Published online Apr 28, 2015. doi: 10.3748/wjg.v21.i16.5081
Peer-review started: August 21, 2014
First decision: September 27 ,2014
Revised: October 28, 2014
Accepted: December 5, 2014
Article in press: December 8, 2014
Published online: April 28, 2015
Processing time: 249 Days and 9.9 Hours
AIM: To evaluate the relationship between apurinic endonuclease 1 (APE1) Asp148Glu polymorphism and the susceptibility to gastrointestinal (GI) cancers.
METHODS: We searched PubMed, ISI Web of Knowledge, and Chinese National Knowledge Infrastructure (CNKI) databases updated on July 15, 2014 for relevant studies. Only case-control studies comparing APE1 Asp148Glu polymorphism and GI cancer risk were included. We excluded studies reporting only standardized incidence ratios without control groups and those without detailed genotyping data. Meta-analysis was performed on 17 studies involving 4856 cancer patients and 6136 cancer-free controls. Review Manager version 5.1 was used to perform the meta-analysis. The pooled odds ratios (ORs) and 95% confidence intervals (CIs) were estimated under the allele contrast, homozygous, heterozygous, dominant and recessive genetic models. We also conducted subgroup analyses stratified by ethnicity and cancer type. Publication bias was evaluated using Begg’s test.
RESULTS: The meta-analysis showed a significant association between APE1 Asp148Glu polymorphism and GI cancer risk in three genetic models in the overall population (G vs T: OR = 1.18; 95%CI: 1.05-1.32; TG vs TT: OR = 1.28; 95%CI: 1.08-1.52; TG + GG vs TT: OR = 1.32; 95%CI: 1.10-1.57). Stratified analysis by ethnicity revealed a statistically increased GI cancer risk in Asians (G vs T: OR = 1.27; 95%CI: 1.07-1.51; GG vs TT: OR = 1.58; 95%CI: 1.05-2.38; TG vs TT: OR = 1.30; 95%CI, 1.01- 1.67; and TG + GG vs TT: OR = 1.38; 95%CI: 1.07-1.78), but not in Caucasians. Further subgroup analysis by cancer type indicated that APE1 Asp148Glu polymorphism may contribute to gastric cancer risk. However, Asp148Glu has no significant association with colorectal or esophageal cancer risk in any genetic model.
CONCLUSION: This meta-analysis suggests that the APE1 Asp148Glu polymorphism G allele is associated with an increased GI cancer risk, especially in gastric cancer.
Core tip: Apurinic endonuclease 1 (APE1) plays an important role in the DNA repair system and therefore has been implicated in human carcinogenesis. Many studies have suggested an association between the APE1 Asp148Glu polymorphism and gastrointestinal cancer susceptibility. However, the results remained inconclusive. We performed a meta-analysis on pooled data from previously published studies. The results showed that the APE1 Asp148Glu polymorphism G allele is associated with an increased gastrointestinal cancer risk, especially in gastric cancer.
- Citation: Dai ZJ, Shao YP, Kang HF, Tang W, Xu D, Zhao Y, Liu D, Wang M, Yang PT, Wang XJ. Relationship between apurinic endonuclease 1 Asp148Glu polymorphism and gastrointestinal cancer risk: An updated meta-analysis. World J Gastroenterol 2015; 21(16): 5081-5089
- URL: https://www.wjgnet.com/1007-9327/full/v21/i16/5081.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i16.5081
Gastrointestinal (GI) cancers, especially esophageal, gastric, and colorectal cancers, are the leading causes of cancer-related death worldwide[1]. GI cancers are multifactorial diseases caused by complex interactions between many genetic and environmental factors[1,2]. Allelic variations in oncogenes are candidate genetic risk factors that may alter the onset and outcome of GI cancers[3].
Apurinic/apyrimidinic endonuclease 1 (APE1) is an essential enzyme in the base excision repair pathway[4]. APE1 plays an important role in the DNA repair system and therefore has been implicated in human carcinogenesis[5]. The human APE1 is located on chromosome 14q11.2 and consists of five exons, spanning roughly 2.5 to 3 kb of DNA[6]. Many single nucleotide polymorphisms in the APE1 gene have been reported, including the commonly occurring Asp148Glu in the fifth exon and -141T/G in the promoter region[7]. These nonconservative amino acid alterations have been reported to reduce the DNA repair activity of APE1 and consequently increase cancer risk[8]. Our previous study has suggested that the APE1 -141T/G but not the Asp148Glu polymorphism may influence the susceptibility to and progression of breast cancer in the Chinese population[9]. However, a recent study reported that the APE1 148 GG genotype is associated with an increased risk of colorectal cancer (CRC)[10].
It is important to summarize inconclusive results from different studies to further validate the association of one polymorphism with cancer risk[11]. To clarify the role of the APE1 Asp148Glu polymorphism in GI cancer risk, we performed a meta-analysis on all eligible case-control studies to estimate the overall cancer risk associated with the APE1 Asp148Glu polymorphism. Furthermore, we conducted subgroup analyses stratified by ethnicity and cancer type.
The procedures performed in this meta-analysis are in accordance with recent guidelines for the reporting of meta-analyses (PRISMA guidelines).
We searched the electronic databases of PubMed, Web of Knowledge and Chinese National Knowledge Infrastructure databases to collect articles reporting case-control studies related to the association of APE1 Asp148Glu polymorphisms with GI cancer risk. The keywords used for search were as follows: apurinic/apyrimidinic endonuclease-1/APE1/APEX/HAP1/REF-1, gastrointestinal/esophageal/gastric/colorectal, cancer/carcinoma/tumor/neoplasm, polymorphism/genotype/SNP/variation. The latest search was updated on July 15, 2014. Furthermore, reference lists of main reports and review articles were also reviewed manually to identify additional relevant publications.
The following criteria were used to select studies for further meta-analysis: (1) case-control studies; (2) studies that evaluated the associations between APE1 Asp148Glu polymorphism and GI cancer risk; (3) studies that contained at least two comparison groups (cancer vs control); and (4) studies that included detailed genotyping data.
Articles were reviewed independently by two reviewers and data with discrepancies were discussed by all authors. For each included study, the following information was collected: first author, year of publication, country of origin, ethnicity, source of control, total numbers of cases and controls, genotyping methods as well as numbers of cases and controls with the different genotypes. Different ethnic groups were categorized as Caucasian, Asian, African, and “mixed”. All the case and control groups were well controlled. The non-cancer controls had no history of gynecologic disease, and there was no present evidence of any malignant disease.
The associations between APE1 Asp148Glu polymorphism and GI cancer risk were measured by odds ratio (OR) with 95% confidence interval (CI). The significance of the pooled OR was determined by the Z-test.
The meta-analysis assessed association by using five different genetic models: (1) allele contrast genetic model - A vs a (where ‘‘a’’ is the wild-type allele and ‘‘A’’ is the variant allele); (2) homozygous genetic model-comparison between the 2 homozygous genotypes (AA vs aa); (3) heterozygous genetic model-comparison between the heterozygous and homozygous wild-type genotype groups (Aa vs aa); (4) dominant genetic model-comparison between the wild-type homozygous genotype vs the variant allele-positive genotypes (AA + Aa vs aa); and (5) recessive genetic model-comparison between the variant homozygous genotype vs the rest (AA vs aa + Aa).
Statistical heterogeneity among studies was assessed with the Q and I2 statistics. If the P-value of heterogeneity test was more than 0.1 (P≥ 0.1), the pooled OR estimate of the study was calculated using the fixed-effects model. Otherwise, the random-effects model was used[11]. The value of the I index was used to assess the degree of heterogeneity (I2 < 25%: no heterogeneity; 25% < I2 < 50%: moderate heterogeneity; 50% < I2 < 75%: high heterogeneity; I2 > 75%: extremely high heterogeneity). Publication bias was evaluated by the funnel plot and further assessed by the method of Egger’s linear regression test. All statistical analyses were carried out with the Review Manager version 5.1 (Revman; The Cochrane Collaboration, Oxford, United Kingdom).
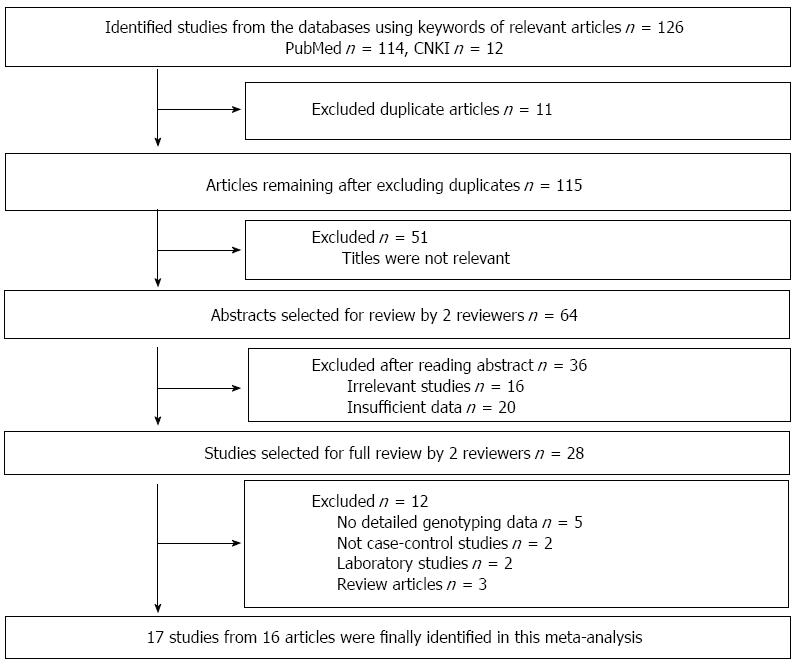
As shown in Figure 1, a total of 126 potential publications were initially extracted. After reading the abstracts, we excluded 67 irrelevant studies, 20 studies with insufficient data, and 11 duplicated studies. After reading the full-texts, we excluded 5 articles with no detailed genotyping data, 2 non-case-control studies, 2 laboratory studies and 3 review articles. Finally, 17 studies from 16 articles were included in this meta-analysis.
Overall, 17 studies on APE1 Asp148Glu polymorphism and GI cancer risk were identified[10,12-26], including a total of 4856 cases and 6136 case-free controls. The characteristics of the included studies are listed in Table 1. Among the eligible 17 studies, nine were carried out in Caucasians from United States, Italy, Czech, Spain, Poland and Turkey, seven were based on Asian background and carried out in China and Japan, and one was based on mixed ethnic groups. All studies were case-controlled, including 11 CRC studies, 4 gastric cancer (GC) studies and 2 esophageal cancer (EC) studies. All GI cancers were confirmed by histology or pathology. The histological type of cancers in the included studies was adenocarcinoma except one EC study[26]. Moreover, controls were matched mainly by age. Eleven studies were population-based and six were hospital-based. Several genotyping methods were used in the studies, including polymerase chain reaction-restriction fragment length polymorphism, PCR-ligase detection reaction, TaqMan, MassARRAY, and Arrayed primer extension.
| Ref. | Year | Country | Ethnicity | Cancer type | Genotyping method | Source of controls | Total sample size (cases/controls) |
| Zhang et al[10] | 2014 | China | Asian | CRC | PCR-CTPP | PB | 247/300 |
| Li et al[12] | 2013 | China | Asian | CRC | PCR-RFLP | HB | 451/631 |
| Canbay et al[13] | 2011 | Turkey | Caucasian | CRC | PCR-RFLP | PB | 79/247 |
| Gu et al[14] | 2011 | China | Asian | GC | PCR-RFLP | PB | 572/547 |
| Li et al[15] | 2011 | China | Asian | GC | PCR-RFLP | PB | 126/156 |
| Canbay et al[16] | 2010 | Turkey | Caucasian | GC | PCR-RFLP | PB | 40/247 |
| Brevik et al[17] | 2010 | United States | Caucasian | CRC | TaqMan | HB | 304/359 |
| Jelonek et al[18] | 2010 | Poland | Caucasian | CRC | PCR-RFLP | PB | 153/273 |
| Palli et al[19] | 2010 | Italy | Caucasian | GC | TaqMan | PB | 314/548 |
| Ye et al[20] | 2010 | China | Asian | CRC | MassARRAY | HB | 123/158 |
| Kasahara et al[21] | 2008 | Japan | Asian | CRC | PCR-RFLP | HB | 68/121 |
| Pardini et al[22] | 2008 | Czech | Caucasian | CRC | TaqMan | HB | 532/532 |
| Tse et al[23] | 2008 | United States | Caucasian | EC | TaqMan | HB | 312/454 |
| Berndt et al[24] | 2007 | United States | Caucasian | CRC | TaqMan | PB | 720/725 |
| Berndt et al[24] | 2007 | United States | Mixed | CRC | TaqMan | PB | 47/48 |
| Moreno et al[25] | 2006 | Spain | Caucasian | CRC | Arrayed primer extension | PB | 359/312 |
| Hao et al[26] | 2004 | China | Asian | EC | PCR-RFLP | PB | 409/478 |
As shown in Table 2, the frequency of the G allele varied widely across the 12 studies, ranging from 0.31 to 0.54. The average frequency of the G allele in the overall population, Caucasian population and Asian population was 0.47, 0.46, and 0.47, respectively. There was no significant difference between Asians and Caucasians (P > 0.05). The average frequency of the G allele in CRC, GC and EC was 0.46, 0.50, and 0.46, respectively.
| Ref. | Genotype (n) | Allele frequency (n) | MAF | ||||||||||
| Cases | Controls | Cases | Controls | ||||||||||
| Total | TT | TG | GG | Total | TT | TG | GG | T | G | T | G | ||
| Zhang et al[10] | 247 | 87 | 90 | 70 | 300 | 121 | 137 | 41 | 264 | 230 | 381 | 219 | 0.47 |
| Li et al[12] | 451 | 123 | 247 | 81 | 631 | 186 | 335 | 110 | 493 | 409 | 707 | 555 | 0.45 |
| Canbay et al[13] | 79 | 28 | 43 | 8 | 247 | 151 | 63 | 33 | 99 | 59 | 365 | 129 | 0.37 |
| Gu et al[14] | 338 | 69 | 185 | 84 | 362 | 110 | 183 | 69 | 323 | 353 | 403 | 321 | 0.52 |
| Li et al[15] | 126 | 26 | 64 | 36 | 156 | 56 | 70 | 30 | 116 | 136 | 182 | 130 | 0.54 |
| Canbay et al[16] | 40 | 14 | 18 | 8 | 247 | 151 | 63 | 33 | 46 | 34 | 365 | 129 | 0.42 |
| Brevik et al[17] | 304 | 102 | 137 | 65 | 359 | 108 | 167 | 84 | 341 | 267 | 383 | 335 | 0.44 |
| Jelonek et al[18] | 113 | 49 | 59 | 5 | 273 | 70 | 141 | 62 | 157 | 69 | 163 | 143 | 0.31 |
| Palli et al[19] | 298 | 103 | 147 | 48 | 546 | 208 | 243 | 95 | 353 | 243 | 659 | 433 | 0.41 |
| Ye et al[20] | 123 | 37 | 86 | 0 | 158 | 52 | 106 | 0 | 160 | 86 | 210 | 106 | 0.35 |
| Kasahara et al[21] | 68 | 23 | 45 | 0 | 121 | 70 | 51 | 0 | 91 | 45 | 191 | 51 | 0.33 |
| Pardini et al[22] | 531 | 140 | 261 | 130 | 530 | 157 | 267 | 106 | 541 | 521 | 581 | 479 | 0.49 |
| Tse et al[23] | 311 | 75 | 162 | 74 | 454 | 123 | 228 | 103 | 312 | 310 | 474 | 434 | 0.50 |
| Berndt et al[24] | 692 | 175 | 364 | 153 | 710 | 204 | 335 | 171 | 714 | 670 | 743 | 677 | 0.48 |
| Berndt et al[24] | 47 | 11 | 23 | 13 | 48 | 18 | 22 | 7 | 45 | 49 | 58 | 36 | 0.48 |
| Moreno et al[25] | 359 | 95 | 177 | 87 | 312 | 99 | 147 | 66 | 367 | 351 | 345 | 279 | 0.49 |
| Hao et al[26] | 409 | 126 | 211 | 72 | 477 | 149 | 243 | 95 | 463 | 355 | 541 | 433 | 0.43 |
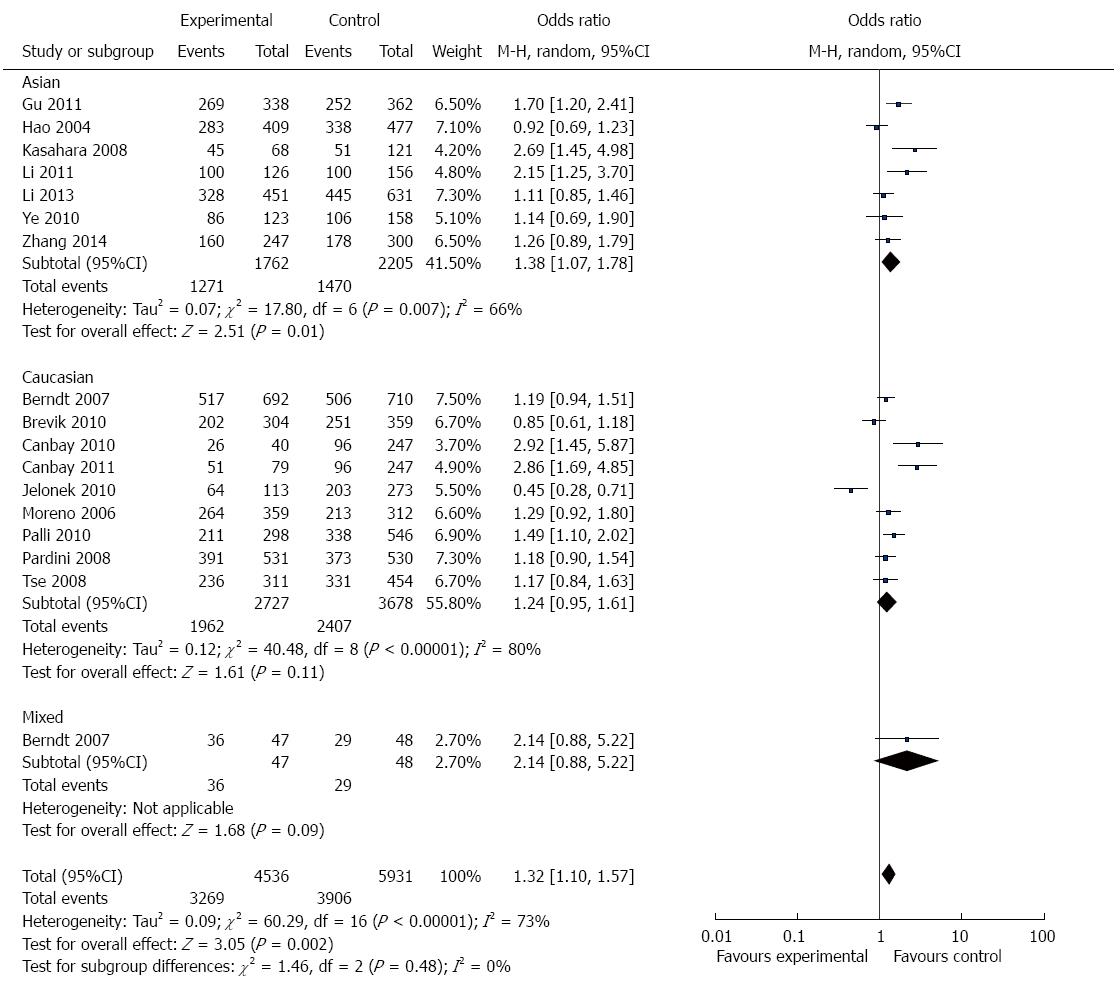
Overall, there was evidence of an association between GI cancer risk and the variant genotypes when all the eligible studies were pooled into the meta-analysis. As show in Figure 2 and Table 3, there was a significant association between APE1 Asp148Glu polymorphism and GI cancer risk in three genetic models in the overall population (G vs T: OR = 1.18, 95%CI = 1.05-1.32, P = 0.004; TG vs TT: OR =1.28, 95%CI = 1.08-1.52, P = 0.004; TG + GG vs TT: OR = 1.32, 95%CI =1.10-1.57, P = 0.002). However, there was no significant association in the other two genetic models (GG vs TT: OR = 1.25, 95%CI = 0.98-1.59, P = 0.07; GG vs TT + TG: OR = 1.10, 95%CI = 0.90-1.34, P = 0.33).
| Comparison | OR | 95%CI | P value | Heterogeneity | Effects model | |
| I2 | P value | |||||
| G vs T | 1.18 | 1.05-1.32 | 0.0041 | 71% | < 0.00001 | Random |
| Caucasian | 1.09 | 0.94-1.26 | 0.24 | 72% | 0.0004 | Random |
| Asian | 1.27 | 1.07-1.51 | 0.0071 | 70% | 0.003 | Random |
| Colorectal cancer | 1.15 | 0.99-1.33 | 0.06 | 73% | < 0.0001 | Random |
| Gastric cancer | 1.41 | 1.09-1.83 | 0.0091 | 70% | 0.02 | Random |
| Esophageal cancer | 1.01 | 0.88-1.16 | 0.87 | 0% | 0.40 | Fixed |
| GG vs TT | 1.25 | 0.98-1.59 | 0.07 | 73% | < 0.00001 | Random |
| Caucasian | 1.04 | 0.77-1.41 | 0.81 | 72% | 0.0003 | Random |
| Asian | 1.58 | 1.05-2.38 | 0.031 | 76% | 0.002 | Random |
| Colorectal cancer | 1.12 | 0.79-1.59 | 0.51 | 79% | < 0.00001 | Random |
| Gastric cancer | 1.77 | 1.11-2.84 | 0.021 | 63% | 0.04 | Random |
| Esophageal cancer | 1.02 | 0.77-1.35 | 0.90 | 0% | 0.34 | Fixed |
| TG vs TT | 1.28 | 1.08-1.52 | 0.0041 | 68% | < 0.0001 | Random |
| Caucasian | 1.26 | 0.98-1.63 | 0.07 | 76% | <0.0001 | Random |
| Asian | 1.30 | 1.01-1.67 | 0.041 | 63% | 0.01 | Random |
| Colorectal cancer | 1.23 | 0.98-1.54 | 0.08 | 73% | < 0.0001 | Random |
| Gastric cancer | 1.66 | 1.20-2.31 | 0.0021 | 51% | < 0.0001 | Random |
| Esophageal cancer | 1.04 | 0.83-1.31 | 0.72 | 0% | 0.41 | Fixed |
| GG+TG vs TT | 1.32 | 1.10-1.57 | 0.0021 | 73% | < 0.00001 | Random |
| Caucasian | 1.24 | 0.95-1.61 | 0.11 | 80% | < 0.00001 | Random |
| Asian | 1.38 | 1.07-1.78 | 0.011 | 66% | 0.007 | Random |
| Colorectal cancer | 1.23 | 0.98-1.55 | 0.08 | 75% | < 0.0001 | Random |
| Gastric cancer | 1.77 | 1.40-2.24 | < 0.00011 | 19% | 0.29 | Fixed |
| Esophageal cancer | 1.02 | 0.81-1.29 | 0.84 | 9% | 0.29 | Fixed |
| GG vs TT+TG | 1.10 | 0.90-1.34 | 0.33 | 70% | < 0.0001 | Random |
| Caucasian | 0.95 | 0.75-1.20 | 0.66 | 64% | 0.004 | Random |
| Asian | 1.36 | 0.95-1.95 | 0.10 | 77% | 0.002 | Random |
| Colorectal cancer | 1.05 | 0.77-1.43 | 0.76 | 79% | < 0.00001 | Random |
| Gastric cancer | 1.25 | 0.99-1.56 | 0.06 | 33% | 0.21 | Fixed |
| Esophageal cancer | 0.96 | 0.75-1.21 | 0.71 | 0% | 0.38 | Fixed |
In the stratified analysis by population, as shown in Figure 2 and Table 3, meta-analysis showed that Asp148Glu polymorphism had no association with GI cancer risk in Caucasians in all genetic models (allele contrast genetic model: OR = 1.09, 95%CI = 0.94-1.26, P = 0.24; homozygote comparison: OR = 1.25, 95%CI = 0.98-1.59, P = 0.81; heterozygote comparison: OR = 1.26, 95%CI = 0.98- 1.63, P = 0.07; the dominant model: OR = 1.24, 95%CI = 0.95-1.61, P = 0.11; and the recessive model: OR = 0.95, 95%CI = 0.75-1.20, P = 0.66).
There were 7 studies with 1587 cases and 1913 controls for assessing the relationship between Asp148Glu polymorphism and GI cancer susceptibility in Asians. As shown in Figure 2 and Table 3, Asp148Glu polymorphism was significantly associated with GI cancer risk in four genetic models (allele contrast genetic model: OR = 1.27, 95%CI = 1.07-1.51, P = 0.007; homozygote comparison: OR = 1.58, 95%CI = 1.05-2038, P = 0.03; heterozygote comparison: OR = 1.30, 95%CI = 1.01-1.67, P = 0.04; and the dominant model: OR = 1.38, 95%CI = 1.07-1.78, P = 0.01.
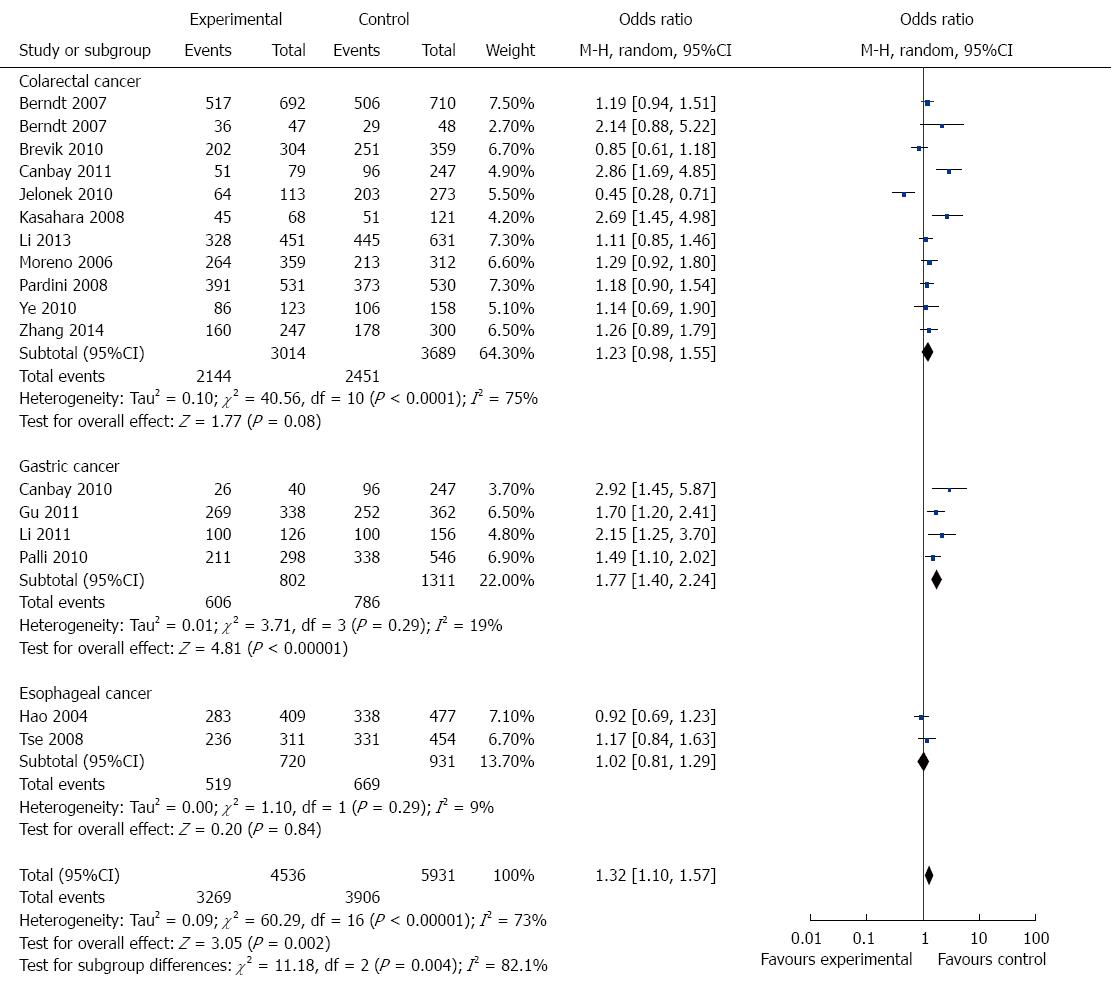
In the stratified analysis by cancer type, 11 studies including 3083 cases and 3706 controls were used to evaluate the relationship between APE1 Asp148Glu polymorphism and CRC risk. As shown in Table 3 and Figure 3, there was no significant association between APE1 Asp148Glu polymorphism and CRC risk under all genetic models (allele contrast genetic model: OR = 1.15, 95%CI = 0.99-1.33, P = 0.06; homozygote comparison: OR = 1.12, 95%CI = 0.79-1.59, P = 0.51; heterozygote comparison: OR = 1.23, 95%CI = 0.98-1.54, P = 0.08; the dominant model: OR = 1.23, 95%CI = 0.98-1.55, P = 0.08; and the recessive model: OR = 1.05, 95%CI = 0.77-1.43, P = 0.76).
There were four studies including 1052 cases and 1498 controls used to evaluate the relationship between APE1 Asp148Glu polymorphism and GC risk. As shown in Table 3 and Figure 3, Asp148Glu polymorphism was significantly associated with an increased risk of GC under all genetic models (allele contrast genetic model: OR = 1.41, 95%CI = 1.09-1.83, P = 0.009; homozygote comparison: OR = 1.77, 95%CI = 1.11-2.84, P = 0.02; heterozygote comparison: OR = 1.66, 95%CI = 1.20-2.31, P = 0.002; and the dominant model: OR = 1.77, 95%CI = 1.40-2.24, P < 0.0001).
There were only two EC studies included in this meta-analysis. As shown in Table 3 and Figure 3, no significant association was detected between Asp148Glu polymorphism and EC risk.
The influence of any single study on the overall estimate was analyzed by excluding one study at a time. No significant difference was observed when any of the studies was excluded. Therefore, our results were statistically reliable.
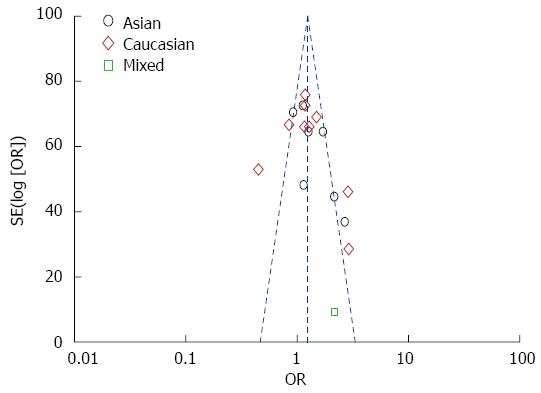
Funnel plot and Egger’s test were performed to evaluate the publication bias. As shown in Figure 4, the funnel plots failed to detect any obvious asymmetry in all genotypes in the overall population, and the Egger’s test revealed no publication bias (P > 0.05). Therefore, no significant publication bias was found in this meta-analysis.
The present meta-analysis, including 4856 cases and 6136 case-free controls from 17 case-control studies, was conducted to evaluate the association between APE1 Asp148Glu polymorphism and GI cancer risk. Our results indicated that the variant genotypes were associated with an increased risk of GI cancers, especially GC.
APE1 is a key multifunctional gene involved in the base excision repair pathway. It was reported that APE1 is associated with aggressive tumor biology and has an impact on survival of GC patients[27]. The Asp148Glu polymorphism is a common non-synonymous APE1 coding region variant. Previous studies on the association between the APE1 Asp148Glu polymorphism and GI cancer risk have shown controversial results[12-26].
In a previous meta-analysis, Gu et al[28] suggested that the APE1 Asp148Glu polymorphism may contribute to genetic susceptibility to cancers, especially CRC. However, a recent meta-analysis by Shen et al[29] failed to detect an association between APE1 Asp148Glu polymorphism and CRC risk. We found several worthwhile queries in Shen’s study. First, the ethnicity of Canbay’s study was identified as Asian in Shen’s meta-analysis[29]. While in the original article, the cases and controls were both based on Caucasians but not Asians[13]. Second, the study by Berndt et al[24] was carried out in Caucasians and a mixed non-Caucasian population, and therefore should probably be divided into two studies. Third, non-English literature database such as the CNKI should also be considered for the search of eligible case-control studies. We have found one eligible study published in Chinese and included it in this study[20].
In our study, 17 case-control studies were included. There was a significant association between APE1 Asp148Glu polymorphism and GI cancers risk in four genetic models in the overall population. Stratified analysis by ethnicity revealed that there was a statistically increased GI cancers risk in Asians. Further subgroup analysis by cancer type indicated that APE1 Asp148Glu polymorphism may contribute to GC risk.
There are some limitations of this meta-analysis that should be noted. First, this meta-analysis was based on pooled data while no individual data were available; thus, we could not assess the risk of cancer based on environmental factors, age, and other risk factors for GI cancers. Second, the small study effect, where the effects reported in small studies are larger, could not be avoided in some studies of a relative small size (< 200). Third, there was no significant association between Asp148Glu polymorphism and EC risk in this meta-analysis. Since only two EC studies with different pathological types[23,26] were included, this negative finding may result from a lack of statistical power. Larger scale multicenter studies are warranted to further validate the association between APE1 Asp148Glu polymorphism and GI cancer risk.
In conclusion, our present meta-analysis provides evidence for the association between the APE1 Asp148Glu polymorphism and GI cancer risk. Results suggest that the APE1 Asp148Glu G allele was associated with an increased GI cancer risk among Asian subjects. Furthermore, the APE1 Asp148Glu polymorphism was associated with an increased risk GC. Further large-population based studies are needed to confirm the association between APE1 Asp148Glu polymorphism and EC risk.
Epidemiological studies have suggested that Asp148Glu polymorphism in the apurinic endonuclease 1 (APE1) gene is associated with gastrointestinal (GI) cancer risk. However, the results are still controversial.
APE1 plays an important role in the DNA repair system and therefore has been implicated in human carcinogenesis. Epidemiologic studies suggested that single nucleotide polymorphisms (SNP) in APE1 may confer individuals’ susceptibility to cancer. Recently, numerous studies have evaluated the association between APE Asp148Glu polymorphism and cancer risk. However, the results remain inconclusive.
The present meta-analysis was performed on all eligible case-control studies to estimate the association of the APE1 Asp148Glu polymorphism with GI cancer risk.
The present meta-analysis showed that the APE1 Asp148Glu G allele was associated with an increased GI cancer risk among Asian subjects. Furthermore, APE1 Asp148Glu polymorphism was associated with an increased risk of GC.
SNP is DNA sequence variation occurring when a single nucleotide in the genome differs between members of a biological species or paired chromosomes. SNP in some genes may cause an increase or decrease in risk for some certain diseases.
This meta-analysis showed that the G allele of APE1 Asp148Glu polymorphism was associated with a higher gastrointestinal tract cancer risk. However, larger scale studies are warranted to further validate the association between this polymorphism and GI cancer risk.
P- Reviewer: Jiang ZY S- Editor: Yu J L- Editor: Wang TQ E- Editor: Zhang DN
| 1. | Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9215] [Cited by in RCA: 9856] [Article Influence: 821.3] [Reference Citation Analysis (4)] |
| 2. | Haq S, Ali S, Mohammad R, Sarkar FH. The complexities of epidemiology and prevention of gastrointestinal cancers. Int J Mol Sci. 2012;13:12556-12572. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 3. | Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513-1530. [PubMed] |
| 4. | Li M, Wilson DM. Human apurinic/apyrimidinic endonuclease 1. Antioxid Redox Signal. 2014;20:678-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 205] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 5. | Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid Redox Signal. 2005;7:367-384. [PubMed] |
| 6. | Tell G, Fantini D, Quadrifoglio F. Understanding different functions of mammalian AP endonuclease (APE1) as a promising tool for cancer treatment. Cell Mol Life Sci. 2010;67:3589-3608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 105] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 7. | Dai ZJ, Wang XJ, Kang AJ, Ma XB, Min WL, Lin S, Zhao Y, Yang PT, Wang M, Kang HF. Association between APE1 Single Nucleotide Polymorphism (rs1760944) and Cancer Risk: a Meta-Analysis Based on 6,419 Cancer Cases and 6,781 Case-free Controls. J Cancer. 2014;5:253-259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 8. | Lo YL, Jou YS, Hsiao CF, Chang GC, Tsai YH, Su WC, Chen KY, Chen YM, Huang MS, Hu CY. A polymorphism in the APE1 gene promoter is associated with lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18:223-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 289] [Reference Citation Analysis (0)] |
| 9. | Kang H, Dai Z, Ma X, Ma L, Jin Y, Liu X, Wang X. A genetic variant in the promoter of APE1 gene (-656 T& gt; G) is associated with breast cancer risk and progression in a Chinese population. Gene. 2013;531:97-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 10. | Zhang SH, Wang LA, Li Z, Peng Y, Cun YP, Dai N, Cheng Y, Xiao H, Xiong YL, Wang D. APE1 polymorphisms are associated with colorectal cancer susceptibility in Chinese Hans. World J Gastroenterol. 2014;20:8700-8708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 14] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 11. | Gao J, Kang AJ, Lin S, Dai ZJ, Zhang SQ, Liu D, Zhao Y, Yang PT, Wang M, Wang XJ. Association between MDM2 rs 2279744 polymorphism and breast cancer susceptibility: a meta-analysis based on 9,788 cases and 11,195 controls. Ther Clin Risk Manag. 2014;10:269-277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 12. | Li Y, Li S, Wu Z, Hu F, Zhu L, Zhao X, Cui B, Dong X, Tian S, Wang F. Polymorphisms in genes of APE1, PARP1, and XRCC1: risk and prognosis of colorectal cancer in a northeast Chinese population. Med Oncol. 2013;30:505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 13. | Canbay E, Cakmakoglu B, Zeybek U, Sozen S, Cacina C, Gulluoglu M, Balik E, Bulut T, Yamaner S, Bugra D. Association of APE1 and hOGG1 polymorphisms with colorectal cancer risk in a Turkish population. Curr Med Res Opin. 2011;27:1295-1302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 14. | Gu D, Wang M, Wang S, Zhang Z, Chen J. The DNA repair gene APE1 T1349G polymorphism and risk of gastric cancer in a Chinese population. PLoS One. 2011;6:e28971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 15. | Li ZH. Association between DNA repair gene polymorphisms and environmental factors and Hp-associated gastric cancer and duodenal ulcer [Master]. Nanchang, China: Nanchang University 2011; . |
| 16. | Canbay E, Agachan B, Gulluoglu M, Isbir T, Balik E, Yamaner S, Bulut T, Cacina C, Eraltan IY, Yilmaz A. Possible associations of APE1 polymorphism with susceptibility and HOGG1 polymorphism with prognosis in gastric cancer. Anticancer Res. 2010;30:1359-1364. [PubMed] |
| 17. | Brevik A, Joshi AD, Corral R, Onland-Moret NC, Siegmund KD, Le Marchand L, Baron JA, Martinez ME, Haile RW, Ahnen DJ. Polymorphisms in base excision repair genes as colorectal cancer risk factors and modifiers of the effect of diets high in red meat. Cancer Epidemiol Biomarkers Prev. 2010;19:3167-3173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 18. | Jelonek K, Gdowicz-Klosok A, Pietrowska M, Borkowska M, Korfanty J, Rzeszowska-Wolny J, Widlak P. Association between single-nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a Polish population. J Appl Genet. 2010;51:343-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 19. | Palli D, Polidoro S, D’Errico M, Saieva C, Guarrera S, Calcagnile AS, Sera F, Allione A, Gemma S, Zanna I. Polymorphic DNA repair and metabolic genes: a multigenic study on gastric cancer. Mutagenesis. 2010;25:569-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 84] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 20. | Ye CC, Huang ZM, Zhou CY. APE1 D148E, PARP1 V762A and XRCC1 R399Q polymorphisms and genetic susceptibility to colorectal cancer. Shijie Huaren Xiaohua Zazhi. 2010;18:1275-1279. |
| 21. | Kasahara M, Osawa K, Yoshida K, Miyaishi A, Osawa Y, Inoue N, Tsutou A, Tabuchi Y, Tanaka K, Yamamoto M. Association of MUTYH Gln324His and APEX1 Asp148Glu with colorectal cancer and smoking in a Japanese population. J Exp Clin Cancer Res. 2008;27:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 82] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 22. | Pardini B, Naccarati A, Novotny J, Smerhovsky Z, Vodickova L, Polakova V, Hanova M, Slyskova J, Tulupova E, Kumar R. DNA repair genetic polymorphisms and risk of colorectal cancer in the Czech Republic. Mutat Res. 2008;638:146-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 95] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 23. | Tse D, Zhai R, Zhou W, Heist RS, Asomaning K, Su L, Lynch TJ, Wain JC, Christiani DC, Liu G. Polymorphisms of the NER pathway genes, ERCC1 and XPD are associated with esophageal adenocarcinoma risk. Cancer Causes Control. 2008;19:1077-1083. [PubMed] |
| 24. | Berndt SI, Huang WY, Fallin MD, Helzlsouer KJ, Platz EA, Weissfeld JL, Church TR, Welch R, Chanock SJ, Hayes RB. Genetic variation in base excision repair genes and the prevalence of advanced colorectal adenoma. Cancer Res. 2007;67:1395-1404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 53] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 25. | Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A, Blanco I, González S, Guino E, Capellà G, Canzian F. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12:2101-2108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 190] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 26. | Hao B, Wang H, Zhou K, Li Y, Chen X, Zhou G, Zhu Y, Miao X, Tan W, Wei Q. Identification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinoma. Cancer Res. 2004;64:4378-4384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 181] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 27. | Zhao Q, Wang W, Zhang Z, Wang S, Wang M, Zhou J, Gong W, Tan Y, Wang B, Chen G. A genetic variation in APE1 is associated with gastric cancer survival in a Chinese population. Cancer Sci. 2011;102:1293-1297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 28. | Gu D, Wang M, Wang M, Zhang Z, Chen J. The DNA repair gene APE1 T1349G polymorphism and cancer risk: a meta-analysis of 27 case-control studies. Mutagenesis. 2009;24:507-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 29. | Shen E, Liu C, Wei L, Hu J, Weng J, Yin Q, Wang Y. The APE1 Asp148Glu polymorphism and colorectal cancer susceptibility: a meta-analysis. Tumour Biol. 2014;35:2529-2535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |