Published online May 7, 2015. doi: 10.3748/wjg.v21.i17.5295
Peer-review started: November 3, 2014
First decision: November 26, 2014
Revised: December 11, 2014
Accepted: January 30, 2015
Article in press: January 30, 2015
Published online: May 7, 2015
Processing time: 193 Days and 20.2 Hours
AIM: To report an acute gastroenteritis outbreak caused by a genogroup 2 genotype 6 (GII.6) strain norovirus in Shanghai, China.
METHODS: Noroviruses are responsible for approximately half of all reported gastroenteritis outbreaks in many countries. Genogroup 2 genotype 4 strains are the most prevalent. Rare outbreaks caused by GII.6 strains have been reported. An acute gastroenteritis outbreak occurred in an elementary school in Shanghai in December of 2013. Field and molecular epidemiologic investigations were conducted.
RESULTS: The outbreak was limited to one class in an elementary school located in southwest Shanghai. The age of the students ranged from 9 to 10 years. The first case emerged on December 10, 2013, and the last case emerged on December 14, 2013. The cases peaked on December 11, 2013, with 21 new cases. Of 45 students in the class, 32 were affected. The main symptom was gastroenteritis, and 15.6% (5/32) of the cases exhibited a fever. A field epidemiologic investigation showed the pathogen may have been transmitted to the elementary school from employees in a delicatessen via the first case student, who had eaten food from the delicatessen one day before the gastroenteritis episodes began. A molecular epidemiologic investigation identified the cause of the gastroenteritis as norovirus strain GII.6; the viral sequence of the student cases showed 100% homology with that of the shop employees. Genetic relatedness analyses showed that the new viral strain is closely related to previously reported GII.6 sequences, especially to a strain reported in Japan.
CONCLUSION: This is the first report to show that norovirus strain GII.6 can cause a gastroenteritis outbreak. Thus, the prevalence of GII.6 noroviruses requires attention.
Core tip: Noroviruses are responsible for approximately half of all reported gastroenteritis outbreaks in many countries. Rare outbreaks caused by genogroup 2 genotype 6 (GII.6) strains have been reported. An acute gastroenteritis outbreak occurred in an elementary school in Shanghai in December of 2013. Molecular epidemiologic investigations showed that the gastroenteritis outbreak was caused by norovirus strain GII.6 infection. Thus, the prevalence of GII.6 noroviruses requires attention.
- Citation: Luo LF, Qiao K, Wang XG, Ding KY, Su HL, Li CZ, Yan HJ. Acute gastroenteritis outbreak caused by a GII.6 norovirus. World J Gastroenterol 2015; 21(17): 5295-5302
- URL: https://www.wjgnet.com/1007-9327/full/v21/i17/5295.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i17.5295
Norovirus infection is recognized as a leading cause of epidemic and sporadic acute gastroenteritis in children and adults worldwide[1]. Most infected individuals get sick within one day of norovirus ingestion[2], though the incubation period ranges from 12 h to 48 h[3]. The symptoms include vomiting, watery diarrhea, or both. Stomach pain and a general feeling of tiredness, headache, and muscle aches are also common[4]. Fever occurs in one-third to one-half of infected individuals[5]. The main transmission routes of noroviruses include oral-fecal, person-to-person, and waterborne[1-5]. Noroviruses cause outbreaks in autumn and winter in over-populated settings such as schools and hospital wards[6].
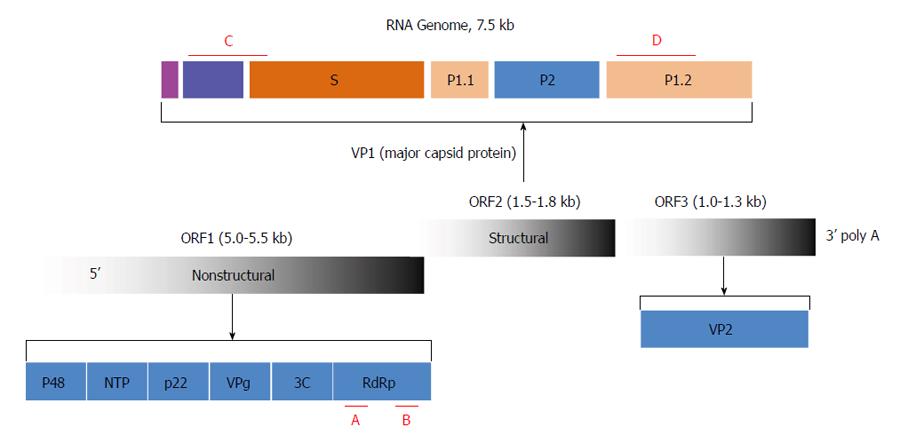
The Norovirus genus belongs to the Caliciviridae family[7]. The genome is comprised of a single-stranded positive-sense RNA molecule approximately 7.5 kb in length with three open reading frames (ORFs)[7]. ORF1 encodes a polypeptide of six nonstructural proteins, including RNA-dependent RNA polymerase (RdRp)[7,8]. ORF2 encodes the major capsid protein VP1, which consists of a shell (S) and two protruding (P) domains, P1 and P2. ORF3 encodes the minor capsid protein VP2[7,8]. Noroviruses are categorized into five distinct genogroups (GI-V); of these, GI, GII, and GIV can infect humans[9]. The genogroups of noroviruses were defined on the basis of the amino acid diversity of the three ORFs, RdRp, and VP1[10,11]. The current five genogroups, which were developed based on VP1 protein divergence, can be further divided into genotypes and subgenotypes[12,13]. The genotypes are defined on the basis of either the RdRp sequence or the capsid sequence[12-14]. Recombination occurs occasionally at the ORF1/ORF2 junction[15,16]; thus, to define a field norovirus strain it is necessary to determine both the polymerase and capsid genotypes[15,16]. This explains why the current nomenclature comprises both polymerase and capsid genotypes. To date, 14 GI and 29 GII polymerase genotypes, and 8 GI and 23 GII capsid genotypes, have been described[14].
The genogroup and genotype of norovirus strains associated with sporadic and epidemic gastroenteritis remain poorly described. Two recent systematic literature reviews demonstrated several important points[14,17]. First, genogroup GII is the most prevalent, accounting for 96% of all sporadic infections. GII genotype 4 (GII.4) is the most prevalent genotype, accounting for 70% of the capsid genotypes and 60% of the polymerase genotypes, followed by capsid genotype GII.3 (16%) and polymerase genotype GII.b (14%)[14]. Second, based on 71724 illnesses, 501 hospitalizations, and 45 deaths, the overall hospitalization and mortality rates are 0.54 and 0.06%, respectively. GII.4 norovirus strains are associated with higher hospitalization and mortality rates[17].
At present, reports on acute gastroenteritis caused by norovirus GII.6 are rare. In a study of 187 fecal specimens collected from non-hospitalized children with acute gastroenteritis in Shizuoka, Japan, between July of 2008 and June of 2009, 55.6% tested positive for noroviruses; of these, 53.8% and 40.4% contained strains GII.4 and GII.6, respectively[18]. Active surveillance for laboratory-confirmed cases of norovirus among children younger than five years of age with acute gastroenteritis in hospitals in the United States showed that GII.6 noroviruses were also detected in fecal specimens[19]. These are the only two reports of GII.6 norovirus infections, and neither report provided evidence showing that GII.6 could cause gastroenteritis outbreaks. In this study, we present an acute gastroenteritis outbreak caused by norovirus strain GII.6 in an elementary school located in Minhang District, Shanghai, China, which occurred in 2013. Our data complement the current understanding of norovirus infection, especially of GII.6 noroviruses.
Fecal specimens from 32 elementary school students in Class 1/Grade 4 were collected for suspected gastroenteritis pathogen detection. Fecal specimens were also collected from two employees of a delicatessen who were suspected of being the source of the pathogen. Data on the hospitalized students, including body temperature and routine blood test results, were also collected.
This study was conducted according to the principles of World Medical Association Declaration of Helsinki. The study was specifically approved by Internal Review Board of the Center for Disease Control and Prevention of Shanghai Minhang District, China (Permit Number: 2013-0012). All participants gave written informed consent for research use of stool samples. We also obtained written informed consent from the parents on the behalf of the minors enrolled in our study. The ethics committee specifically approved the consent procedure for the participants between nine and ten years of age (Permit Number: 2013-0012m).
The detection of rotavirus was performed as described by Jothikumar et al[20]. For bacterial evidence, the isolation and culture of Campylobacter spp, Escherichia coli (E. coli), Salmonella, and Shigella were performed as reported previously[21,22].
The fecal specimens were prepared as a 10% (w/v) suspension in distilled water and then centrifuged for 10 min at 10000 ×g. Viral RNA was extracted from the suspensions using a QIAamp Viral RNA Mini Kit (Qiagen, Venlo, Limburg, Netherlands), according to the manufacturer’s instructions. The RNA pellet was re-dissolved in 10 μL of 10 mmol/L dithiothreitol containing 5% (v/v) RNasin (40 U/μL; Promega, Madison, WI, United States) and stored at -80 °C until use.
Based on routine disease surveillance data and the characteristics of the acute gastroenteritis outbreak, a norovirus was suspected to be the causative pathogen. For norovirus detection, the regions encoding RdRp and VP1 were amplified by one-step reverse transcription (RT)-PCR (Figure 1). The primers used for the VP1 region were GIIF1: 5’-GGHCCMBMDTTYTACAGCAA-3’; GIIF2: 5’-GGHCCMBMDTTYTACAAGAA-3’; GIIF3: 5’-GGHCCMBMDTTYTACARNAA-3’; and GIIR: 5’-CCRCCNGCATRHCCRTTRTACAT-3’. The estimated amplicon size was 468 bp. The primers used for the RdRp region were GF: 5’-TCATCATCACCATAGAAIGAG-3’ and GR: 5’-ATACCACTATGATGCAGAYTA-3’. The estimated amplicon size for RdRp was 327 bp.
The reaction mix contained 1 μL of RNA, 2.5 μL of 10 μmol/L primer solution, 21.5 μL of H2O, and 25 μL of 2× One-Step Fast RT-PCR Mix (BiovisuaLab, Shanghai, China). PCR was performed using a Robocycler thermal cycler (Stratagene, La Jolla, CA, United States) with the following cycling parameters: RT at 42 °C for 0.5 h, followed by 35 cycles of denaturation at 95 °C for 35 s, annealing at 60 °C for 30 s, and elongation at 70 °C for 30 s.
In positive samples, nucleotide sequencing was performed directly on the purified RT-PCR products using internal primers and an ABI PRISM BigDye Terminator Cycle Sequencing Kit (Applied Biosystems of Thermo Fisher Scientific, Waltham, MA, United States) in an ABI PRISM 3130XL DNA Analyzer (Applied Biosystems).
The sequences of the RdRp and VP1 regions were input into the Basic Local Alignment Search Tool (BLAST, http://blast.ncbi.nlm.nih.gov/Blast.cgi), and the genogroup and genotype definition, as well as the possibility of recombination, were decided according to the “Sequences producing significant alignments of 100 Blast Hits on the Query Sequence.” A distance tree of the “100 Blast Hits on the Query Sequence” was drawn using the “Fast Minimum Evolution” method.
On December 10, 2013, the first case of acute gastroenteritis emerged in a Class 1/Grade 4 of 45 students (9-10 years of age) in an elementary school in Shanghai. The case was hospitalized for diarrhea and was reported to the local Center for Disease Control and Prevention. On the second day, December 11, 2013, the number of cases of acute gastroenteritis in the same class increased to 22; among the 21 new cases, 8 visited a hospital and 4 had a fever (mean body temperature: 39.0 ± 0.4 °C). On the third day, December 12, 2013, the number of new cases decreased to 9 (Table 1). No new case emerged on December 13, 2013. The last case emerged on December 14, 2013. The main symptoms of gastroenteritis were vomiting, diarrhea, abdominal pain, and fever; of the 32 cases, the overall frequencies of vomiting, diarrhea, abdominal pain, and fever were 100%, 56.3%, 43.8%, and 15.6%, respectively. Among the ten cases who visited a hospital, five underwent a routine blood examination; the average white blood cell count was 17.2 × 109/L ± 2.3 × 109/L. Hospitalization was restricted to outpatient treatment, including routine blood tests and an intravenous infusion with saline and glucose.
| Characteristic | Day | |||
| 10-Dec-13 | 11-Dec-13 | 12-Dec-13 | 14-Dec-13 | |
| Cases | 1 | 21 | 9 | 1 |
| Symptom | ||||
| Vomiting | 1 | 21 | 9 | 1 |
| Diarrhea | 1 | 12 | 5 | 0 |
| Abdominal pain | 1 | 11 | 2 | 0 |
| Fever | 0 | 4 | 1 | 0 |
| Hospital visit | 1 | 8 | 1 | 0 |
On the second day (December 12, 2013) of the outbreak, specialists from the Center for Disease Control and Prevention of Shanghai’s Minhang District performed a field epidemiologic investigation. The school was located in southwest Shanghai in an area with both urban and rural areas; Class 1/Grade 4 was located at the west corner on the fourth floor of a five-story building. There were three classes on the same floor, with 45 students in each class. Extracurricular activities among classes at the school are rare; the only sites for cross-class interaction are the lavatories, of which one is for male students and the other is for female students. All of the students at the school eat lunch in school. The lunches provided to the elementary school students are tightly controlled by the local department of public health. Based on evidence showing that the gastroenteritis outbreak was limited to Class 1/Grade 4, the possibility that the gastroenteritis outbreak was caused by having lunch in school was excluded. All students in the school drink purified water from a bottled-water supplier located in each classroom. The water was sampled for pathogen detection. A questionnaire survey for diarrheal diseases showed that the first case had a history of ingesting possibly contaminated food from a delicatessen. A field epidemiologic investigation was then performed in the delicatessen. None of the foods were kept in a refrigerated space. There were four employees in the shop. Fecal and food samples were collected for pathogen detection. Any sporadic gastroenteritis case related to the delicatessen would be difficult to identify owing to the high population mobility in the area.
Based on routine testing for diarrheal pathogens, the season, and the course of the disease and its transmission features, the pathogen was initially suspected to be a gastroenteritis virus, including a norovirus. To confirm this speculation, two sets of norovirus-specific primers were used for PCR-based detection. One of the primer sets, located in region C (Figure 1), targeted the VP1 region of all noroviruses belonging to GII. The other primer set, which covered region B (Figure 1), targeted the RdRp region of all norovirus genogroups. In total, 26/32 students with acute gastroenteritis and 2/4 employees of the delicatessen tested positive for norovirus RNA in their feces. No norovirus RNA was detected in the other samples, including the food samples from the delicatessen and water samples.
Rotavirus was not detected in any of the samples. Testing for C. jejuni, E. coli, Salmonella, Shigella, and Campylobacter spp produced a low-positive rate.
Together, the laboratory data suggested that the pathogen responsible for this acute gastroenteritis outbreak was a GII norovirus.
As described in the introduction, recombination occurs occasionally between two noroviruses at the ORF1/ORF2 junction (Figure 1); thus, in order to define a field norovirus strain, determination of both the polymerase and capsid genotypes is necessary. The regions currently used for norovirus definition are located within RdRp and VP1. To identify the detected norovirus, the PCR fragments amplified from the 26 students and 2 employees for both RdRp and VP1 were sequenced. The sequences were spliced and aligned using Sequencher 4.9. The consensus sequences for each subject at each site were used for homology comparisons. The sequences derived from all 28 subjects displayed complete homology in the RdRp and VP1 regions, suggesting that the subjects were infected by the same norovirus strain, and that the norovirus was transmitted from the employees at the delicatessen to the first case student and then to the rest of the affected students.
The consensus sequences for RdRp and VP1 were input into the BLAST. The “Sequences producing significant alignments of 100 Blast Hits on the Query Sequence” showed 92%-99% homology with the RdRp and VP1 sequences reported for GII.6, suggesting that the strain was a GII.6 norovirus, and that no recombination had occurred.
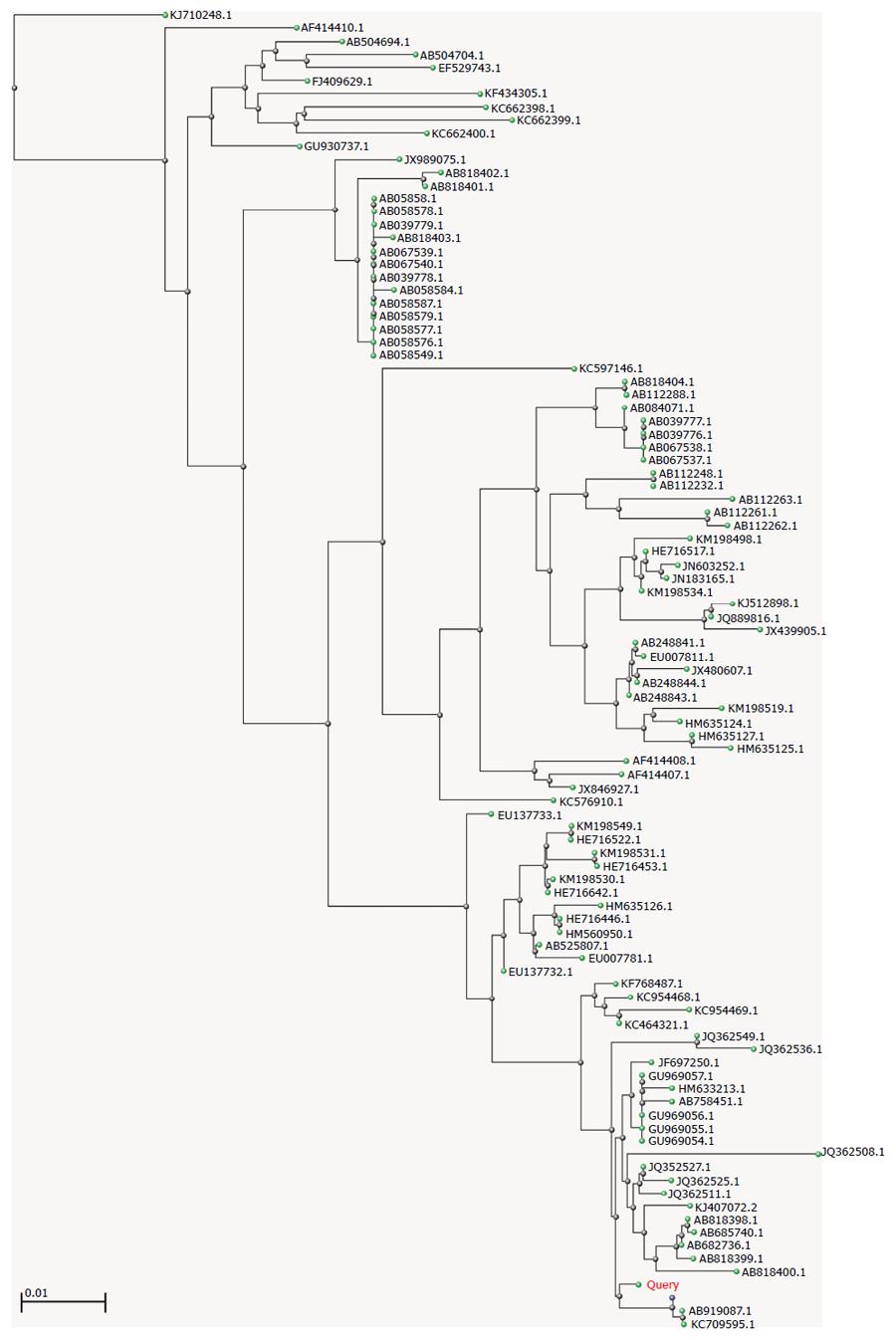
To study the genetic relatedness among GII.6 noroviruses, the consensus sequence was input into the BLAST and a distance tree of the “100 Blast Hits on the Query Sequence” was drawn using the “Fast Minimum Evolution” method. The lowest homology among the 100 hits was 92%, suggesting that they belonged to the same genotype. As shown in Figure 2, GII.6 strains are widely distributed in Japan, Vietnam, Korea, Ireland, Sweden, and the United States. Interestingly, in addition to humans, viral RNA could be isolated from oysters and effluent (e.g., JQ362549, JQ362508.1, JQ362536.1, KC954469.1, and KC954468.1) (Figure 2). The closest strains were AB919087.1 (a human isolate detected from 2013 to 2014 in Okinawa, Japan), KC709595.1 (an isolate from an outpatient in Beijing, China), and AB818397.1 (a human isolate detected in 2009 in Ehime, Japan), suggesting that, although outbreaks of gastroenteritis caused by GII.6 are rare or are rarely reported, closely related GII.6 strains have actively circulated in these areas for a certain period of time.
In this study, we investigated a gastroenteritis outbreak that occurred in an elementary school in Shanghai, China, in 2013. Molecular epidemiologic data showed that the outbreak was caused by a GII.6 norovirus strain. As far as we know, GII.6 is a rare cause of gastroenteritis outbreaks; the existing literature indicates that GII.4 is the most prevalent norovirus genotype in the world[14,17]. A report from Japan showed that GII.6, the second most prevalent strain after GII.4, exclusively caused sporadic gastroenteritis in Shizuoka, Japan[18]. The closest strain from Japan to the strain reported in this study was isolated from Okinawa, not Shizuoka. These findings suggest that GII.6 noroviruses have spread across Japan for a long period of time. In the NCBI database, only one strain (KC709595.1) reported in 2013 from Beijing, China, belonged to GII.6[23]. Although strain KC709595.1 showed strong homology with the strain in this study, 84.6% of the identified noroviruses in that report were GII.4, while only 1/26 sporadic norovirus cases was related to GII.6. In China, molecular epidemiologic surveillance of viral diseases has been carried out for < 20 years. The resulting lack of lengthwise data have made it difficult to trail viral evolution, and the lack of transverse data have made it difficult to assess viral transmission[24-26]. Although we identified a GII.6 strain in this study, we are not certain whether it is indigenous or imported. Even so, this is the first report to show that norovirus GII.6 can cause a gastroenteritis outbreak; thus, the prevalence of GII.6 noroviruses requires attention.
Noroviruses are recognized as the leading cause of acute viral gastroenteritis worldwide, and the outbreaks always occur in an enclosed environment, such as school, hospital ward, and even cruise ships[1,27]. In the NCBI database, almost one-third of the GII.6 norovirus sequences are derived from oysters and effluent, suggesting that the source and route of norovirus transmission might be mussels and contaminated water[28]. This makes it difficult for norovirus infection control and prevention, especially in settings with a high population density. In China, due to the high population density and weak hygienic conditions, outbreaks or epidemics of infectious diseases are common[26]. This might be why GII.6 causes only sporadic gastroenteritis in Japan, but can produce outbreaks in China.
In developing countries, noroviruses are estimated to cause more than 0.2 million deaths annually among children younger than five years of age, and noroviruses are predicted to become the predominant cause of diarrhea in all age groups worldwide once rotavirus infection is controlled through vaccination[19]. In China, although the vaccine for rotavirus has not been widely applied, the situation for many viral diseases transmitted by the fecal-oral route, including those caused by enteroviruses[29], rotaviruses[30], and noroviruses[31], is severe. Thus, surveillance for norovirus infections is important for the control and prevention of these viral diseases. In this outbreak, fourteen students in the same class succumbed to a norovirus infection. Evidence indicates that individuals can be infected by noroviruses repeatedly; thus, pre-existing immunity did not protect these students from infection. Also, although norovirus RNA was detected in the fecal samples of the two delicatessen employees, the two adults showed no symptoms of gastroenteritis before, during, or after the outbreak. The above phenomenon might be explained by individual resistance to noroviruses or an inapparent infection.
The authors would like to thank all staff in the Center for Disease Control and Prevention of Minghang District, for their technical support.
Noroviruses are responsible for approximately half of all reported gastroenteritis outbreaks in many countries.
Genogroup 2 genotype 4 (GII.4) strains of noroviruses are the most prevalent. Rare outbreaks caused by genotype 6 (GII.6) strains have been reported.
This is the first report to show that norovirus strain GII.6 can cause a gastroenteritis outbreak.
The prevalence and perniciousness of GII.6 noroviruses requires attention.
This article presents the first report of a norovirus outbreak due to a GII.6 strain of norovirus. Norovirus-caused gastroenteritis is a disease of high burden and is the leading cause of medically attended childhood gastroenteritis in areas where rotavirus vaccine uptake is high. Knowledge of noroviruses, including molecular epidemiology, is the key to the prevention for this disease. This paper discusses an important issue and is well written.
P- Reviewer: Gualano MR, Takeuchi M S- Editor: Ma YJ L- Editor: AmEditor E- Editor: Wang CH
| 1. | Division of Viral Diseases, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention. Updated norovirus outbreak management and disease prevention guidelines. MMWR Recomm Rep. 2011;60:1-18. [PubMed] |
| 2. | Stock I. [Norovirus infections]. Med Monatsschr Pharm. 2007;30:362-370; quiz 371-372. [PubMed] |
| 3. | MacCannell T, Umscheid CA, Agarwal RK, Lee I, Kuntz G, Stevenson KB; Healthcare Infection Control Practices Advisory Committee-HICPAC. Guideline for the prevention and control of norovirus gastroenteritis outbreaks in healthcare settings. Infect Control Hosp Epidemiol. 2011;32:939-969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 109] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 4. | Kirby A, Iturriza-Gómara M. Norovirus diagnostics: options, applications and interpretations. Expert Rev Anti Infect Ther. 2012;10:423-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 5. | Kanda A. [Norovirus gastroenteritis in adult]. Nihon Rinsho. 2012;70:1371-1375. [PubMed] |
| 6. | Vega E, Barclay L, Gregoricus N, Shirley SH, Lee D, Vinjé J. Genotypic and epidemiologic trends of norovirus outbreaks in the United States, 2009 to 2013. J Clin Microbiol. 2014;52:147-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 245] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 7. | Clarke IN, Lambden PR. Organization and expression of calicivirus genes. J Infect Dis. 2000;181 Suppl 2:S309-S316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 121] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 8. | Bok K, Green KY. Norovirus gastroenteritis in immunocompromised patients. N Engl J Med. 2012;367:2126-2132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 263] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 9. | Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. Norovirus classification and proposed strain nomenclature. Virology. 2006;346:312-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 864] [Cited by in RCA: 812] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 10. | Dingle KE, Lambden PR, Caul EO, Clarke IN. Human enteric Caliciviridae: the complete genome sequence and expression of virus-like particles from a genetic group II small round structured virus. J Gen Virol. 1995;76:2349-2355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 116] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 11. | Green SM, Dingle KE, Lambden PR, Caul EO, Ashley CR, Clarke IN. Human enteric Caliciviridae: a new prevalent small round-structured virus group defined by RNA-dependent RNA polymerase and capsid diversity. J Gen Virol. 1994;75:1883-1888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 88] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Vinjé J, Hamidjaja RA, Sobsey MD. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J Virol Methods. 2004;116:109-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 271] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 13. | Kageyama T, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Kojima S, Takai R, Oka T, Takeda N, Katayama K. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to Norovirus in Japan. J Clin Microbiol. 2004;42:2988-2995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 299] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 14. | Hoa Tran TN, Trainor E, Nakagomi T, Cunliffe NA, Nakagomi O. Molecular epidemiology of noroviruses associated with acute sporadic gastroenteritis in children: global distribution of genogroups, genotypes and GII.4 variants. J Clin Virol. 2013;56:185-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 219] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 15. | Buesa J, Collado B, López-Andújar P, Abu-Mallouh R, Rodríguez Díaz J, García Díaz A, Prat J, Guix S, Llovet T, Prats G. Molecular epidemiology of caliciviruses causing outbreaks and sporadic cases of acute gastroenteritis in Spain. J Clin Microbiol. 2002;40:2854-2859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 144] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 16. | Bull RA, Tanaka MM, White PA. Norovirus recombination. J Gen Virol. 2007;88:3347-3359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 268] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 17. | Desai R, Hembree CD, Handel A, Matthews JE, Dickey BW, McDonald S, Hall AJ, Parashar UD, Leon JS, Lopman B. Severe outcomes are associated with genogroup 2 genotype 4 norovirus outbreaks: a systematic literature review. Clin Infect Dis. 2012;55:189-193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 132] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 18. | Chan-It W, Thongprachum A, Khamrin P, Kobayashi M, Okitsu S, Mizuguchi M, Ushijima H. Emergence of a new norovirus GII.6 variant in Japan, 2008-2009. J Med Virol. 2012;84:1089-1096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 19. | Payne DC, Vinjé J, Szilagyi PG, Edwards KM, Staat MA, Weinberg GA, Hall CB, Chappell J, Bernstein DI, Curns AT. Norovirus and medically attended gastroenteritis in U.S. children. N Engl J Med. 2013;368:1121-1130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 469] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 20. | Jothikumar N, Kang G, Hill VR. Broadly reactive TaqMan assay for real-time RT-PCR detection of rotavirus in clinical and environmental samples. JIN2@cdc.gov. J Virol Methods. 2009;155:126-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 90] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 21. | Zhang Y, Zhao Y, Ding K, Wang X, Chen X, Liu Y, Chen Y. Analysis of bacterial pathogens causing acute diarrhea on the basis of sentinel surveillance in Shanghai, China, 2006-2011. Jpn J Infect Dis. 2014;67:264-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 22. | Wang XG, Zhang YH, Wang P, Chen XH, Luo LF, Liu Y, Liu JQ, Song CP, Ou YL, Chen GQ. [Establishment and application of multiplex PCR for non-O157 H7 STEC virulence genes detection]. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2013;27:388-391. [PubMed] |
| 23. | Mai H, Jin M, Guo X, Liu J, Liu N, Cong X, Gao Y, Wei L. Clinical and epidemiologic characteristics of norovirus GII.4 Sydney during winter 2012-13 in Beijing, China following its global emergence. PLoS One. 2013;8:e71483. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Wang W, Peng H, Tao Q, Zhao X, Tang H, Tang Z, Wang Y, Wang Y, Zhao P, Qi Z. Serologic assay for avian-origin influenza A (H7N9) virus in adults of Shanghai, Guangzhou and Yunnan, China. J Clin Virol. 2014;60:305-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 25. | Liu X, Liu Y, Zhang Y, Chen Z, Tang Z, Xu Q, Wang Y, Zhao P, Qi Z. Pre-existing immunity with high neutralizing activity to 2009 pandemic H1N1 influenza virus in Shanghai population. PLoS One. 2013;8:e58810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 26. | Lu J, Zhou Y, Lin X, Jiang Y, Tian R, Zhang Y, Wu J, Zhang F, Zhang Y, Wang Y. General epidemiological parameters of viral hepatitis A, B, C, and E in six regions of China: a cross-sectional study in 2007. PLoS One. 2009;4:e8467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 87] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 27. | Bert F, Scaioli G, Gualano MR, Passi S, Specchia ML, Cadeddu C, Viglianchino C, Siliquini R. Norovirus outbreaks on commercial cruise ships: a systematic review and new targets for the public health agenda. Food Environ Virol. 2014;6:67-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 28. | Rajko-Nenow P, Waters A, Keaveney S, Flannery J, Tuite G, Coughlan S, O’Flaherty V, Doré W. Norovirus genotypes present in oysters and in effluent from a wastewater treatment plant during the seasonal peak of infections in Ireland in 2010. Appl Environ Microbiol. 2013;79:2578-2587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 29. | Xu J, Qian Y, Wang S, Serrano JM, Li W, Huang Z, Lu S. EV71: an emerging infectious disease vaccine target in the Far East? Vaccine. 2010;28:3516-3521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 107] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 30. | Kawai K, O’Brien MA, Goveia MG, Mast TC, El Khoury AC. Burden of rotavirus gastroenteritis and distribution of rotavirus strains in Asia: a systematic review. Vaccine. 2012;30:1244-1254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 141] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 31. | Tian G, Jin M, Li H, Li Q, Wang J, Duan ZJ. Clinical characteristics and genetic diversity of noroviruses in adults with acute gastroenteritis in Beijing, China in 2008-2009. J Med Virol. 2014;86:1235-1242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |