression promoted a significant survival advantage that persisted in multivariate analysis. Our prognostic results are in accordance with a study performed in gastric cancer patients. The previous study by Wen et al[18] demonstrated a significant correlation between RASSF6 expression and tumor differentiation, but our findings are different. In our data, RASSF6 expression tended to be negative in larger tumors or higher T stage ones, but not positive in poorly differentiated tissues. Coupled with the finding that RASSF6 expression was decreased in metastatic tissues revealed by Western blot and that RASSF6 stained strongly in chronic pancreatitis tissues, we proposed that RASSF6 expression might decrease along with progression of cancer.
Interestingly, tumors with perineural invasion showed a significant decrease in the RASSF6 expression level compared to those without perineural invasion (P = 0.026) in our study, and RASSF6 protein expression in tumor cells that invaded nerve tissues was also decreased compared with adjacent PDAC tumor cells. Perineural invasion has been reported in certain types of cancer, including pancreatic, prostate, colorectal, etc.[24,25]. PDACs have the highest incidences of perineural invasion among cancers[26,27], and the presence of perineural invasion was reported as a strong poor prognostic factor for both OS and disease-free survival[28,29]. Meanwhile, perineural invasion is considered the primary cause of abdominal pain in pancreatic cancer patients[25]. Therefore, targeting perineural invasion is an attractive therapeutic approach for pancreatic cancer. Until now, most of the studies on perineural invasion focused on the interaction between cancer cells and nerves. Therefore, research on the biological mechanism of perineural invasion was limited to the ligand-receptor inter-attraction between tumor cells and nerve tissues[25]. Few studies have examined the characteristics of the tumor cells themselves and the roles of tumor stromal components, such as fibroblasts, infiltrating immune cells and extracellular matrix proteins in perineural invasion. Our findings regarding the relationship between altered RASSF6 expression and perineural invasion indicated a potential role of RASSF6 in the process of perineural invasion. The effect of the RASSF6 protein in perineural invasion and whether the expression of RASSF6 is regulated by an endopathic approach or external stimuli need further study. Recently, a study reported that RASSF6 expression in adipocytes was down-regulated by interacting with macrophages[30], indicating a potential role of infiltrating macrophages in perineural invasion though regulating RASSF6 expression.
The other notable structural feature of RASSF6 is the C-terminal Salvador-RASSF-Hippo (SARAH) domain, which mediates the antagonism effect of RASSF on the Drosophila Hippo pathway[31]. The Hippo pathway is a key regulator of organ size and cell density through regulating cell proliferation, apoptosis, and stem cell/progenitor cell expansion[32]. The Hippo signaling cascade antagonizes the transcriptional coactivator yes-associated protein (YAP) whose overexpression has been reported in different human tumors, including colon, prostate, ovarian, and lung cancers[33]. Two recent studies reported a crucial requirement for the Hippo signaling pathway in regulating the development of the pancreas[33,34]. However, the role of the Hippo pathway in pancreatic cancer remains unclear. Ikeda et al[32] recently found that RASSF6 antagonizes Hippo signaling and mediates apoptosis through a pathway that is parallel to the canonical Hippo pathway. In the present study, we also defined a crucial role of the RASSF6 protein in the development of PDAC. These findings suggested that the Hippo pathway and/or YAP might play a potentially important role in pancreatic cancer.
In conclusion, this is the first report to demonstrate the clinical significance of RASSF6 expression in PDAC. RASSF6 expression appears to be decreased with the progression of pancreatic cancer and is significantly associated with poor prognosis. Our results suggest that the presence of RASSF6 expression in PDAC may have a protective effect independent of tumor stage in resectable tumors. In addition, we believe that these results provide a basis for further, more detailed studies on perineural invasion and the Hippo pathway in pancreatic cancer.
Pancreatic ductal adenocarcinoma (PDAC) is a highly malignant digestive tumor with a very poor prognosis. Inappropriate activation of Ras proteins is a hallmark of PDAC, which promotes a variety of malignant phenotypes, including enhanced growth, loss of contact inhibition, reduced requirement for growth factors, enhanced motility and invasion and resistance to apoptosis.
Recently, RASSF6 was identified as a new negative effector of the RAS protein, which demonstrates a tumor suppressor function. In 239-T cells, a dramatic synergistic activation of cell death was observed when the cells were transfected with activated K-ras and RASSF6 together, and siRNA-mediated knockdown of RASSF6 expression in human lung tumor cells increases tumorigenicity. However, the role of RASSF6 in PDAC is still unknown. In this study, the authors demonstrated that RASSF6 expression is decreased in PDAC tissues and lymph node metastases, and the expression of RASSF6 has a significant impact on the survival of patients with PDAC. These results might indicate that RASSF6 could be a potential prognostic factor for PDAC patients.
This is the first study to report that RASSF6 is decreased in PDAC and that the expression of RASSF6 might decrease with the development of cancer.
This study showed that the RASSF6 expression level may be used as a potential predictor of prognosis in pancreatic cancer.
RASSF6 is the abbreviation of Ras association (RalGDS/AF-6) domain family member 6. This gene encodes a member of the Ras-association domain family (RASSF). Members of this family form the core of a highly conserved tumor suppressor network, the Salvador-Warts-Hippo pathway. The protein encoded by this gene is a Ras effector protein that induces apoptosis. A genomic region containing this gene has been linked to susceptibility to viral bronchiolitis. Alternative splicing results in multiple transcript variants and protein isoforms.
This is an interesting study with valuable information regarding the expression and clinical impact of RASSF6 in pancreatic ductal adenocarcinoma.
P- Reviewer: Mosteiro L, Sun YW S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Liu XM
| 1. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11128] [Cited by in RCA: 11831] [Article Influence: 845.1] [Reference Citation Analysis (4)] |
| 2. | Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801-1806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3216] [Cited by in RCA: 3022] [Article Influence: 177.8] [Reference Citation Analysis (0)] |
| 3. | Sun C, Rosendahl AH, Ansari D, Andersson R. Proteome-based biomarkers in pancreatic cancer. World J Gastroenterol. 2011;17:4845-4852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 4. | Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1690] [Cited by in RCA: 1638] [Article Influence: 126.0] [Reference Citation Analysis (0)] |
| 5. | Iacobuzio-Donahue CA. Genetic evolution of pancreatic cancer: lessons learnt from the pancreatic cancer genome sequencing project. Gut. 2012;61:1085-1094. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 110] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 6. | van Heek T, Rader AE, Offerhaus GJ, McCarthy DM, Goggins M, Hruban RH, Wilentz RE. K-ras, p53, and DPC4 (MAD4) alterations in fine-needle aspirates of the pancreas: a molecular panel correlates with and supplements cytologic diagnosis. Am J Clin Pathol. 2002;117:755-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 60] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 7. | Haeno H, Gonen M, Davis MB, Herman JM, Iacobuzio-Donahue CA, Michor F. Computational modeling of pancreatic cancer reveals kinetics of metastasis suggesting optimum treatment strategies. Cell. 2012;148:362-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 307] [Article Influence: 23.6] [Reference Citation Analysis (2)] |
| 8. | di Magliano MP, Logsdon CD. Roles for KRAS in pancreatic tumor development and progression. Gastroenterology. 2013;144:1220-1229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 315] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 9. | Reichert M, Rustgi AK. Pancreatic ductal cells in development, regeneration, and neoplasia. J Clin Invest. 2011;121:4572-4578. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 177] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 10. | Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1287] [Cited by in RCA: 1375] [Article Influence: 62.5] [Reference Citation Analysis (0)] |
| 11. | Frame S, Balmain A. Integration of positive and negative growth signals during ras pathway activation in vivo. Curr Opin Genet Dev. 2000;10:106-113. [PubMed] |
| 12. | Cox AD, Der CJ. The dark side of Ras: regulation of apoptosis. Oncogene. 2003;22:8999-9006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 343] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 13. | Nicke B, Bastien J, Khanna SJ, Warne PH, Cowling V, Cook SJ, Peters G, Delpuech O, Schulze A, Berns K. Involvement of MINK, a Ste20 family kinase, in Ras oncogene-induced growth arrest in human ovarian surface epithelial cells. Mol Cell. 2005;20:673-685. [PubMed] |
| 14. | Sherwood V, Recino A, Jeffries A, Ward A, Chalmers AD. The N-terminal RASSF family: a new group of Ras-association-domain-containing proteins, with emerging links to cancer formation. Biochem J. 2010;425:303-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 15. | Allen NP, Donninger H, Vos MD, Eckfeld K, Hesson L, Gordon L, Birrer MJ, Latif F, Clark GJ. RASSF6 is a novel member of the RASSF family of tumor suppressors. Oncogene. 2007;26:6203-6211. [PubMed] |
| 16. | Hesson LB, Dunwell TL, Cooper WN, Catchpoole D, Brini AT, Chiaramonte R, Griffiths M, Chalmers AD, Maher ER, Latif F. The novel RASSF6 and RASSF10 candidate tumour suppressor genes are frequently epigenetically inactivated in childhood leukaemias. Mol Cancer. 2009;8:42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 87] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 17. | Sherwood V, Manbodh R, Sheppard C, Chalmers AD. RASSF7 is a member of a new family of RAS association domain-containing proteins and is required for completing mitosis. Mol Biol Cell. 2008;19:1772-1782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | Wen Y, Wang Q, Zhou C, Yan D, Qiu G, Yang C, Tang H, Peng Z. Decreased expression of RASSF6 is a novel independent prognostic marker of a worse outcome in gastric cancer patients after curative surgery. Ann Surg Oncol. 2011;18:3858-3867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 19. | Vos MD, Ellis CA, Bell A, Birrer MJ, Clark GJ. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J Biol Chem. 2000;275:35669-35672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 209] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 20. | Vos MD, Ellis CA, Elam C, Ulku AS, Taylor BJ, Clark GJ. RASSF2 is a novel K-Ras-specific effector and potential tumor suppressor. J Biol Chem. 2003;278:28045-28051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 142] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 21. | Eckfeld K, Hesson L, Vos MD, Bieche I, Latif F, Clark GJ. RASSF4/AD037 is a potential ras effector/tumor suppressor of the RASSF family. Cancer Res. 2004;64:8688-8693. [PubMed] |
| 22. | Agathanggelou A, Cooper WN, Latif F. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005;65:3497-3508. [PubMed] |
| 23. | Djos A, Martinsson T, Kogner P, Carén H. The RASSF gene family members RASSF5, RASSF6 and RASSF7 show frequent DNA methylation in neuroblastoma. Mol Cancer. 2012;11:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 24. | Marchesi F, Piemonti L, Mantovani A, Allavena P. Molecular mechanisms of perineural invasion, a forgotten pathway of dissemination and metastasis. Cytokine Growth Factor Rev. 2010;21:77-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 195] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 25. | Bapat AA, Hostetter G, Von Hoff DD, Han H. Perineural invasion and associated pain in pancreatic cancer. Nat Rev Cancer. 2011;11:695-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 335] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 26. | Liu B, Lu KY. Neural invasion in pancreatic carcinoma. Hepatobiliary Pancreat Dis Int. 2002;1:469-476. [PubMed] |
| 27. | Pour PM, Bell RH, Batra SK. Neural invasion in the staging of pancreatic cancer. Pancreas. 2003;26:322-325. [PubMed] |
| 28. | Chen JW, Bhandari M, Astill DS, Wilson TG, Kow L, Brooke-Smith M, Toouli J, Padbury RT. Predicting patient survival after pancreaticoduodenectomy for malignancy: histopathological criteria based on perineural infiltration and lymphovascular invasion. HPB (Oxford). 2010;12:101-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 96] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 29. | di Mola FF, di Sebastiano P. Pain and pain generation in pancreatic cancer. Langenbecks Arch Surg. 2008;393:919-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 30. | Sanada Y, Kumoto T, Suehiro H, Nishimura F, Kato N, Hata Y, Sorisky A, Yanaka N. RASSF6 expression in adipocytes is down-regulated by interaction with macrophages. PLoS One. 2013;8:e61931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 32. | Ikeda M, Kawata A, Nishikawa M, Tateishi Y, Yamaguchi M, Nakagawa K, Hirabayashi S, Bao Y, Hidaka S, Hirata Y. Hippo pathway-dependent and -independent roles of RASSF6. Sci Signal. 2009;2:ra59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 81] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 33. | George NM, Day CE, Boerner BP, Johnson RL, Sarvetnick NE. Hippo signaling regulates pancreas development through inactivation of Yap. Mol Cell Biol. 2012;32:5116-5128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 109] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 34. | Gao T, Zhou D, Yang C, Singh T, Penzo-Méndez A, Maddipati R, Tzatsos A, Bardeesy N, Avruch J, Stanger BZ. Hippo signaling regulates differentiation and maintenance in the exocrine pancreas. Gastroenterology. 2013;144:1543-1553, 1553.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 119] [Article Influence: 9.9] [Reference Citation Analysis (0)] |