Published online May 7, 2015. doi: 10.3748/wjg.v21.i17.5220
Peer-review started: November 20, 2014
First decision: December 26, 2014
Revised: January 18, 2015
Accepted: March 27, 2015
Article in press: March 27, 2015
Published online: May 7, 2015
Processing time: 174 Days and 17.7 Hours
Gastric cancer is the second leading cause of cancer-related deaths. Metastasis, which is an important element of gastric cancer, leads to a high mortality rate and to a poor prognosis. Gastric cancer metastasis has a complex progression that involves multiple biological processes. The comprehensive mechanisms of metastasis remain unclear, though traditional regulation modulates the molecular functions associated with metastasis. Long non-coding RNAs (lncRNAs) have a role in different gene regulatory pathways by epigenetic modification and by transcriptional and post-transcription regulation. lncRNAs participate in various diseases, including Alzheimer’s disease, cardiovascular disease, and cancer. The altered expressions of certain lncRNAs are linked to gastric cancer metastasis and invasion, as with tumor suppressor genes or oncogenes. Studies have partly elucidated the roles of lncRNAs as biomarkers and in therapies, as well as their gene regulatory mechanisms. However, comprehensive knowledge regarding the functional mechanisms of gene regulation in metastatic gastric cancer remains scarce. To provide a theoretical basis for therapeutic intervention in metastatic gastric cancer, we reviewed the functions of lncRNAs and their regulatory roles in gastric cancer metastasis.
Core tip: Long non-coding RNAs (lncRNAs), are emerging as players in multiple biological processes, and are involved in many diseases via the regulation of gene expression at the chromatin, transcriptional, or posttranscriptional level. Their roles in gastric carcinoma metastasis are quite complex; however, the comprehensive study of metastasis will provide us with new perspectives to develop future therapeutic treatments. lncRNAs, which play pivotal roles in gastric cancer metastasis, may help provide future treatments to improve the quality of life of patients with metastatic gastric cancer.
- Citation: Yang ZG, Gao L, Guo XB, Shi YL. Roles of long non-coding RNAs in gastric cancer metastasis. World J Gastroenterol 2015; 21(17): 5220-5230
- URL: https://www.wjgnet.com/1007-9327/full/v21/i17/5220.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i17.5220
Gastric cancer (GC) is one of the most common malignancies worldwide. With its high lethality, GC ranks second in cancer-associated deaths, although the morbidity associated with this malignancy has decreased in most countries[1,2]. Because specific biomarkers of GC are currently limited, patients are typically diagnosed after metastasis, which is associated with recurrence, GC-related death, and poor prognoses. The following important changes occur during GC metastasis: (1) GC cells invade the primary protective barriers and migrate to adjacent tissue; (2) GC cells migrate to the circulatory system to a distant location; (3) GC cells invade the lymphatic system and disseminate to a secondary site; and (4) GC cells leave the primary tumor and adhere to a sensitive location. Recent studies have revealed that the processes of angiogenesis and adhesion, the cell-to-cell tight junctions, and the extracellular matrix play essential roles in GC metastasis[3-6]. However, the mechanisms of GC metastasis have yet to be comprehensively elucidated. GC tumorigenesis and metastasis are multistep processes that involve a myriad of signaling pathways and gene regulation in which oncogenic and tumor-suppressive factors, such as long non-coding RNAs (lncRNAs) play pivotal roles[7].
lncRNAs are transcriptional products longer than 200 nucleotides that primarily have a regulatory role rather than encode proteins[8-10]. In some studies, lncRNAs encode short, evolutionarily divergent proteins[11,12]. Additionally, lncRNAs play a pivotal role in many biological mechanisms, including gene imprinting, activation and repression[13-15]. At the transcriptional level, lncRNAs can bind to DNA or proteins, regulate the subcellular localization of transcription factors, and inhibit transcript elongation[13,16,17]. lncRNAs affect many posttranscriptional processes, including alternative splicing, RNA editing, transport, degradation, translation, and miR-mediated regulation[18-20], and are involved in diseases such as Alzheimer’s disease, cardiovascular diseases, and malignancies, including lung cancer[21], renal cell carcinoma[22], bladder cancer[23], esophageal carcinoma[24], hepatocellular carcinoma[25], and acute leukemia[26]. lncRNAs also play important roles in the proliferation, migration, and invasion of human cancer cells, and are associated with cancer metastasis and therapeutic sensitivity, implicating lncRNAs as potential novel biomarkers for diagnosis and as targets for therapeutic approaches[27,28].
Studies of GC metastasis related to lncRNAs will provide us with a greater understanding of their regulatory roles in GC tumorigenesis, progression, and metastasis. In the present review, we summarize the lncRNAs involved in GC metastasis, and the mechanisms of their regulatory functions.
Angiogenesis in cancer, including GC, is important for proliferation, metastasis, and drug sensitivity. Angiogenesis not only provides a wealth of nutrients and oxygen for GC cells and transports tumor metabolites, but also provides favorable conditions for GC vessel metastasis, increasing the opportunity for cancer cells to enter the blood circulation and to seed in secondary locations. Thus, antiangiogenic treatment can improve therapeutic efficiency[29,30]. As a newly discovered group of regulatory genes, lncRNAs play essential roles in angiogenesis; however, their mechanisms of action in GC angiogenesis and metastasis have not been fully elucidated.
lncRNAs promote carcinoma metastasis by activating the angiogenic system in which phosphoglycerate kinase (PGK) participates. PGK, which is a disulfide reductase conventionally thought to be involved in glycolysis, also functions as a carcinoma metastasis inhibitor by modulating angiogenesis. The hepatocellular carcinoma overexpressed lncRNA, MVIH, is associated with microvascular invasion, TNM stage, recurrence-free survival, and overall survival, and promotes distant metastasis by inhibiting PGK1 secretion and by activating angiogenesis[31,32]. Certain GC-associated lncRNAs modulate angiogenesis. For example, H19 promotes vasculogenesis by activating tumor necrosis factor-α, which indirectly induces angiogenic factors[3]. Other lncRNAs regulate basal sprouting and migration, endothelial cell proliferation, capillary density, vascular endothelial growth factor α and its receptor, which participate in angiogenesis[33-35].
Tight junctions (TJs) between cells play pivotal roles in regulating the diffusion of ions and specific molecules and in maintaining the integrity of the cell-to-cell protective barrier. Recent studies have revealed that cell-to-cell junctions function as protectors of GC metastasis. The aberrant expression or distribution of TJ proteins leads to the loss of cell-to-cell adhesion and tissue integrity, assisting cancer cell invasion and promoting metastasis[4,5].
TUC339 is a 1198-bp ultraconserved lncRNA that regulates cancer cell growth and adhesion[36]. Ephrins are cell surface protein ligands that can mediate cellular adhesion and function via interactions with the Eph receptor tyrosine kinases. Hypoxia induces the expression of hypoxia-inducible factor, leading to EFNA3 lncRNA expression, Ephrin-A3 protein accumulation, and metastatic dissemination in breast cancer[37]. As the interactions between lncRNAs and mRNAs are accepted, the important function of lncRNAs in cell-to-cell junctions and adhesion can be recognized by studying the association between lncRNAs and adhesion-associated genes. Zhao et al[38] demonstrated that Down Syndrome Cell Adhesion Molecule (DSCAM) is a member of the cell adhesion immunoglobulin superfamily. Its antisense lncRNA (DSCAM-AS1) is overexpressed in lung adenocarcinoma and may play a pivotal role in regulating cell-to-cell adhesion because this type of antisense lncRNA can interact with its host genes.
Epithelial-to-mesenchymal transition (EMT) and its inverse process, mesenchymal-to-epithelial transition, play critical roles in embryonic growth, stem cell biology, and tumorigenesis progression, which involves the invasion and migration of carcinoma cells with multiple participating signaling pathways[39,40]. The upregulated breast cancer-associated lncRNA, linc-ROR, can induce EMT, enhancing migration and invasion. However, silencing its expression represses lung metastasis. Linc-ROR functions as a miR sponge, binding with miR-205 and inhibiting the degradation of its target genes, including ZEB2, which induces EMT[41]. The Hox transcript antisense intergenic RNA (HOTAIR) is overexpressed in GC carcinoma metastatic lymph nodes, and its silencing downregulates the expression of the transcription factor Snail, with tighter cell-to-cell adhesion and rounder morphology, as well as decreased expression of the mesenchymal markers vimentin and N-cadherin, and increased expression of the epithelial markers E-cadherin and ZO-1, indicating that HOTAIR silencing can reverse EMT in GC cells and inhibit distal metastasis[42]. The lncRNA, MALAT1, modulates the expression of the transcription factors ZEB1, ZEB2, Slug, and E-cadherin, and promotes EMT by activating the Wnt signaling pathway, which regulates gene expression at the transcriptional level[43]. Other lncRNAs, such as the lncRNA, LEIGC, which is slightly expressed in GC, also regulate EMT[44]. Therefore, EMT regulation provides a target for the therapeutic intervention of GC metastasis.
The extracellular matrix prevents GC invasion and metastasis. Its damage degrades the protective barrier against GC metastasis, and provides a favorable environment. Pericellular proteases can degrade matrix proteins and modulate cancer metastasis via regulating the cleavage of proteins such as cell adhesion molecules[6]. Matrix metalloproteinases (MMPs) are a subgroup of proteases that degrade the extracellular matrix and modulate GC invasion and metastasis[45].
lncRNAs have great importance in stabilizing or degrading the extracellular matrix. H19 is downregulated in prostate cancer, and H19-derived miR-675 inhibits the expression of the extracellular matrix protein transforming growth factor β-induced protein mRNA by binding to its 3’-untranslated region (3’-UTR), thereby inhibiting prostate cancer metastasis[46]. Park et al[47] found that the lncRNA, BM742401, was downregulated in cancer, and its overexpression inhibited GC cell migration, invasion and metastasis by regulating MMP9 secretion. Other lncRNAs aberrantly expressed in GC, including HOTAIR and FENDRR, correlate with GC metastasis; these lncRNAs regulate extracellular matrix degradation by modulating the expression of cancer metastasis-associated genes such as ICAM-1, MMP1, MMP2, MMP3 and MMP9[48,49].
One difference between cancer and normal tissue is that cancer cells can proliferate infinitely, damage normal tissue under suitable conditions, migrate to adjacent normal tissue, vessels, and lymphatic tubes, and promote cancer metastasis and carcinoma-related death. lncRNAs, which are emerging regulators of gene expression in various biological progresses, play important modulatory roles in cancer cell invasion and metastasis, and in patient prognosis.
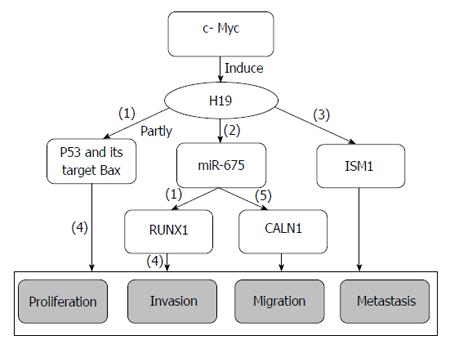
GC metastasis-associated lncRNA H19: H19, which is a 2.3-kb lncRNA transcribed from the paternally imprinted gene H19 on chromosome 11p15.5, is highly expressed during embryogenesis; however, its expression almost disappears from all tissues after birth[50] (Figure 1). H19 plays pivotal roles in tumorigenesis and metastasis in the same manner as oncogenes or tumor suppressor genes. In bladder cancer, H19 activates Wnt/β-catenin by associating with enhancer of zeste homolog 2 (EZH2), increasing its metastasis[51]. H19 partly promotes HMGA2-mediated EMT by antagonizing let-7 in pancreatic cancer, and increasing the migration and invasion potential of pancreatic ductal adenocarcinoma cells[52]. However, H19 represses prostate cancer metastasis through the H19-miR-675 axis[46].
H19 is also overexpressed in GC, and can promote cell proliferation by partly inactivating p53, decreasing its activity, and suppressing the expression of the p53 target protein Bax. In contrast, H19 silencing results in GC cell apoptosis, suggesting that H19 promotes GC tumorigenesis by activating p53[53]. H19 is the precursor of miR-675[50], and the H19/miR-675 signaling axis plays a critical role in the carcinogenesis of colorectal cancer[54]. miR-675 serves as a tumor suppressor by targeting the 3’-UTR of runt domain transcription factor1 (RUNX1), downregulating its expression at the mRNA and protein levels in GC cells and restoring the proliferation inhibitory effects induced by siRNA-H19, indicating that H19 acts as a oncogene by indirectly regulating RUNX1 expression[55]. However, entirely different conclusions suggest that H19 regulates the binding protein ISM1, but that miR-675 promotes cell proliferation, invasion and migration by targeting CALN1 in GC[56]. In other studies, c-Myc associates with H19-induced GC tumorigenesis, development, and metastasis[57]. Taken together, the inhibition of H19 expression and the suppression of its effects on carcinoma progression and metastasis may improve GC therapeutic treatment and prognosis.
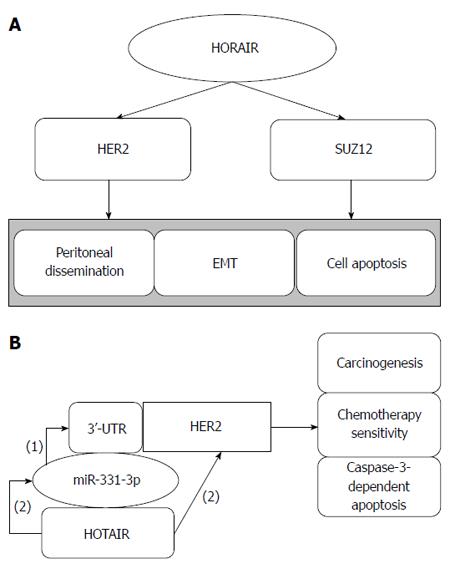
GC metastasis-associated lncRNA HOTAIR: HOTAIR, which is a 2.2-kb ncRNA transcribed from the HOXC locus of chromosome 12q13.13 in the opposite direction of HOXC, acts as a scaffold. Its 5’ domain binds to polycomb repressive complex 2 (PRC2), and its 3’ domain binds the LSD1/CoREST/REST complex, coordinating PRC2 and LSD1 to target chromatin, which leads to the methylation of histone H3 lysine 27 and to the demethylation of lysine 4, respectively (Figure 2). Thus, HOTAIR transcription affects distant gene silencing[58,59]. HOTAIR is overexpressed in primary breast cancer and can promote cancer invasiveness and metastasis depending on PRC2, making HOTAIR a possible predictor of cancer metastasis[60]. Increased expression of HOTAIR is also observed in hepatocellular carcinoma, and its expression is associated with MMP9 and vascular endothelial growth factor protein, which affect cell activity and cancer metastasis[61]. HOTAIR expression is increased in colon cancer and is related to the depth of tumor invasion, lymph node metastasis, organ metastasis, histological differentiation, vascular invasion, advanced tumor stage, higher recurrence rate and poorer overall survival. HORAIR is a potential target for therapeutic intervention by modulating EMT[62].
HOTAIR is overexpressed in GC metastatic lymph nodes, and its upregulation is associated with peritoneal dissemination, poor prognosis and EMT, which is related to cancer metastasis[48,63]. Human epithelial growth factor receptor 2 (HER2) encodes a protein associated with trastuzumab-based therapy in carcinogenesis and resistance. Liu et al[10] showed that HOTAIR inhibition enhances caspase-3-dependent apoptosis and increases early apoptosis. Furthermore, miR-331-3p inhibits the expression of HER2 by targeting its 3’-UTR, and HOTAIR regulates the expression of HER2 by competitive binding to miR-331-3p, acting as a competing endogenous RNAs (ceRNA). Other results have shown that HOTAIR correlates with SUZ12 and affects epigenetics[64]. Taken together, we conclude that HOTAIR can be treated as a therapeutic target that participates in GC tumorigenesis and metastasis via the HOTAIR/EMT and HOTAIR/SUZ12 pathways, and in drug sensitivity by regulating endogenous competition, providing a new path for inhibiting GC metastasis.
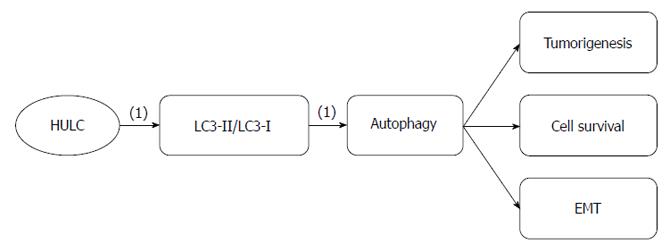
GC metastasis associated-lncRNA highly upregulated in liver cancer: Highly upregulated in liver cancer (HULC) is an lncRNA that was first identified by its aberrant expression in hepatocellular carcinoma (Figure 3). In subsequent studies, its ectopic expression was found in other cancers, including GC and pancreatic cancer[65-67].
Increased HULC expression has been observed in SGC7901, BGC823 and AGS GC lines; however, no significant difference in HULC expression has been observed between MKN28, MKN45 and the gastric epithelial mucosa cell line GES-1. The expression level of HULC correlates with GC lymph node metastasis, distant metastasis and TNM stage. HULC expression promotes GC cell proliferation, migration and invasion via modulating EMT, which is important for metastasis[66]. Autophagy, which is a form of programmed cell death that differs from apoptosis, regulates tumorigenesis and cancer progression. Some studies have shown that upregulated autophagy in cancer cells leads to cancer cell survival and EMT regulation[40,68,69]. HULC upregulation increases the ratio of the microtubule-associated protein 1 light chain 3 (LC3)-II, a marker of autophagosomes, to LC3-I, suggesting that HULC promotes GC cell proliferation by activating autophagy[66]. Therefore, we can repress GC cancer progression and metastasis by downregulating HULC and, correspondingly, modulating EMT and autophagy.
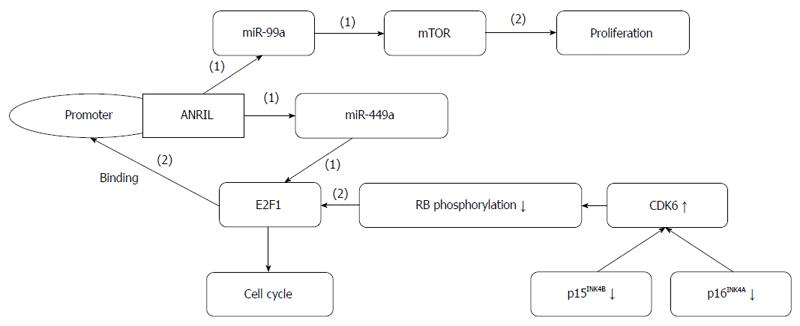
GC metastasis-associated lncRNA ANRIL (CDKN2B-AS1): ANRIL is a 3.8-kb lncRNA transcribed from the INK4A-ARF-INK4B gene cluster in the opposite direction[70] (Figure 4). ANRIL binds to PRC2 with its component SUZ12 and recruits PRC2, thereby decreasing the expression of p15/INK4B, which is a tumor suppressor gene[71]. ANRIL and PRC2 play repressive roles at the epigenetic transcriptional level by binding to chromobox 7 within PRC1 and recruiting PRC1 to the p16 (INK4A)/p14 (ARF) locus, subsequently leading to its silencing[70]. The lncRNA ANRIL is also related to atherosclerosis, periodontitis and certain cancers[72].
ANRIL is aberrantly expressed in some cancers and used to predict lung cancer metastasis[73]. ANRIL expression is increased in GC, and its upregulation is related to higher TNM stage, larger tumor size and poorer prognosis. ANRIL promotes GC cell proliferation and growth partly by the epigenetic silencing of p15/INK4B and p16/INK4A in Cis and by the regulation of miR-99a/miR-449a expression by binding to PRC2 with EZH2 and SUZ12 in Trans. This regulation forms a positive feedback loop and establishes prognostic roles for ANRIL in GC diagnosis and therapeutic treatment[74].
GC metastasis-associated lncRNA maternally expressed gene 3: Maternally expressed gene 3 (MEG3) is an imprinted gene on human chromosome 14q32.3 that can encode a lncRNA[75] (Figure 5). LncRNA MEG3 expression is downregulated in multiple cancers, and its downregulation is associated with advanced pathological stage, larger tumor size, and poor prognosis. MEG3 induces G2/M cell cycle arrest and promotes apoptosis by partially activating p53, thereby inhibiting cell proliferation and growth[76,77].
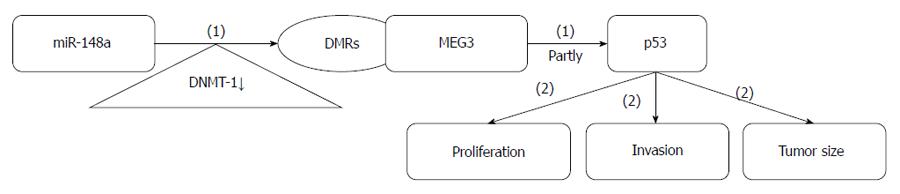
In GC, MEG3 expression is significantly lower than in adjacent normal tissue, and its downregulation correlates with higher TNM stage, deeper invasion. and larger tumor size. MEG3 acts as a tumor suppressor partly through activation of p53. Methylation of the MEG3 regulatory regions (MEG3-DMRs) is involved in MEG3 modulation. miR-148a increases the expression of MEG3 by downregulating DNA methyltransferase-1, which is traditionally thought to be essential for DNA methylation and genome stability[78,79]. miR-148a is a therapeutic agent that can repress GC metastasis by indirectly upregulating MEG3, which functions as a tumor suppressor gene.
GC metastasis associated lncRNA FENDRR: The lncRNA, FENDRR, which is transcribed 1250 bp upstream of the 5’ end of the gene encoding the transcription factor FOXF1, plays important roles in the differentiation of the lateral mesoderm and in the development of the heart and the body wall (Figure 6). FENDRR modulates chromatin signatures through binding to both the PRC2 and the Trithorax group/Mixed lineage leukemia complexes[80].
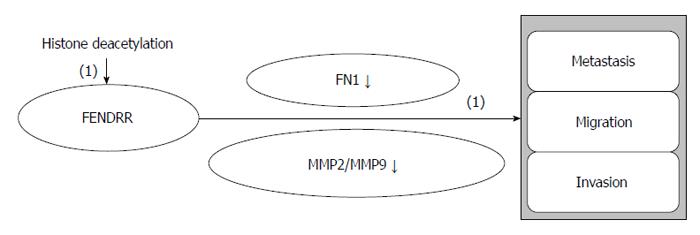
FENDRR is downregulated in GC, and its low expression is related to deeper tumor invasion, higher tumor stage, lymphatic metastasis, and poor prognosis. FENDRR negatively regulates fibronectin 1 and MMP2/MMP9, indicating that FENDRR is a mediator of cell differentiation, growth, and migration and of GC progression and metastasis. FENDRR expression is downregulated by histone deacetylation in GC[49]. FENDRR will help us to comprehensively understand the biological mechanisms of GC metastasis and to discover novel therapeutic treatments for GC.
Other aberrantly expressed lncRNAs in GC are closely related to TNM stage, invasion depth, lymph node metastasis, and patient prognosis, and some of their mechanisms have been partially elucidated[81-83].
Colon cancer-associated transcript (CCAT1) is an lncRNA that is ectopically expressed in colon adenocarcinoma and is highly expressed in GC, being associated with early GC growth, lymphatic node metastasis, and distal metastasis. c-Myc binds to the E-box element in the promoter region of CCAT1, promoting GC activity, proliferation and migration by upregulating proliferating cell nuclear antigen[84,85]. Under hypoxic conditions, the lncRNA, AK058003, decreases the CpG island methylation of SNCG, which is a metastasis-related gene, correspondingly modulating the hypoxia-induced metastasis of GC cells[86]. The lncRNA, MALAT1, is ectopically expressed in GC, promoting cancer cell proliferation by recruiting and partly modulating SF2/ASF[87]. Other lncRNAs, such as GHET1 and GAS5, are aberrantly expressed in GC and are related to tumor size, TNM stage, invasion, and prognosis. These lncRNAs function as GC metastasis regulators by modulating the interaction between insulin-like growth factor 2 binding protein-1 and c-Myc mRNA, consequently upregulating the expression of c-Myc mRNA and increasing its stability, thereby promoting proliferation or enhancing caspase-3-dependent apoptosis[88,89]. These regulatory mechanisms of lncRNAs in gastric carcinogenesis and metastasis pave the way for future molecular target therapy for GC.
In the human transcriptome, aberrant sequence expression involves both protein-coding RNAs and non-protein-coding transcripts. However, non-coding elements were neglected previously, and even considered as noise with no biological functions. Recent research has paid increasing attention to a traditionally ignored area, namely lncRNAs, which are a new cluster of non-coding RNAs transcribed from non-protein-coding sequences. lncRNAs can not only regulate the subcellular localization and activity of other RNAs and proteins, but also regulate local or global gene expression in trans or cis by influencing chromatin modification, and transcriptional or posttranscriptional gene regulation[90]. Recently, the functions and expression levels of lncRNAs were shown to correlate with diseases such as neurological disorders, Mendelian disorders, cardiovascular disease, and cancer[14]. lncRNAs play different biological and physical roles in normal individuals. lncRNAs are involved in gene expression programs by binding to various chromatin regulatory proteins and modulating the pluripotency and differentiation of embryonic stem cells[91]. lncRNAs play critical roles in the development of life and organs, such as the brain[92], heart[93], and lungs[94]. The aberrant expression and regulatory roles of lncRNAs in clinical diseases were demonstrated simultaneously with their functions to some extent. In recent years, their functions and expression levels have been shown to be associated with neurological disorders, Mendelian disorders, cardiovascular disease, and cancer[14,95]. In cancer, lncRNAs participate in a multitude of processes, including cancer cell proliferation, migration, invasiveness, and metastasis[96-98]. The lncRNA, H19, is highly expressed in hepatic metastatic individuals, giving it prognostic value for cancer metastasis[96]. The lncRNA, MALAT1, modulates gene expression, thus changing the metastasis phenotype of lung cancer cells[99]. The upregulated expression of MALAT1 in gallbladder carcinoma suggests that this lncRNA might function as an oncogene, activating the ERK/MAPK pathway and promoting gallbladder cancer cell proliferation and metastasis[97].
lncRNAs also modulate GC tumorigenesis, growth, and metastasis by affecting GC cell proliferation, cell migration and invasion, tumor suppressor genes and oncogenes such as p53[53,78] and c-Myc[89], mediating EMT[66], acting as ceRNAs[10], and being processed into other small molecular RNAs[50,55]. Increasing evidence has shed light on the role of lncRNAs in diverse cancers, particularly GC; however, their functional links to gene regulation, disease occurrence and, specifically, GC metastasis have yet to be thoroughly studied. lncRNAs serve as a group of multiple genes involved in the regulation of GC metastasis[86,100]. Studies have shown that angiogenesis and the stroma play pivotal roles in cancer metastasis[101]. Researchers have demonstrated that lncRNAs participate in neovasculature formation[31,32], TJ and adhesion regulation[37,38,42], and extracellular matrix degradation[47-49]. Some lncRNAs are aberrantly expressed or play essential roles in cell activity and cancer metastasis, indicating that lncRNAs may promote or inhibit GC metastasis.
Unfortunately, few studies have systematically described a functional overview of the mechanisms underlying the regulatory roles of lncRNAs and their association with GC metastasis. Provision of reliable approaches for predicting and identifying the functions of the versatile lncRNAs, and exploring this mysterious field in a comprehensive manner remains necessary[8]. For GC metastasis-related lncRNAs, a stepwise approach to the comprehensive understanding of GC metastasis-related lncRNAs is also required. Uncompleted tasks include the following: (1) screening for more GC metastasis-associated lncRNAs; (2) exploring the mechanisms of lncRNAs involved in GC carcinogenesis and metastasis; (3) searching for lncRNAs with metastasis value and constructing a lncRNA regulatory network; (4) seeking lncRNA pathways participating in GC invasion and metastasis and selecting the key pathway; and (5) intervening in GC at the gene level to inhibit metastasis. Taken together, the study of newly discovered lncRNAs is a promising field for future molecular targeted therapy of GC via metastasis intervention.
P- Reviewer: Park WS S- Editor: Ma YJ L- Editor: Cant MR E- Editor: Wang CH
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25541] [Article Influence: 1824.4] [Reference Citation Analysis (7)] |
| 2. | Bertuccio P, Chatenoud L, Levi F, Praud D, Ferlay J, Negri E, Malvezzi M, La Vecchia C. Recent patterns in gastric cancer: a global overview. Int J Cancer. 2009;125:666-673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 446] [Cited by in RCA: 484] [Article Influence: 30.3] [Reference Citation Analysis (1)] |
| 3. | Ayesh S, Matouk I, Schneider T, Ohana P, Laster M, Al-Sharef W, De-Groot N, Hochberg A. Possible physiological role of H19 RNA. Mol Carcinog. 2002;35:63-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 94] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 4. | Martin TA. The role of tight junctions in cancer metastasis. Semin Cell Dev Biol. 2014;36:224-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 97] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 5. | Martin TA, Jiang WG. Loss of tight junction barrier function and its role in cancer metastasis. Biochim Biophys Acta. 2009;1788:872-891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 360] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 6. | Sevenich L, Joyce JA. Pericellular proteolysis in cancer. Genes Dev. 2014;28:2331-2347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 146] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 7. | Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1127] [Cited by in RCA: 1340] [Article Influence: 95.7] [Reference Citation Analysis (0)] |
| 8. | Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 2014;30:439-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 214] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 9. | Clark BS, Blackshaw S. Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front Genet. 2014;5:164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 122] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 10. | Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 659] [Cited by in RCA: 789] [Article Influence: 71.7] [Reference Citation Analysis (0)] |
| 11. | Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 288] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 12. | Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014;33:981-993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 509] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 13. | Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4004] [Cited by in RCA: 3940] [Article Influence: 246.3] [Reference Citation Analysis (0)] |
| 14. | Maass PG, Luft FC, Bähring S. Long non-coding RNA in health and disease. J Mol Med (Berl). 2014;92:337-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 199] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 15. | Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 727] [Cited by in RCA: 759] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 16. | Wang X, Song X, Glass CK, Rosenfeld MG. The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb Perspect Biol. 2011;3:a003756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 119] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 17. | Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30:348-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 350] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 18. | Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 823] [Cited by in RCA: 954] [Article Influence: 79.5] [Reference Citation Analysis (0)] |
| 19. | Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925-938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1521] [Cited by in RCA: 1737] [Article Influence: 115.8] [Reference Citation Analysis (0)] |
| 20. | Mourtada-Maarabouni M, Williams GT. Growth arrest on inhibition of nonsense-mediated decay is mediated by noncoding RNA GAS5. Biomed Res Int. 2013;2013:358015. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 21. | White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15:429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 22. | Malouf GG, Zhang J, Yuan Y, Compérat E, Rouprêt M, Cussenot O, Chen Y, Thompson EJ, Tannir NM, Weinstein JN. Characterization of long non-coding RNA transcriptome in clear-cell renal cell carcinoma by next-generation deep sequencing. Mol Oncol. 2015;9:32-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 23. | Peter S, Borkowska E, Drayton RM, Rakhit CP, Noon A, Chen W, Catto JW. Identification of differentially expressed long noncoding RNAs in bladder cancer. Clin Cancer Res. 2014;20:5311-5321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 24. | Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY, Xue WQ, Chen YB, Zhang Y, Jia WH. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 2013;104:1675-1682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 221] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 25. | Yuan SX, Tao QF, Wang J, Yang F, Liu L, Wang LL, Zhang J, Yang Y, Liu H, Wang F. Antisense long non-coding RNA PCNA-AS1 promotes tumor growth by regulating proliferating cell nuclear antigen in hepatocellular carcinoma. Cancer Lett. 2014;349:87-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 26. | Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, Aifantis I. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 357] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 27. | Malek E, Jagannathan S, Driscoll JJ. Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget. 2014;5:8027-8038. [PubMed] |
| 28. | Serviss JT, Johnsson P, Grandér D. An emerging role for long non-coding RNAs in cancer metastasis. Front Genet. 2014;5:234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 91] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 29. | Ishiwata T, Matsuda Y, Naito Z. Nestin in gastrointestinal and other cancers: effects on cells and tumor angiogenesis. World J Gastroenterol. 2011;17:409-418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 98] [Cited by in RCA: 113] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 30. | Lazăr D, Raica M, Sporea I, Tăban S, Goldiş A, Cornianu M. Tumor angiogenesis in gastric cancer. Rom J Morphol Embryol. 2006;47:5-13. [PubMed] |
| 31. | Lay AJ, Jiang XM, Kisker O, Flynn E, Underwood A, Condron R, Hogg PJ. Phosphoglycerate kinase acts in tumour angiogenesis as a disulphide reductase. Nature. 2000;408:869-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 218] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 32. | Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, Yang Y, Wang RY, Yang S, Huo XS. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231-2241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 331] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 33. | Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114:1389-1397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 652] [Cited by in RCA: 760] [Article Influence: 69.1] [Reference Citation Analysis (0)] |
| 34. | Gordon FE, Nutt CL, Cheunsuchon P, Nakayama Y, Provencher KA, Rice KA, Zhou Y, Zhang X, Klibanski A. Increased expression of angiogenic genes in the brains of mouse meg3-null embryos. Endocrinology. 2010;151:2443-2452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 137] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 35. | Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48:R45-R53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 524] [Cited by in RCA: 607] [Article Influence: 46.7] [Reference Citation Analysis (0)] |
| 36. | Kogure T, Yan IK, Lin WL, Patel T. Extracellular Vesicle-Mediated Transfer of a Novel Long Noncoding RNA TUC339: A Mechanism of Intercellular Signaling in Human Hepatocellular Cancer. Genes Cancer. 2013;4:261-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 262] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 37. | Gómez-Maldonado L, Tiana M, Roche O, Prado-Cabrero A, Jensen L, Fernandez-Barral A, Guijarro-Muñoz I, Favaro E, Moreno-Bueno G, Sanz L. EFNA3 long noncoding RNAs induced by hypoxia promote metastatic dissemination. Oncogene. 2014;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 88] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 38. | Zhao W, Luo J, Jiao S. Comprehensive characterization of cancer subtype associated long non-coding RNAs and their clinical implications. Sci Rep. 2014;4:6591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 94] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 39. | Guo F, Parker Kerrigan BC, Yang D, Hu L, Shmulevich I, Sood AK, Xue F, Zhang W. Post-transcriptional regulatory network of epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions. J Hematol Oncol. 2014;7:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 113] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 40. | Lv Q, Hua F, Hu ZW. DEDD, a novel tumor repressor, reverses epithelial-mesenchymal transition by activating selective autophagy. Autophagy. 2012;8:1675-1676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 41. | Hou P, Zhao Y, Li Z, Yao R, Ma M, Gao Y, Zhao L, Zhang Y, Huang B, Lu J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 274] [Cited by in RCA: 293] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 42. | Sun W, Wu Y, Yu X, Liu Y, Song H, Xia T, Xiao B, Guo J. Decreased expression of long noncoding RNA AC096655.1-002 in gastric cancer and its clinical significance. Tumour Biol. 2013;34:2697-2701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 43. | Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289-2294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 284] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 44. | Han Y, Ye J, Wu D, Wu P, Chen Z, Chen J, Gao S, Huang J. LEIGC long non-coding RNA acts as a tumor suppressor in gastric carcinoma by inhibiting the epithelial-to-mesenchymal transition. BMC Cancer. 2014;14:932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 45. | Westermarck J, Kähäri VM. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J. 1999;13:781-792. [PubMed] |
| 46. | Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766-3775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 258] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 47. | Park SM, Park SJ, Kim HJ, Kwon OH, Kang TW, Sohn HA, Kim SK, Moo Noh S, Song KS, Jang SJ. A known expressed sequence tag, BM742401, is a potent lincRNA inhibiting cancer metastasis. Exp Mol Med. 2013;45:e31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 48. | Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ, Huang L, Yu PF, Cheng XD. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci. 2013;9:587-597. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 200] [Cited by in RCA: 224] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 49. | Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L, Chen WM, Han L, Zhang EB, Kong R. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 226] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 50. | Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 458] [Cited by in RCA: 488] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 51. | Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 400] [Cited by in RCA: 461] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 52. | Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z, Ai K. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014;35:9163-9169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 234] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 53. | Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159-3165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 376] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 54. | Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, Kwok TT. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 374] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 55. | Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 171] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 56. | Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318-2329. [PubMed] |
| 57. | Zhang EB, Han L, Yin DD, Kong R, De W, Chen J. c-Myc-induced, long, noncoding H19 affects cell proliferation and predicts a poor prognosis in patients with gastric cancer. Med Oncol. 2014;31:914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 107] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 58. | Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689-693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2754] [Cited by in RCA: 2649] [Article Influence: 176.6] [Reference Citation Analysis (0)] |
| 59. | Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311-1323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3533] [Cited by in RCA: 3390] [Article Influence: 188.3] [Reference Citation Analysis (0)] |
| 60. | Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071-1076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4202] [Cited by in RCA: 4230] [Article Influence: 282.0] [Reference Citation Analysis (0)] |
| 61. | Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39:2119-2128. [PubMed] |
| 62. | Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S, Qiu GQ, Peng ZH, Yan DW. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;32:395-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 173] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 63. | Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731-2739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 228] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 64. | Hajjari M, Behmanesh M, Sadeghizadeh M, Zeinoddini M. Up-regulation of HOTAIR long non-coding RNA in human gastric adenocarcinoma tissues. Med Oncol. 2013;30:670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 92] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 65. | Strohmaier HM, Panzitt K, Moustafa T, Stradner M, Guelly C, Buck CR, Zatloukal K. A novel non-coding RNA implicated in liver cancer. Proc Annu Meet Am Assoc Cancer Res. 2004;45:622-623. |
| 66. | Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep. 2014;31:358-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 221] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 67. | Peng W, Gao W, Feng J. Long noncoding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Med Oncol. 2014;31:346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 127] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 68. | Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679-2688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1271] [Cited by in RCA: 1326] [Article Influence: 66.3] [Reference Citation Analysis (0)] |
| 69. | Enomoto M, Tsuchida A, Miyazawa K, Yokoyama T, Kawakita H, Tokita H, Naito M, Itoh M, Ohyashiki K, Aoki T. Vitamin K2-induced cell growth inhibition via autophagy formation in cholangiocellular carcinoma cell lines. Int J Mol Med. 2007;20:801-808. [PubMed] |
| 70. | Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662-674. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1163] [Cited by in RCA: 1090] [Article Influence: 72.7] [Reference Citation Analysis (0)] |
| 71. | Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956-1962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 830] [Cited by in RCA: 812] [Article Influence: 58.0] [Reference Citation Analysis (0)] |
| 72. | Bochenek G, Häsler R, El Mokhtari NE, König IR, Loos BG, Jepsen S, Rosenstiel P, Schreiber S, Schaefer AS. The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum Mol Genet. 2013;22:4516-4527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 164] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 73. | Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031-8041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1584] [Cited by in RCA: 1797] [Article Influence: 81.7] [Reference Citation Analysis (0)] |
| 74. | Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276-2292. [PubMed] |
| 75. | Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer. 2011;129:773-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 225] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 76. | Qin R, Chen Z, Ding Y, Hao J, Hu J, Guo F. Long non-coding RNA MEG3 inhibits the proliferation of cervical carcinoma cells through the induction of cell cycle arrest and apoptosis. Neoplasma. 2013;60:486-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 134] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 77. | Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 288] [Cited by in RCA: 350] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 78. | Sun M, Xia R, Jin F, Xu T, Liu Z, De W, Liu X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;35:1065-1073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 236] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 79. | Yan J, Guo X, Xia J, Shan T, Gu C, Liang Z, Zhao W, Jin S. MiR-148a regulates MEG3 in gastric cancer by targeting DNA methyltransferase 1. Med Oncol. 2014;31:879. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 109] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 80. | Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 766] [Cited by in RCA: 779] [Article Influence: 64.9] [Reference Citation Analysis (0)] |
| 81. | Mei D, Song H, Wang K, Lou Y, Sun W, Liu Z, Ding X, Guo J. Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med Oncol. 2013;30:709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 103] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 82. | Xiao B, Guo J. Long noncoding RNA AC096655.1-002 has been officially named as gastric cancer-associated transcript 1, GACAT1. Tumour Biol. 2013;34:3271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 83. | Ding J, Li D, Gong M, Wang J, Huang X, Wu T, Wang C. Expression and clinical significance of the long non-coding RNA PVT1 in human gastric cancer. Onco Targets Ther. 2014;7:1625-1630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 107] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 84. | Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437-445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 228] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 85. | Mizrahi I, Mazeh H, Grinbaum R, Beglaibter N, Wilschanski M, Pavlov V, Adileh M, Stojadinovic A, Avital I, Gure AO. Colon Cancer Associated Transcript-1 (CCAT1) Expression in Adenocarcinoma of the Stomach. J Cancer. 2015;6:105-110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 72] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 86. | Wang Y, Liu X, Zhang H, Sun L, Zhou Y, Jin H, Zhang H, Zhang H, Liu J, Guo H. Hypoxia-inducible lncRNA-AK058003 promotes gastric cancer metastasis by targeting γ-synuclein. Neoplasia. 2014;16:1094-1106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 90] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 87. | Wang J, Su L, Chen X, Li P, Cai Q, Yu B, Liu B, Wu W, Zhu Z. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. 2014;68:557-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 139] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 88. | Sun M, Jin FY, Xia R, Kong R, Li JH, Xu TP, Liu YW, Zhang EB, Liu XH, De W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 265] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 89. | Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K, Gu Y, Fang G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281:802-813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 191] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 90. | Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1668] [Cited by in RCA: 1870] [Article Influence: 116.9] [Reference Citation Analysis (0)] |
| 91. | Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295-300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1477] [Cited by in RCA: 1574] [Article Influence: 112.4] [Reference Citation Analysis (0)] |
| 92. | Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 527] [Cited by in RCA: 560] [Article Influence: 46.7] [Reference Citation Analysis (0)] |
| 93. | Ounzain S, Pezzuto I, Micheletti R, Burdet F, Sheta R, Nemir M, Gonzales C, Sarre A, Alexanian M, Blow MJ. Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease. J Mol Cell Cardiol. 2014;76:55-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 97] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 94. | Herriges MJ, Swarr DT, Morley MP, Rathi KS, Peng T, Stewart KM, Morrisey EE. Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev. 2014;28:1363-1379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 128] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 95. | Li G, Zhang H, Wan X, Yang X, Zhu C, Wang A, He L, Miao R, Chen S, Zhao H. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed Res Int. 2014;2014:780521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 96. | Fellig Y, Ariel I, Ohana P, Schachter P, Sinelnikov I, Birman T, Ayesh S, Schneider T, de Groot N, Czerniak A. H19 expression in hepatic metastases from a range of human carcinomas. J Clin Pathol. 2005;58:1064-1068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 64] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 97. | Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding Q, Weng H, Shu YJ, Liu TY, Jiang L. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther. 2014;15:806-814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 196] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 98. | Zhang L, Zhou XF, Pan GF, Zhao JP. Enhanced expression of long non-coding RNA ZXF1 promoted the invasion and metastasis in lung adenocarcinoma. Biomed Pharmacother. 2014;68:401-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 99. | Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180-1189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1087] [Cited by in RCA: 1300] [Article Influence: 100.0] [Reference Citation Analysis (0)] |
| 100. | Fang XY, Pan HF, Leng RX, Ye DQ. Long noncoding RNAs: novel insights into gastric cancer. Cancer Lett. 2015;356:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 166] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 101. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47127] [Article Influence: 3366.2] [Reference Citation Analysis (5)] |