Published online May 7, 2015. doi: 10.3748/wjg.v21.i17.5250
Peer-review started: November 3, 2014
First decision: November 26, 2014
Revised: December 22, 2014
Accepted: February 11, 2015
Article in press: February 11, 2015
Published online: May 7, 2015
Processing time: 193 Days and 17.7 Hours
AIM: To investigate the role of autophagy in the anti-apoptotic effect of augmenter of liver regeneration (ALR).
METHODS: Autophagy was induced through serum deprivation. An ALR-expressing plasmid was transfected into HepG2 cells, and autophagic flux was determined using fluorescence microscopy, electron microscopy, Western blot and quantitative polymerase chain reaction (qPCR) assays. After ALR-expressing plasmid transfection, an autophagy inhibitor [3-methyladenine (3-MA)] was added to HepG2 cells, and apoptosis was observed using fluorescence microscopy and flow cytometry.
RESULTS: Autophagy was activated in HepG2 cells, peaking at 24 h after serum deprivation. Microtubule-associated protein light chain three-II levels were higher in HepG2 cells treated with ALR than in control cells, fluorescence microscopy, electron microscopy and qPCR studies showed the similar trend, and p62 levels showed the opposite trend, which indicated that ALR may play an important role in increasing autophagy flux. The numbers of apoptotic cells were substantially higher in HepG2 cells treated with both ALR and 3-MA than in cells treated with ALR alone. Therefore, the protective effect of ALR was significantly attenuated or abolished when autophagy was inhibited, indicating that the anti-apoptotic effect of ALR may be related to autophagy.
CONCLUSION: ALR protects cells from apoptosis partly through increased autophagy in HepG2 cells and may be valuable as a new therapeutic treatment for liver disease.
Core tip: Recent studies have found that augmenter of liver regeneration (ALR) plays an important role in suppressing hepatocyte apoptosis, although the mechanism remains unknown. Our study shows that ALR plays an important role in increasing the level of autophagy in HepG2 cells. The protective effect of ALR was significantly attenuated or abolished when autophagy was inhibited, indicating that the anti-apoptotic effect of ALR may be related to autophagy. Hence, we conclude that ALR protects cells against apoptosis partly by increasing autophagic activity in HepG2 cells and may be valuable for developing new therapeutic treatments for liver disease.
- Citation: Shi HB, Sun HQ, Shi HL, Ren F, Chen Y, Chen DX, Lou JL, Duan ZP. Autophagy in anti-apoptotic effect of augmenter of liver regeneration in HepG2 cells. World J Gastroenterol 2015; 21(17): 5250-5258
- URL: https://www.wjgnet.com/1007-9327/full/v21/i17/5250.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i17.5250
In 1975, LaBrecque et al[1] discovered hepatic stimulator substance (HSS), which specifically stimulates hepatocyte DNA synthesis, in livers regenerating from partial hepatectomy and in the cytosol of neonatal rat livers. In 1994, Hagiya et al[2] cloned the cDNA of HSS, clarified its primary structure and renamed it augmenter of liver regeneration (ALR). Recent studies found that the ALR protein is a flavin-containing sulfhydryl oxidase that localizes to the mitochondrial intermembrane space and plays an important role in suppressing hepatocyte apoptosis[3-5]. Polimeno et al[3] demonstrated that ALR can control apoptosis in regenerating livers in rats treated with ALR 72 h after a partial hepatectomy. However, the mechanism underlying the anti-apoptotic or protective effect of ALR remains unknown.
Multiple lines of direct and indirect evidence suggest a mechanistic overlap and interaction between the apoptotic machinery and autophagic proteins[6,7], which can be mutually regulated by nuclear factor (NF)-κB, c-Jun N-terminal kinase, p62, Beclin-1, Bcl-2, Caspase, and p53 signaling[8,9]. For example, Mei et al[10] found that tumor necrosis factor (TNF)-α/ActD can inhibit autophagy through the Caspase-3-mediated cleavage of Beclin-1, which produces a truncated protein that is unable to promote autophagy. In addition, when the Caspase inhibitor Z-VAD-FMK was used to inhibit TNF-α/ActD-induced apoptosis, autophagy increased significantly due to the inhibition of Beclin-1 cleavage. Thus, we hypothesized that autophagy may play an important role in the anti-apoptotic effect of ALR.
To test our hypothesis, we determined the effect of ALR on autophagy by transfecting an ALR-expressing plasmid into HepG2 cells. We then monitored hepatocyte apoptosis in HepG2 cells treated with an ALR-expressing plasmid and an autophagy inhibitor to uncover the role of autophagy in the anti-apoptotic effect of ALR. This study provides new insight into mechanisms underlying the protective effect of ALR in the liver, which may be valuable for developing new therapeutic treatments for liver disease.
HepG2 cells, a human hepatoma cell line (Xiangfu Biological Company, Shanghai), were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum in a 37 °C incubator with 5% CO2. All culture reagents were purchased from Hyclone. Autophagy was observed at different time points (0, 12, 24, and 48 h) after starvation caused by serum deprivation. For 3-methyladenine (3-MA, Sigma) treatment, 3-MA (final concentration, 750 ng/mL) was added to the medium when the medium was changed to serum-free medium.
The GFP-LC3B[11] and ALR-expressing plasmids[5] (1ug) were transfected into HepG2 cells using the X-tremeGENE HP DNA transfection reagent (Roche) according to the manufacturer’s instructions. Cells were then cultured in DMEM for 6 h, followed by treatment with serum-free medium to induce autophagy.
For fluorescence microscopy, cells were cultured in 24-well plates with a microscope glass coverslip. After the designated treatments, the cells were fixed with 4% paraformaldehyde in PBS. All images were obtained on an inverted fluorescence microscope (Nikon Eclipse E800). To quantify autophagic cells, GFP-LC3 puncta were determined in triplicate by counting more than 30 cells[10]. Apoptotic cells were visualized by staining with Annexin V-FITC and propidium iodide (KeyGEN BioTECH).
HepG2 cells were fixed with 2.5% glutaraldehyde in 0.1 mol/L phosphate buffer (pH 7.4). After dehydration, thin sections were cut and stained with uranyl acetate and lead citrate. Digital images were obtained using an electron microscope (H-7650, Japan). The numbers of typical autophagosomes and autolysosomes from each cell section were counted randomly in more than 30 cells[10].
Cells were washed in PBS, and cell pellets were lysed with RIPA buffer supplemented with phosphatase inhibitors and protease inhibitors. Total proteins (50 μg) were separated via 12% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were incubated overnight with rabbit anti-LC3B and anti-p62 antibodies (Sigma) at 4 °C. Then, the membranes were treated with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Cell Signaling) and developed with a chemiluminescent substrate (Pierce). Densitometry analysis was performed using ImageJ software, and the relative levels of microtubule-associated protein light chain three (LC3)-II in each group were normalized to the loading control.
After the designated treatments, the cells were digested with EDTA-free trypsin. The single-cell solution was washed with cold PBS, and approximately 105 cells were collected. Binding buffer was added to suspend the cells, which were then stained with Annexin V-FITC and propidium iodide (KeyGEN BioTECH) at 4 °C for 15 min. The cells were analyzed using flow cytometry (BD FACS Calibur, United States).
Total RNA was extracted using a Trizol kit (Invitrogen, United States) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 5 μg RNA (Superscript III cDNA Synthesis Kit, Invitrogen), and ATG5, ATG7, Beclin-1 and GADPH mRNA levels were estimated by quantitative polymerase chain reaction (qPCR) using the SYBR Green PCR Kit (Invitrogen) on a real-time PCR system (ABI PRISM7300, United States). The relative quantity of the cycle threshold value in each primer was normalized to an internal primer.
All data are expressed as the mean ± SD. Independent-sample t-tests were performed to compare differences between two groups. One-way analysis of variance (ANOVA) followed by the post hoc LSD test was used to compare differences among multiple groups. P < 0.05 was considered significant. All data were analyzed using SPSS 11.5 software.
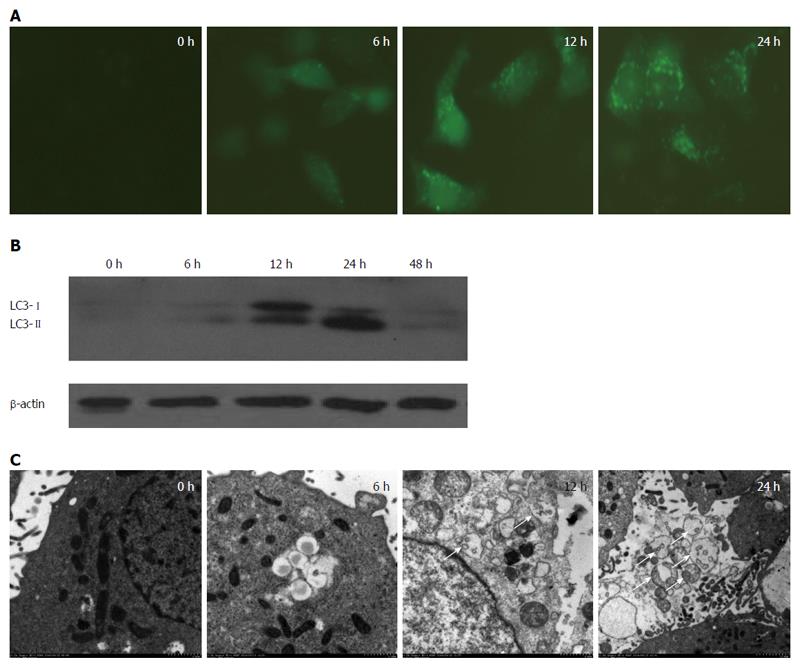
We first assessed the induction of autophagy in HepG2 cells using a classic method of inducing autophagy, which is to starve cells via serum deprivation[12]. To assess autophagy, a GFP-LC3 plasmid was transfected into HepG2 cells. GFP-LC3 behaves similarly to endogenous LC3 and has been widely used to monitor autophagy[13]. Upon autophagy induction, GFP-LC3 translocates to autophagosome membranes, displaying punctate green fluorescence after conjugation with phosphatidylethanolamine[14]. The number of green puncta can then be quantified as an indication of the number of autophagosomes and autolysosomes.
After transfection, HepG2 cells displayed an increase in the number of green puncta beginning at 6 h and peaking at 24 h after serum deprivation (Figure 1A). To verify this result, we monitored the changes in the levels of LC3. The transformation of LC3 from type I to II is generally regarded as a marker of autophagosome formation[15]. LC3-II expression also reached its highest point after 24 h (Figure 1B). To further confirm the induction of autophagy, we performed electron microscopy studies of the HepG2 cells. Serum deprivation significantly increased the number of double-membrane autophagosomes in HepG2 cells, especially after 24 h, and most of these had enveloped cytosolic contents (Figure 1C, arrows). Thus, autophagy was successfully activated in HepG2 cells after serum deprivation.
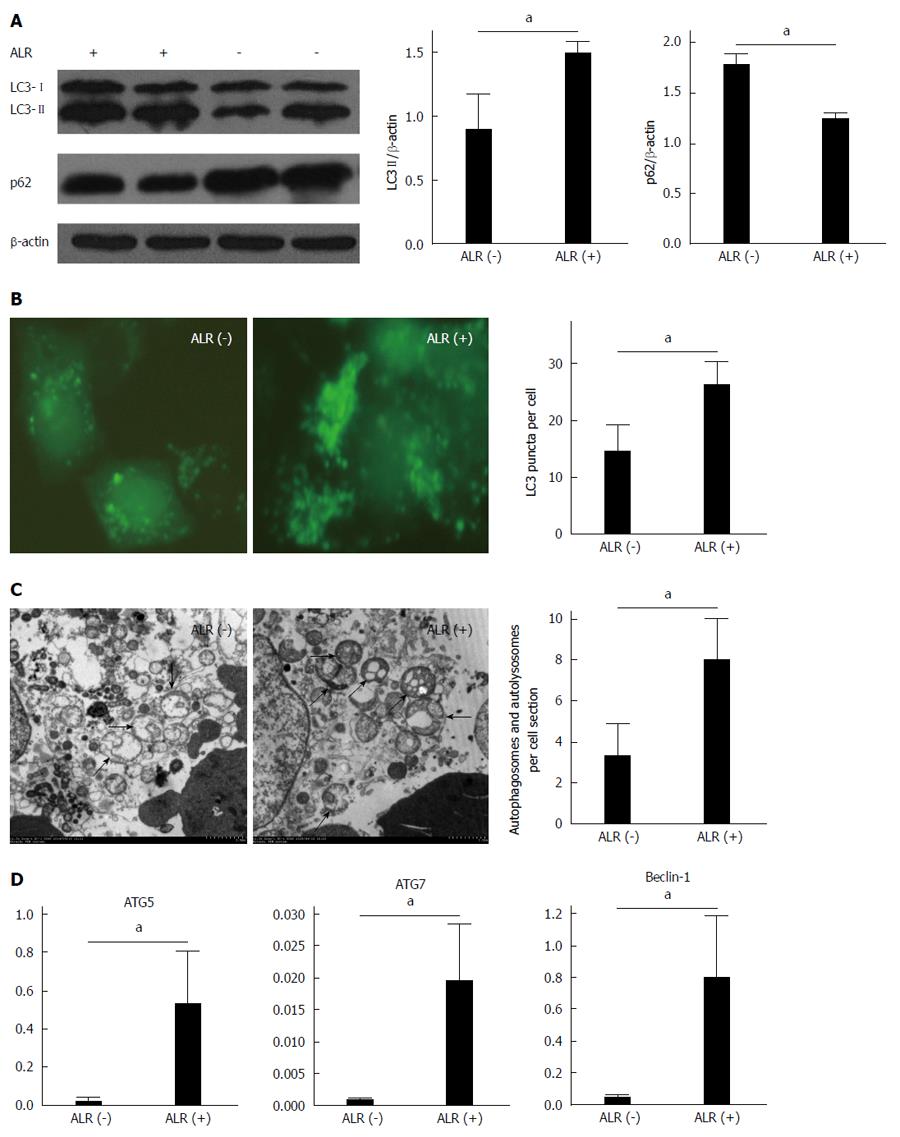
HepG2 cells were transfected with an ALR-expressing plasmid to observe the effect of ALR on autophagy. In the LC3 turnover assay, LC3-II levels were substantially higher in ALR-treated cells than in control cells (Figure 2A, P < 0.05). To further confirm this result, GFP-LC3 and ALR-expressing plasmids were cotransfected into HepG2 cells. After a 24-h starvation, we observed a greater number of green puncta in HepG2 cells treated with ALR than in control cells (Figure 2B, P < 0.05). Electron microscopy also revealed a similar trend as the fluorescence microscopy studies, namely, the number of autophagosomes in HepG2 cells treated with ALR was higher than that in control cells (Figure 2C, P < 0.05). However, LC3-II levels indicate only upstream level of autophagic processes, and it is important to distinguish whether autophagosome accumulation is due to the induction of autophagy or the inhibition of downstream steps, for instance, reduced fusion of autophagosomes with lysosomes. p62, which is a lysosome substrate, is degraded when autophagosomes fuse with lysosomes. In our study, p62 levels were lower in HepG2 cells treated with ALR than in control cells (Figure 2A, P < 0.05), suggesting that ALR treatment did not block the fusion of autophagosomes with lysosomes. Taken together, these results indicate that ALR increases autophagic flux in HepG2 cells 24 h after serum deprivation.
Of the autophagy-related genes (ATGs) that are known to regulate autophagy, ATG6/Beclin-1 plays a critical role in regulating the initial nucleation of autophagic vesicles, whereas ATG5 and ATG7 are involved in the extension and closure of autophagic vesicles[16]. The levels of ATG5, ATG7 and Beclin-1 mRNAs in HepG2 cells treated with the ALR-expressing plasmid were higher than that in control cells (Figure 2D, P < 0.05), indicating that ALR plays an important role in increasing overall autophagic activity.
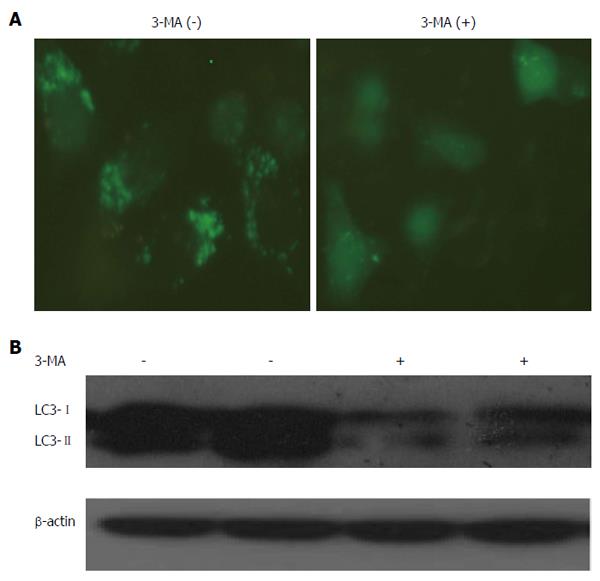
The class-III PI3 kinase inhibitor 3-MA is widely used to inhibit autophagic formation because the Beclin-1/class-III PI3 kinase complex is important for the regulation of autophagic vesicle nucleation[17]. In our study, we observed fewer green puncta in HepG2 cells treated with 3-MA than in control cells (Figure 3A). In the LC3 turnover assay, LC3-II levels were significantly decreased upon 3-MA treatment (Figure 3B). These results suggested that 3-MA was able to suppress autophagic activity in HepG2 cells.
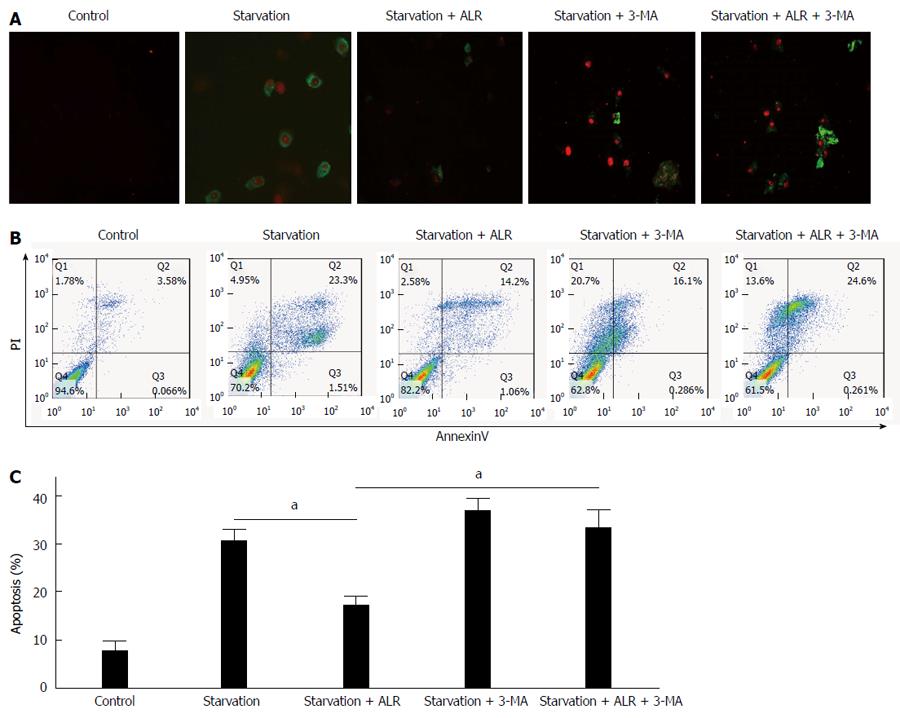
We monitored apoptosis in HepG2 cells using Annexin V-FITC and propidium iodide, which can differentiate between early- (green) and late-stage (red) apoptosis. Most apoptotic cells were found to be in the early stage in HepG2 cells treated by starvation only, whereas most were in the late stage or were necrotic when treated with 3-MA (Figure 4). 3-MA treatment significantly increased the number of late-stage apoptotic cells, indicating that inhibiting autophagy promoted apoptosis in HepG2 cells.
Similar to the results from other studies[18-20], fewer apoptotic cells were observed when HepG2 cells were treated both by starvation and with ALR compared with cells treated by starvation only, suggesting that ALR plays an important role in protecting HepG2 cells from apoptosis (Figure 4). The number of apoptotic cells was also substantially higher in HepG2 cells treated with both ALR and 3-MA than in cells treated with ALR alone, indicating that the protective effect of ALR was significantly attenuated or completely abolished when autophagy was inhibited. Hence, the anti-apoptotic effect of ALR may be related to autophagy (Figure 4). ALR was therefore able to protect cells against apoptosis partly via increased autophagic activity in HepG2 cells.
Autophagy is defined as self-eating, which is a self-degradation process that depends on lysosomes in eukaryotes. The first step in autophagy involves the formation of lipid bilayer structures that engulf cellular macromolecules and organelles to form autophagosomes. These autophagosomes then fuse with lysosomes to form mature autolysosomes, in which, the sequestered components are digested to provide the cell with molecular building blocks and energy[21]. Autophagy functions as a mechanism to survive cellular stresses such as nutrient deprivation, which plays an important role in cell self-protection.
Increasing evidence shows that autophagy exhibits a protective effect in hepatocytes[22-24]. Ni et al[23] determined the cell viability of primary mouse hepatocytes treated with various concentrations of APAP in the presence of 3-MA or CQ and found that the induction of autophagy protects against APAP-induced necrosis, whereas the inhibition of autophagy further exacerbates it. Ding et al[24] also reached a similar conclusion using an ethanol-induced model, wherein the suppression of macroautophagy using CQ or siRNAs significantly increased hepatocyte apoptosis; thus, macroautophagy protected cells from the toxic effects of ethanol.
Based upon this premise, we explored the hypothesis that autophagy is involved in the anti-apoptotic effect of ALR in hepatocytes. We found that starvation induced the activation of autophagy in HepG2 cells, which peaked at 24 h after serum deprivation. Li et al[25] reported that peak ALR expression in hepatocytes occurred 24-36 h after plasmid transfection. Hence, we chose to transfect the ALR-expressing plasmid into HepG2 cells approximately 30 h before observing the effects of ALR on autophagy.
We found that ALR significantly increased autophagic flux in HepG2 cells after serum deprivation. Although very few studies similar to ours have been reported, some studies of liver regeneration have provided indirect confirmation of our results. Toshima et al[26] found that hepatocyte growth factor (HGF) increased autophagic activity in primary hepatocytes and that autophagy was activated in the regenerating liver after PHx. It is known that ALR levels in the serum and liver increase quickly in a 70% PHx mouse model[27], consistent with the trend of autophagic flux, thereby indicating a potential relationship between ALR levels and autophagic expression. As growth factors, ALR and HGF play similar roles in promoting cell proliferation via the MAPK and ERK pathways; therefore, ALR may also play a role similar to HGF in regulating autophagy.
Similar to other studies, we found that inhibiting autophagy promoted apoptosis in HepG2 cells and that ALR played an important role in protecting HepG2 cells against apoptosis. In addition, we found that the anti-apoptotic effect of ALR may be related to autophagy, indicating that ALR may play a protective role partially by increasing autophagic activity in HepG2 cells. However, the mechanism or pathway by which ALR modulates autophagy is still unknown. One possibility is that ALR promotes autophagy by inhibiting the cleavage of Beclin-1, which is mediated by the Caspase family. In fact, ALR has been suggested to inhibit Caspase-3 activation by promoting ATP synthesis and reducing the release of cytochrome c[19]. Elucidating these mechanisms will contribute to our understanding of liver recovery and regeneration, which holds promise for the treatment of liver diseases in the future.
Recent studies found that augmenter of liver regeneration (ALR) plays an important role in suppressing hepatocyte apoptosis, but the mechanism is still unknown. Increasing evidence suggests that autophagy has a protective effect in the liver. There may also be a mechanistic overlap between the apoptotic machinery and autophagy proteins. Thus, the present study hypothesized that autophagy may play an important role in the anti-apoptotic effect of ALR.
Direct and indirect evidence suggests a mechanistic overlap and interaction between the apoptotic machinery and autophagy proteins, which are mutually regulated through nuclear factor-κB, c-Jun N-terminal kinase, p62, Beclin-1, Bcl-2, Caspase, and p53. For example, tumor necrosis factor (TNF)-α/ActD can inhibit autophagy through the Caspase-3-mediated cleavage of Beclin-1, which produces a truncated protein that is unable to promote autophagy. In addition, when the Caspase inhibitor Z-VAD-FMK was used to inhibit TNF-α/ActD-induced apoptosis, autophagy increased significantly.
Recent studies have shown that ALR plays an important role in suppressing hepatocyte apoptosis, though the mechanism remains unknown. This study shows that ALR significantly increases the level of autophagy in HepG2 cells following serum deprivation and that the anti-apoptotic effect of ALR can be significantly attenuated or abolished when autophagy was inhibited. Thus, ALR is able to protect cells against apoptosis partly by increasing autophagic activity in HepG2 cells.
The results of this study highlight a novel mechanism for the anti-apoptotic effect of ALR, which may be valuable for developing new therapeutic treatments for liver disease in the future.
Autophagy is defined as self-eating, which is a self-degradation process that is dependent on lysosomes in eukaryotes. The first step of autophagy involves the formation of lipid bilayer structures that engulf cellular macromolecules and organelles to form autophagosomes. These autophagosomes then fuse with lysosomes to form mature autolysosomes, wherein the sequestered components are digested to provide the cell with molecular building blocks and energy.
This is a good study in which the authors analyzed the role of autophagy in the anti-apoptosis effect of augmenter of liver regeneration in HepG2 cells. The results are interesting and suggest that ALR was able to protect cells against apoptosis partly through increased autophagic activity in HepG2 cells, which may be valuable for new therapeutic treatments for liver disease in the future.
P- Reviewer: Berg T, Dai ZJ, D'Orazi G, Kim JS, Osna NA S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Wang CH
| 1. | LaBrecque DR, Pesch LA. Preparation and partial characterization of hepatic regenerative stimulator substance (SS) from rat liver. J Physiol. 1975;248:273-284. [PubMed] |
| 2. | Hagiya M, Francavilla A, Polimeno L, Ihara I, Sakai H, Seki T, Shimonishi M, Porter KA, Starzl TE. Cloning and sequence analysis of the rat augmenter of liver regeneration (ALR) gene: expression of biologically active recombinant ALR and demonstration of tissue distribution. Proc Natl Acad Sci USA. 1994;91:8142-8146. [PubMed] |
| 3. | Polimeno L, Pesetti B, Annoscia E, Giorgio F, Francavilla R, Lisowsky T, Gentile A, Rossi R, Bucci A, Francavilla A. Alrp, a survival factor that controls the apoptotic process of regenerating liver after partial hepatectomy in rats. Free Radic Res. 2011;45:534-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 4. | Zhang J, Li Y, Jiang S, Yu H, An W. Enhanced endoplasmic reticulum SERCA activity by overexpression of hepatic stimulator substance gene prevents hepatic cells from ER stress-induced apoptosis. Am J Physiol Cell Physiol. 2014;306:C279-C290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 5. | Jiang Y, Zhao M, An W. Increased hepatic apoptosis in high-fat diet-induced NASH in rats may be associated with downregulation of hepatic stimulator substance. J Mol Med (Berl). 2011;89:1207-1217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 6. | Li S, Zhang HY, Wang T, Meng X, Zong ZH, Kong DH, Wang HQ, Du ZX. BAG3 promoted starvation-induced apoptosis of thyroid cancer cells via attenuation of autophagy. J Clin Endocrinol Metab. 2014;99:E2298-E2307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 7. | Hu R, Chen ZF, Yan J, Li QF, Huang Y, Xu H, Zhang X, Jiang H. Complement C5a exacerbates acute lung injury induced through autophagy-mediated alveolar macrophage apoptosis. Cell Death Dis. 2014;5:e1330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 8. | Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1546] [Cited by in RCA: 1912] [Article Influence: 127.5] [Reference Citation Analysis (0)] |
| 9. | Gump JM, Thorburn A. Autophagy and apoptosis: what is the connection? Trends Cell Biol. 2011;21:387-392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 416] [Cited by in RCA: 399] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 10. | Mei S, Ni HM, Manley S, Bockus A, Kassel KM, Luyendyk JP, Copple BL, Ding WX. Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J Pharmacol Exp Ther. 2011;339:487-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 248] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 11. | Jiao M, Ren F, Zhou L, Zhang X, Zhang L, Wen T, Wei L, Wang X, Shi H, Bai L. Peroxisome proliferator-activated receptor α activation attenuates the inflammatory response to protect the liver from acute failure by promoting the autophagy pathway. Cell Death Dis. 2014;5:e1397. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 80] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 12. | Rautou PE, Mansouri A, Lebrec D, Durand F, Valla D, Moreau R. Autophagy in liver diseases. J Hepatol. 2010;53:1123-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 323] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 13. | Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313-326. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3876] [Cited by in RCA: 3802] [Article Influence: 253.5] [Reference Citation Analysis (0)] |
| 14. | Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728-741. [PubMed] |
| 15. | Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323-335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2701] [Cited by in RCA: 2575] [Article Influence: 183.9] [Reference Citation Analysis (1)] |
| 16. | Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764-776. [PubMed] |
| 17. | González-Rodríguez A, Mayoral R, Agra N, Valdecantos MP, Pardo V, Miquilena-Colina ME, Vargas-Castrillón J, Lo Iacono O, Corazzari M, Fimia GM. Impaired autophagic flux is associated with increased endoplasmic reticulum stress during the development of NAFLD. Cell Death Dis. 2014;5:e1179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 491] [Cited by in RCA: 485] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 18. | Ilowski M, Kleespies A, de Toni EN, Donabauer B, Jauch KW, Hengstler JG, Thasler WE. Augmenter of liver regeneration (ALR) protects human hepatocytes against apoptosis. Biochem Biophys Res Commun. 2011;404:148-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 19. | Thirunavukkarasu C, Wang LF, Harvey SA, Watkins SC, Chaillet JR, Prelich J, Starzl TE, Gandhi CR. Augmenter of liver regeneration: an important intracellular survival factor for hepatocytes. J Hepatol. 2008;48:578-588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 69] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 20. | Polimeno L, Pesetti B, De Santis F, Resta L, Rossi R, De Palma A, Girardi B, Amoruso A, Francavilla A. Decreased expression of the augmenter of liver regeneration results in increased apoptosis and oxidative damage in human-derived glioma cells. Cell Death Dis. 2012;3:e289. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 21. | Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999-2010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 475] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 22. | Cai N, Zhao X, Jing Y, Sun K, Jiao S, Chen X, Yang H, Zhou Y, Wei L. Autophagy protects against palmitate-induced apoptosis in hepatocytes. Cell Biosci. 2014;4:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 23. | Ni HM, Bockus A, Boggess N, Jaeschke H, Ding WX. Activation of autophagy protects against acetaminophen-induced hepatotoxicity. Hepatology. 2012;55:222-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 355] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 24. | Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao W, Lu B, Stolz DB, Clemens DL, Yin XM. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740-1752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 435] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 25. | Li S, Tang Z, Yu H, Li W, Jiang Y, Wang Y, An W. Administration of naked plasmid encoding hepatic stimulator substance by hydrodynamic tail vein injection protects mice from hepatic failure by suppressing the mitochondrial permeability transition. J Pharmacol Exp Ther. 2011;338:750-757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 26. | Toshima T, Shirabe K, Fukuhara T, Ikegami T, Yoshizumi T, Soejima Y, Ikeda T, Okano S, Maehara Y. Suppression of autophagy during liver regeneration impairs energy charge and hepatocyte senescence in mice. Hepatology. 2014;60:290-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 27. | Gandhi CR, Kuddus R, Subbotin VM, Prelich J, Murase N, Rao AS, Nalesnik MA, Watkins SC, DeLeo A, Trucco M. A fresh look at augmenter of liver regeneration in rats. Hepatology. 1999;29:1435-1445. [PubMed] |