Published online Apr 7, 2015. doi: 10.3748/wjg.v21.i13.3983
Peer-review started: September 23, 2014
First decision: October 14, 2014
Revised: November 1, 2014
Accepted: December 5, 2014
Article in press: December 8, 2014
Published online: April 7, 2015
Processing time: 197 Days and 5.2 Hours
AIM: To investigate the role of signal transduction and activation of transcription 4 (STAT4) in the development and progression of human hepatocellular carcinoma (HCC).
METHODS: Recent genetic investigations have identified that a genetic variant of STAT4 is associated with hepatitis B virus (HBV)-related HCC. The level of STAT4 in 90 HCC patients was examined via Western blot and immunohistochemical analyses. The correlation between STAT4 expression and the clinicopathological characteristics of the patients was analyzed. The level of STAT4 expression in the HCC liver tissues was significantly lower than that in the non-HCC liver tissues and correlated with tumor size, histological grade of HCC and serum hepatitis B surface antigen level in HCC patients. The data were statistically analyzed using SPSS. Furthermore, siRNA oligos targeting STAT4 were employed to investigate the influence of STAT4 RNA interference on HCC cell physiology. Based on Cell Counting Kit-8 and flow cytometric assays, we found that depletion of STAT4 expression significantly enhanced the proliferation of L02 cells.
RESULTS: STAT4 protein expression was significantly lower in HCC tissues than in normal liver tissues. Immunohistochemistry followed by statistical analysis revealed that the expression of STAT4 negatively correlated with Ki67 expression (r = 0.851; P < 0.05) and positively correlated with maximal tumor size (P < 0.05), HBV (P = 0.012) and histological grade (P < 0.05). Kaplan-Meier analysis revealed significant differences in the survival curves between HCC patients expressing low and high levels of STAT4 and Ki67 (P < 0.05). Based on a multivariate Cox proportional hazard model, STAT4 expression was an independent prognostic indicator for HCC patients who underwent curative resection. In vitro, following the release of L02 cell lines from serum starvation, the expression of STAT4 was downregulated, and transfection of L02 cells with siRNA targeting STAT4 inhibited cell proliferation.
CONCLUSION: Our data indicate that STAT4 may inhibit HCC development by modulating HCC cell proliferation.
Core tip: In this study, we assessed the role of signal transduction and activation of transcription 4 in hepatocellular carcinoma (HCC) and discussed the possible function of this protein in the development of HCC.
- Citation: Wang G, Chen JH, Qiang Y, Wang DZ, Chen Z. Decreased STAT4 indicates poor prognosis and enhanced cell proliferation in hepatocellular carcinoma. World J Gastroenterol 2015; 21(13): 3983-3993
- URL: https://www.wjgnet.com/1007-9327/full/v21/i13/3983.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i13.3983
Hepatocellular carcinoma (HCC) is among the most common cancers and is the third most common cause of cancer mortality worldwide[1]. Worldwide, more than 600000 new HCC cases are diagnosed annually, among which about 55% are in China[2]. Although intensive efforts have been made by clinical practitioners and basic researchers to identify prognostic markers of and therapeutic targets for HCC, the molecular mechanisms underlying HCC progression remain largely elusive[3,4]. Moreover, effective treatments for HCC are essentially absent. This deficiency emphasizes the urgency to develop new diagnostic and therapeutic strategies for HCC[5].
Signal transducers and activators of transcription (STATs) are members of a well-conserved family of transcription factors that play integral roles in various cellular processes[6,7]. STATs are latent cytoplasmic proteins that are promptly activated by tyrosine phosphorylation by receptor-associated JAK (Janus) kinases in response to cytokine or growth factor exposure. The resulting functional STATs are capable of entering the nucleus, where they directly bind to DNA and activate the transcription of a variety of target genes[8,9]. Generally, STAT proteins regulate cytokine-mediated cell proliferation by modulating the expression of crucial cell cycle regulators, such as cyclin D1, p21 and p27[6]. In addition to cell cycle regulation, STATs modulate various other cellular processes, such as apoptosis, differentiation and migration, via the transcription of various target genes, including Bcl-2 family members, cytokines, matrix metalloproteinases and miRNAs. Accordingly, dysregulated STAT proteins are closely associated with the pathogenesis of human cancers. The hyperactivation of STAT signaling has been widely documented in various cancer types, including ovarian cancer, breast cancer, brain tumors, gastric cancer and colon cancer[10-13]. Consistent with these findings, STATs have been considered as promising therapeutic targets in cancer drug discovery. However, much remains unclear with respect to the expression profiles and roles of STATs in HCC development[14].
Studies have indicated that several members of the STAT family play crucial roles in the pathology of liver diseases. STAT1 has been demonstrated to play a key role in antiviral defense, inflammation, and injury in the liver of STAT1 knockout mice[15-18], and STAT1 negatively regulates HCC cell proliferation[19]. STAT2-deficient mice exhibit an increased susceptibility to viral infections, and the loss of a type I IFN autocrine/paracrine loop indicates that STAT2 performs an antiviral defense function in the liver[20]. STAT3, which is activated by a variety of extracellular signals, has been shown to play key roles in the acute phase response, protection against liver injury, the promotion of liver regeneration, glucose homeostasis, and hepatic lipid metabolism[14]. STAT5 is primarily activated by growth hormone, which regulates the expression of a wide range of hepatic genes, including cytochrome P450, glutathione S-transferase, sulfotransferase enzyme, the growth hormone receptor, serine protease inhibitor Sp12.1, insulin-growth factor I, and hepatocyte growth factor[21,22], which suggests that STAT5 regulates HCC cell proliferation[23,24]. STAT6, which is primarily activated by interleukin (IL)-12, IL-4, and IL-13, plays an important role in Th2 differentiation[6]. The study of STAT4 in liver diseases, compared with other members of the STAT family, is limited. STAT4 was primarily regarded as a transducer of IL-12 signaling, affecting a broad range of immune cell physiology[25,26]. A very recent study by Jiang and colleagues reported that STAT4 might prevent HBV-related hepatocarcinogenesis[27]. However, the precise involvement of STAT4 in HCC development remains unclear.
Our study aimed to investigate the possible involvement of STAT4 in HCC pathology and to evaluate the prognostic value of STAT4 expression for HCC development. We found that STAT4 was significantly downregulated in HCC specimens compared with adjacent nontumorous specimens. Furthermore, we showed that the expression of STAT4 correlated with HBV, the maximal tumor size, the histological grade and Ki67 expression. These findings provide novel insight into the mechanisms underlying HCC development.
Paired samples of tumor and adjacent nontumor tissues were obtained from 90 HCC patients who underwent curative surgery at the Affiliated Cancer Hospital of Nantong University. After surgical removal, a portion of the paired tissue samples was snap-frozen in liquid nitrogen and then maintained at -80 °C until use for protein extraction, and another portion of the paired tissue samples was immediately fixed in formalin and embedded in wax for immunohistochemistry.
The inclusion criteria for all patients in this study were: (1) HCC diagnosis as confirmed by experienced pathologists based on histological examination of HE-stained biopsy sections; (2) no anticancer treatment before surgery; (3) curative resection, defined as the macroscopically complete removal of the tumor and histologically demonstrated tumor-free margins; and (4) the availability of complete clinicopathologic and follow-up data. Informed consent was obtained from each patient, and the study protocols were approved by the Institutional Review Board of the Affiliated Cancer Hospital of Nantong University. The patients included 57 males and 33 females with a mean age of 47.3 years (range: 21-75 years). The primary clinicopathological characteristics of the subjects were recorded. The differentiated tumors were histologically classified as grade I-II (n = 49) or grade III-IV (n = 41). The follow-up duration ranged from 1 to 96 mo.
Tissues were formalin-fixed and paraffin-embedded for immunohistochemical analysis. The sections were dewaxed in xylene and rehydrated in graded ethanol solutions. Then, the sections were processed in 10 mmol/L citrate buffer (pH 6.0) and heated in a microwave at high power (750 W) in 10 mm for three cycles of 5 min each for antigen retrieval. Then, endogenous peroxidase activity was blocked by soaking the sections in 0.3% hydrogen peroxide for 15 min after cooling at room temperature for 1 h. Goat serum was applied for 15 min to block nonspecific reactivity. The sections were incubated overnight at 4 °C in a rabbit anti-human STAT4 polyclonal antibody (diluted 1:100; Santa Cruz Biotechnology), and an anti-Ki-67 mouse monoclonal antibody (diluted 1:100; clone 7B11; Zymed Laboratories, San Francisco, CA, United States). Negative control samples were processed in parallel using a nonspecific immunoglobulin IgG (Sigma Chemical Co, St. Louis, MO) at the same concentration as the primary antibody. All sections were processed using the peroxidase-antiperoxidase method (Dako, Hamburg, Germany). After rinsing in water, the sections were counterstained with hematoxylin, dehydrated, and coverslipped. All of the immunostained sections were evaluated in a blinded manner with respect to the clinical and pathological characteristics of the patients. For the assessment of STAT4 expression, five high-power fields for each specimen were randomly selected, and nuclear (cytoplasmic) staining was examined. More than 500 cells were counted to determine the LI (labeling index) which represented the percentage of immunostained cells relative to the total number of cells. In half of the samples, the staining was repeated twice to avoid possible technical errors, but similar results were obtained for these samples. The results obtained were confirmed by another investigator (J.Y.X.) using a multihead microscope, and a consensus was achieved.
One normal hepatocyte cell line (L02) and 3 HCC cell lines (HepG2, HuH7, and Hep1) were obtained from the Institute of Cell Biology of the Chinese Academy of Sciences and were cultured in RPMI 1640 and Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin (all media were from Invitrogen, Carlsbad, CA, United States) in 5% - at 37 °C.
Two human STAT4 siRNA expression vectors and pSilencer siRNA were constructed. The siRNA sequences targeting the nucleotide residues AAATCCGGCATCTGCTAGCTC and AATTGGATGAACAGTTGGGGC were termed siRNA-1 and -2, respectively. L02 cells were seeded the day before transfection using DMEM containing 10% FBS lacking antibiotics. Transfection was performed using the transfection reagent Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. The cells were incubated in pSilencer vector-Lipofectamine 2000 complexes for 4-6 h at 37 °C, and FBS was added to the DMEM to a final concentration of 10%. The cells were used for subsequent experiments at 48 h after transfection.
Tissue and cellular protein samples were promptly homogenized in homogenization buffer containing 1 mol/L Tris HCl pH7.5, 1% Triton X-100, 1% Nonidet p-40 (NP-40), 10% sodium dodecyl sulfate (SDS), 0.5% sodium deoxycholate, 0.5 mol/L EDTA, 10 μg/mL leupeptin, 10 μg/mL aprotinin, and 1 m mol/L PMSF, followed by centrifugation at 10000 ×g for 30 min to collect the supernatants. The protein concentrations were determined via a Bio-Rad protein assay (Bio-Rad, Hercules, CA, United States). The total cellular protein extracts were separated via SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. The membranes were blocked with 5% nonfat dry milk in PBS for approximately 2 h at room temperature and then incubated in antibodies against STAT4 (1:1000; Santa Cruz Biotechnology) and GAPDH (1:1000; Sigma) in PBS containing 5% milk for approximately 1 h at room temperature. The membranes were washed three times in PBS buffer, followed by incubation in the appropriate horseradish peroxidase-conjugated secondary antibodies (1:500; Santa Cruz Biotechnology). Specific protein bands in the membranes were visualized using an enhanced chemiluminescence reagent (NEN, Boston, MA). Three independent experiments were performed.
Cell proliferation was determined via a Cell Counting Kit-8 (CCK)-8 assay according to the manufacturer’s instructions. In brief, cells which were transfected with siRNA were seeded at a density of 2 × 104 cells/well in a 96-well cell culture dish (Corning, Corning, NY, United States) in 100 μL of culture medium and incubated overnight. CCK-8 (Dojindo, Kumamoto, Japan) reagents were added to a subset of the wells, and the cells were incubated for 2 h at 37 °C. The absorbance was recorded using an automated plate reader. Each experiment was performed in triplicate and repeated at least three times.
For cell cycle analysis, the cells were fixed in 70% ethanol for 1 h at 4 °C and then incubated in 1 mg/mL RNase A for 30 min at 37 °C. Subsequently, the cells were stained with propidium iodide (PI; 50 μg/mL) (Becton-Dickinson, San Jose, CA, United States) in PBS containing 0.5% Tween-20, followed by flow cytometry using a Becton-Dickinson FACScan and Cell Quest acquisition and analysis software. Gating was applied to exclude cell debris, cell doublets, and cell clumps.
Statistical analysis was performed using the SPSS software package. The association between STAT4 expression and the clinicopathological characteristics was analyzed using the χ2 test. The relationship between the STAT4 and Ki-67 expression levels was evaluated using the Spearman rank correlation test because the data were not normally distributed. Survival analysis was performed using the Kaplan-Meier method, and the difference in the survival rates was assessed using the generalized log-rank test. Multivariate analysis was performed using Cox proportional hazard models. The data were expressed as the means ± SE, and P < 0.05 was considered to be significant.
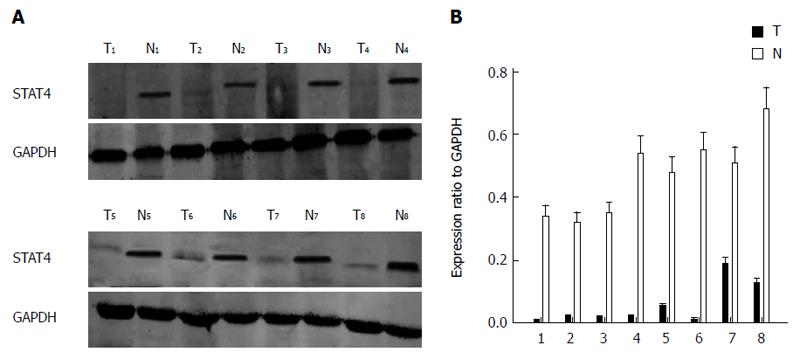
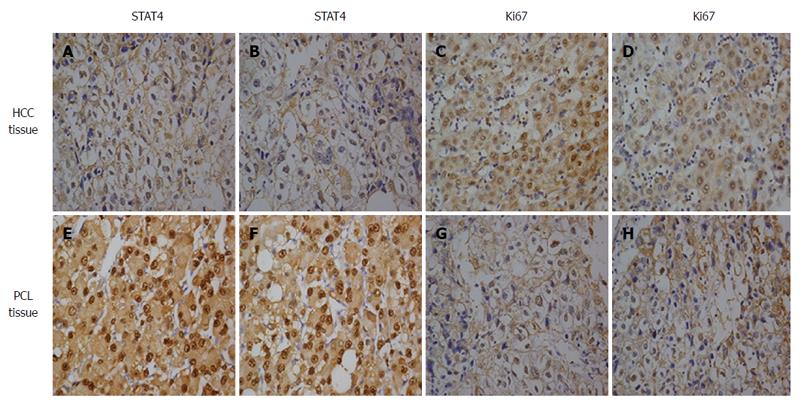
To determine the role of STAT4 in HCC development, we first investigated the expression profile of STAT4 in HCC and non-tumorous tissues via Western blot and immunohistochemical analyses. Western blot analysis of eight paired tumor and adjacent non-tumorous tissues confirmed that STAT4 expression was lower in tumor tissues than in adjacent non-tumorous tissues (Figure 1). Immunohistochemical analysis indicated that STAT4 expression was low or undetectable in most tumorous tissues and was highly expressed in most adjacent nontumor tissues (Figure 2). In addition, the proliferation index of Ki67 was used as a control to indicate the tumorous and non-tumorous tissues.
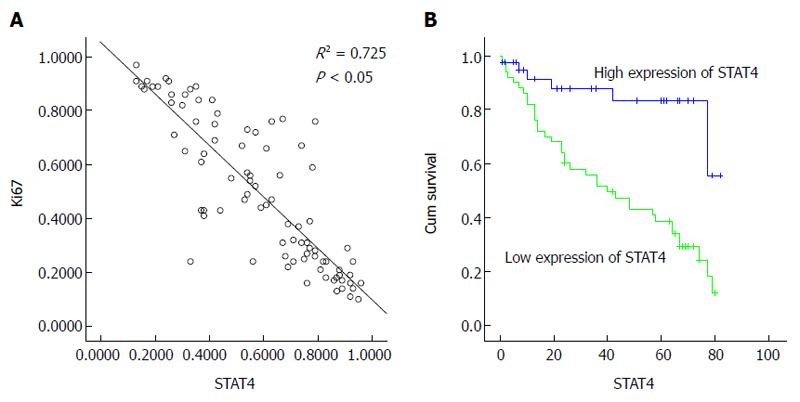
Next, we analyzed the relationship between STAT4 expression and the clinicopathological characteristics of HCC patients. To this end, the patients were stratified into those exhibiting high and low STAT4 expression according to the immunostaining intensity score (high > 0.53, low ≤ 0.53; 0.53 was the mean proportion of STAT4-expressing cells) The clinicopathological and demographic characteristics of the HCC patients are listed in Table 1. Statistical analysis indicated that STAT4 expression significantly correlated with histological grade, HBV infection and the tumor size (P < 0.05) but not to other characteristics, including gender, age, metastasis, the tumor count, the serum AFP level, cirrhosis, and vascular invasion. Furthermore, the patients were stratified into those exhibiting high and low Ki67 expression depending on the expression score for Ki67. We found that Ki67 expression significantly correlated with histological grade and tumor size (P < 0.05) but not with the other characteristics. Very importantly, there was a negative correlation between STAT4 and Ki67 expression (R = 0.85; Figure 3A). These findings suggested that STAT4 downregulation might contribute to HCC progression.
| Clinicopathological features | Total | Stat4 | Ki67 | ||||||
| Low < 0.53 | High > 0.53 | P value | χ2value | Low < 0.48 | High > 0.48 | P value | χ2value | ||
| n = 40 | n = 50 | n = 49 | n = 41 | ||||||
| Age (yr) | |||||||||
| < 45 | 33 | 13 | 20 | 0.463 | 0.538 | 17 | 16 | 0.671 | 0.180 |
| ≥ 45 | 57 | 27 | 30 | 32 | 25 | ||||
| Gender | |||||||||
| Male | 57 | 26 | 31 | 0.769 | 0.086 | 31 | 26 | 0.988 | 0.000 |
| Female | 33 | 14 | 19 | 18 | 15 | ||||
| Serum AFP level (ng/mL) | |||||||||
| < 25 | 34 | 19 | 15 | 0.089 | 2.895 | 18 | 16 | 0.823 | 0.050 |
| ≥ 25 | 56 | 21 | 35 | 31 | 25 | ||||
| Liver cirrhosis | |||||||||
| No | 26 | 13 | 13 | 0.499 | 0.457 | 13 | 13 | 0.589 | 0.291 |
| Yes | 64 | 27 | 37 | 36 | 28 | ||||
| No. of tumor nodes | |||||||||
| Single | 51 | 24 | 27 | 0.568 | 0.326 | 29 | 22 | 0.598 | 0.278 |
| Multiple | 39 | 16 | 23 | 20 | 19 | ||||
| HBV | |||||||||
| Negative | 43 | 25 | 18 | 0.0121 | 6.255 | 27 | 16 | 0.128 | 2.313 |
| Positive | 47 | 15 | 32 | 22 | 25 | ||||
| Maximal tumor size (cm) | |||||||||
| < 4.5 | 40 | 30 | 9 | 0.0001 | 29.403 | 31 | 8 | 0.0001 | 17.402 |
| ≥ 4.5 | 50 | 10 | 41 | 18 | 33 | ||||
| Tumor metastasis | |||||||||
| No | 77 | 36 | 41 | 0.283 | 1.151 | 42 | 35 | 0.963 | 0.002 |
| Yes | 13 | 4 | 9 | 7 | 6 | ||||
| Microvascular invasion | |||||||||
| No | 66 | 30 | 36 | 0.749 | 0.102 | 36 | 30 | 0.975 | 0.001 |
| Yes | 24 | 10 | 14 | 13 | 11 | ||||
| Histological grade | |||||||||
| I-II | 49 | 36 | 13 | 0.0001 | 36.699 | 35 | 14 | 0.000 | 12.510 |
| III-IV | 41 | 4 | 37 | 14 | 27 | ||||
| Ki67 expression | |||||||||
| Low | 49 | 33 | 16 | 0.0001 | 22.849 | ||||
| High | 41 | 7 | 34 | ||||||
The above data indicated that STAT4 downregulation might commonly occur in HCC specimens, indicating the potential value of STAT4 expression for predicting the prognosis of HCC. Therefore, we analyzed whether the STAT4 expression level was indicative of HCC prognosis. Survival analysis was restricted to 90 cases for which complete follow-up data and STAT4 expression scores were available. Kaplan-Meier analysis of the overall survival of these HCC patients demonstrated that the patients displaying low STAT4 expression exhibited significantly worse overall survival than those displaying high STAT4 expression (P < 0.05; Figure 3B). Multivariate analysis using a Cox proportional hazard model showed that STAT4 was an independent prognostic indicator of the overall survival of HCC patients (P < 0.05; Table 2). As shown in Table 3, STAT4 was an independent prognostic indicator based on a multivariate Cox proportional hazard model. These findings convincingly demonstrated that STAT4 may serve as a valuable prognostic marker for HCC prognosis.
| Parameters | Total | Survival Status | P value | χ2value | |
| Dead | Alive | ||||
| n = 43 | n = 47 | ||||
| Age (yr) | |||||
| < 45 | 33 | 16 | 17 | 0.919 | 0.010 |
| ≥ 45 | 57 | 27 | 30 | ||
| Gender | |||||
| Male | 57 | 26 | 31 | 0.589 | 0.292 |
| Female | 33 | 17 | 16 | ||
| Serum AFP level (ug/mL) | |||||
| < 25 | 34 | 15 | 19 | 0.588 | 0.293 |
| ≥ 25 | 56 | 28 | 28 | ||
| Cirrhosis | |||||
| No | 26 | 13 | 13 | 0.788 | 0.072 |
| Yes | 64 | 30 | 34 | ||
| No. of tumor nodes | |||||
| Single | 51 | 23 | 28 | 0.561 | 0.339 |
| Multiple | 39 | 20 | 19 | ||
| HBV | |||||
| Negative | 43 | 14 | 29 | 0.006 | 7.644 |
| Positive | 47 | 29 | 18 | ||
| Maximal tumor size (cm) | |||||
| < 4.5 | 39 | 7 | 32 | < 0.051 | 24.543 |
| ≥ 4.5 | 51 | 36 | 15 | ||
| Tumor metastasis | |||||
| No | 77 | 36 | 41 | 0.636 | 0.224 |
| Yes | 13 | 7 | 6 | ||
| Microvascular invasion | |||||
| No | 66 | 31 | 35 | 0.799 | 0.065 |
| Yes | 24 | 12 | 12 | ||
| Histological grade | |||||
| I-II | 49 | 14 | 35 | < 0.051 | 15.902 |
| III-IV | 41 | 29 | 12 | ||
| Stat4 expression | |||||
| Low | 40 | 6 | 34 | < 0.051 | 31.003 |
| High | 50 | 37 | 13 | ||
| Ki67 expression | |||||
| Low | 49 | 10 | 39 | < 0.051 | 32.293 |
| High | 41 | 33 | 8 | ||
| Parameters | RR | 95%CI | P value |
| Age (yr) | 0.156 | 0.060-0.405 | 0.069 |
| Gender | 1.629 | 0.843-3.147 | 0.147 |
| Serum AFP Level (ng/mL) | 0.885 | 0.476-1.647 | 0.701 |
| Cirrhosis | 2.066 | 0.934-4.572 | 0.073 |
| Tumor numbers | 1.166 | 0.595-2.288 | 0.654 |
| HBV | 0.742 | 0.546-1.013 | 0.057 |
| Maximal tumor size (cm) | 0.448 | 0.460-0.951 | 0.014 |
| Tumor metastasis | 1.732 | 0.644-4.658 | 0.277 |
| Microvascular Invasion | 0.874 | 0.430-1.778 | 0.710 |
| Histological grade | 0.343 | 0.599-0.817 | 0.0231 |
| STAT4 expression | 0.411 | 0.222-0.758 | < 0.0011 |
| Ki67 expression | 0.454 | 0.193-1.065 | 0.0041 |
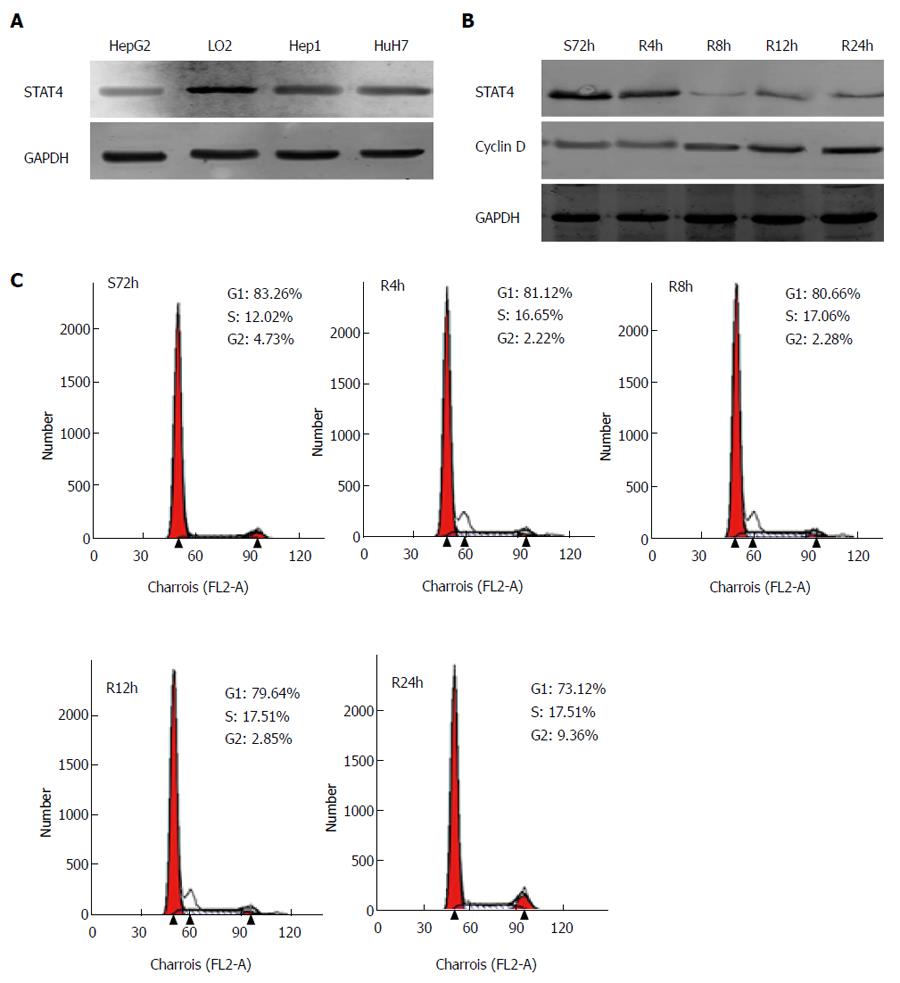
Next, HCC cell lines and L02 hepatocytes were employed to analyze the role of STAT4 in HCC cell physiology. The expression of STAT4 in these cell lines was initially determined via Western blot analysis. L02 cells displayed the highest expression of STAT4 of all of the cell lines examined (Figure 4A). Given that STAT4 expression negatively correlated with histological grade and Ki67 expression in the HCC specimens, we hypothesized that STAT4 might play an inhibitory role in the proliferation of HCC cells. Thus, we analyzed whether the expression of STAT4 was altered in HCC cells during various proliferation statuses using a serum-starvation and refeeding experiment. After serum depletion for 72 h, HepG2 cells were arrested at the G1 phase (Figure 4B). Then, after serum refeeding, the cells progressively entered the S phase. Accordingly, we detected the progressive accumulation of cyclin D1 in HepG2 cells after serum refeeding, whereas the expression level of STAT4 decreased (Figure 4C). These data demonstrated that the expression of STAT4 was associated with HCC cell proliferation.
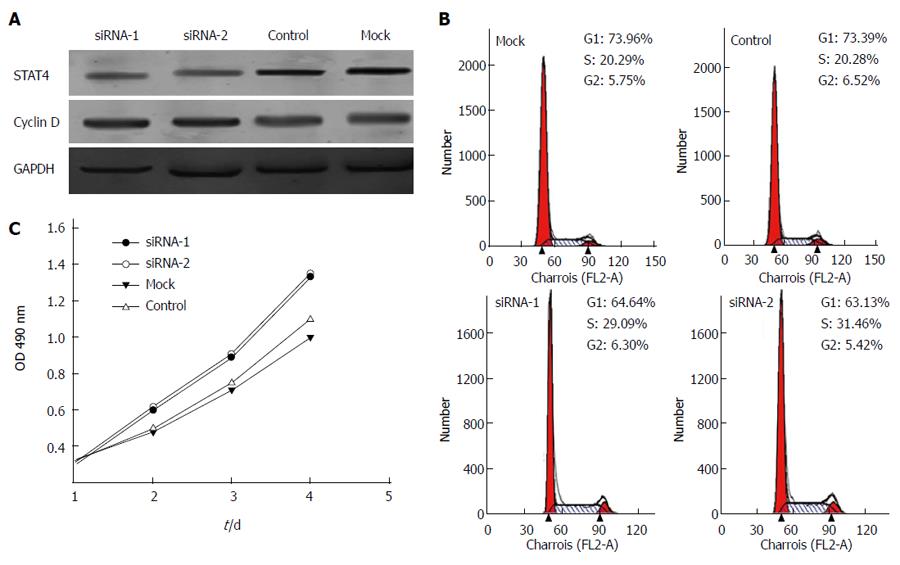
To further examine the functional role of STAT4 in HCC proliferation, STAT4-siRNA oligos were employed to silence STAT4 expression in L02 cells. After transfection with two different STAT4-targeting siRNAs, L02 cells were subjected to Western blot analysis to determine the interference efficiency of each siRNA oligo. As shown in Figure 5A, STAT4 was significantly decreased after transfection with the STAT4-siRNAs. In addition, the expression level of cyclin D1 was elevated after STAT4 depletion, implying that STAT4 negatively regulates the expression of cyclin D1 in L02 cells.
Next, we analyzed the impact of STAT4 depletion on the proliferation of L02 cells. Flow cytometry was performed to determine the cell cycle distribution of L02 cells at 72 h after mock transfection or transfection with control or STAT-targeted siRNA. As predicted, the STAT4-depleted L02 cells consisted of a markedly higher proportion of cells in the S phase (29.09% and 31.46%) than the control cells (20.29% and 20.28%), whereas the percentage of cells in the G1 phase (64.64% and 63.13%) after transfection with STAT4 siRNA-1 and 2 was significantly lower than that in the control cells (73.96% and 73.39%) (Figure 5B). Moreover, the impact of STAT4 depletion on HepG2 cell proliferation was determined using the CCK-8 assay. As shown in Figure 5C, the STAT4-depleted cells exhibited enhanced cell growth compared to the mock transfected and control siRNA-transfected cells. These data suggested that the downregulation of STAT4 might be associated with enhanced HCC cell proliferation.
HCC prognosis remains unsatisfactory because of its high recurrence and metastasis rates, despite significant improvements in surveillance and clinical treatment strategies[28]. The efficacy of traditional therapeutic methods, such as chemotherapy and surgical operation, remains limited. Therefore, it is critical to identify patients exhibiting poor prognosis for timely intervention and to develop novel targeted therapeutic strategies. Our current study showed that the decreased expression of STAT4 was significantly associated with a poor prognosis among HCC patients, which is partially attributed to uncontrolled HCC cell proliferation. Thus, our findings may promote the development of novel therapeutic strategies for HCC patients based on STAT4.
The STAT family members were originally identified as cytokine-related signaling factors and have emerged as promising molecular targets for cancer therapy[29]. Among these proteins, STAT4 plays a critical role in the regulation of diverse biological actions, including anti-viral defense, the induction of cell death and growth arrest[24,30]. In the present study, we detected significantly lower levels of STAT4 expression in HCC tissues. Furthermore, we found that the level of STAT4 in HCC tissues positively correlated with the degree of HCC differentiation and the serum hepatitis B surface antigen levels in HCC patients. It is well known that HBV infection is a risk factor for the development of HCC, contributing to the progression of HCC. Hence, our data suggest that STAT4 may serve as a negative regulator of HCC development and progression. Importantly, recent reports found that genetic alteration of STAT4 was a key risk factor for HBV-related HCC, which is consistent with our data showing that STAT4 downregulation was associated with HBV infection in HCC specimens[27]. However, another recent study reported that the mRNA level of STAT4 was not correlated with HBV infection in HCC patients[31]. Given that, STAT4 may be regulated at both the transcriptional and posttranscriptional levels. However, the detailed relevance of STAT4 to HBV-related hepatocarcinogenesis remains virtually unknown and requires clarification in future studies.
We showed that the expression of STAT4 correlated with the histological degree of HCC and the prognosis of HCC patients. Therefore, STAT4 may serve as an effective biomarker to evaluate the prognosis of HCC in Chinese patients. However, ubiquitous activation of JAK/STAT pathways is detected in human HCC tissues[32]. This discrepancy may be due to the different genetic backgrounds and the various pathologic factors that contribute to the development of HCC. Although chronic viral hepatitis is the predominant factor for the development of HCC in the Chinese population, hyperlipidemia-related and alcoholic liver diseases are crucial for the development of HCC in Western countries. Indeed, the level of STAT4 expression varies for different types of cancers, and even for the same type of cancer in different genetic backgrounds and in patients from different geographic regions. We are interested in further investigating how these factors modulate STAT4 expression and activation, thereby contributing to the development and progression of HCC.
As a crucial member of the STAT family, STAT4 was widely considered to be primarily expressed in immune cells, including T helper cells, natural T killer cells, dendritic cells and macrophages, to mediate IL-12-dependent signaling. However, the function of STAT4 in non-immune cells remains poorly understood. We and other groups recently revealed a role of STAT4 in HCC development. In this regard, it was unexpectedly found that STAT4 was highly expressed in normal liver cells and was dramatically downregulated in HCC cells. In addition, the low expression of STAT4 was verified in HCC cell lines and was found to be associated with the proliferation of HCC cells. Therefore, STAT4 in hepatocytes may exert a suppressive effect on the development of HCC tumors. Aside from HCC, STAT4 has been reported to be expressed in breast cancer cells and to play an important role in the regulation of breast cancer physiology[33]. Moreover, varying degrees of STAT4 activation have been detected in both prostate cancer and normal prostate tissues[34]. Evidence has also indicated that STAT4 might be expressed in several other cancer types, including gastric cancer and ovarian cancer[10,35]. These findings may provide novel insight into the role of STAT4 in the pathogenesis of various human cancers. However, a majority of recent studies inferred that the role of STAT4 in the prevention of liver disease was related to inflammatory pathways[31,36,37]. Therefore, it remains unclear to what extent an inflammation-independent role of STAT4 may contribute to HCC prevention. Further investigation should be performed to resolve this intriguing issue.
In summary, our findings suggest that STAT4 represents a novel and promising therapeutic target and prognostic biomarker for HCC. Our data may be of important clinical value for estimating prognosis and for determining the treatment of HCC patients. Technological development may lead to new treatments based on STAT4 that improve the therapies against HCC.
Signal transducers and activators of transcription (STATs) are members of a well-conserved family of transcription factors that play integral roles in various cellular processes. It is well known that STAT4 is expressed in many cancers, such as breast cancer. However, the expression of STAT4 in hepatocellular carcinoma (HCC) patients has yet to be reported.
The STAT family includes many prominent members, such as STAT1, STAT3, which contribute to the early diagnosis of many cancers. Few studies of STAT4, which inhibits HCC, are available.
Tissue microarray was performed to analyze STAT4 and Ki67 expression and other clinical characteristics, revealing that low STAT4 expression indicates a poor clinical prognosis. Transfection and flow cytometry were used to analyze the proliferation of HCC cells after transfection of siRNA targeting STAT4, demonstrating that high expression of STAT4 inhibits HCC cell proliferation.
STAT4 expression may serve as an indicator of clinical prognosis.
This is a very interesting study of STAT4 and its possible effects on HCC. The expression of STAT4 in HCC with different methods is very good.
P- Reviewer: Lampri ES, Lee JH, Luo XY S- Editor: Ma YJ L- Editor: O’Neill M E- Editor: Liu XM
| 1. | Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10002] [Cited by in RCA: 10453] [Article Influence: 696.9] [Reference Citation Analysis (0)] |
| 2. | Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13286] [Cited by in RCA: 13558] [Article Influence: 677.9] [Reference Citation Analysis (1)] |
| 3. | Makuuchi M, Imamura H, Sugawara Y, Takayama T. Progress in surgical treatment of hepatocellular carcinoma. Oncology. 2002;62 Suppl 1:74-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 4. | Wang XM, Yang LY, Guo L, Fan C, Wu F. p53-induced RING-H2 protein, a novel marker for poor survival in hepatocellular carcinoma after hepatic resection. Cancer. 2009;115:4554-4563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Hung JH, Lu YS, Wang YC, Ma YH, Wang DS, Kulp SK, Muthusamy N, Byrd JC, Cheng AL, Chen CS. FTY720 induces apoptosis in hepatocellular carcinoma cells through activation of protein kinase C delta signaling. Cancer Res. 2008;68:1204-1212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 87] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 6. | Wurster AL, Tanaka T, Grusby MJ. The biology of Stat4 and Stat6. Oncogene. 2000;19:2577-2584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 243] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 7. | Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1357] [Cited by in RCA: 1387] [Article Influence: 51.4] [Reference Citation Analysis (0)] |
| 8. | Darnell JE, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415-1421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4322] [Cited by in RCA: 4587] [Article Influence: 148.0] [Reference Citation Analysis (0)] |
| 9. | Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene. 2002;285:1-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 793] [Cited by in RCA: 814] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 10. | Zhou X, Xia Y, Su J, Zhang G. Down-regulation of miR-141 induced by helicobacter pylori promotes the invasion of gastric cancer by targeting STAT4. Cell Physiol Biochem. 2014;33:1003-1012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 11. | Slattery ML, Lundgreen A, Hines LM, Torres-Mejia G, Wolff RK, Stern MC, John EM. Genetic variation in the JAK/STAT/SOCS signaling pathway influences breast cancer-specific mortality through interaction with cigarette smoking and use of aspirin/NSAIDs: the Breast Cancer Health Disparities Study. Breast Cancer Res Treat. 2014;147:145-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 12. | Lupov IP, Voiles L, Han L, Schwartz A, De La Rosa M, Oza K, Pelloso D, Sahu RP, Travers JB, Robertson MJ. Acquired STAT4 deficiency as a consequence of cancer chemotherapy. Blood. 2011;118:6097-6106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 13. | Slattery ML, Lundgreen A, Kadlubar SA, Bondurant KL, Wolff RK. JAK/STAT/SOCS-signaling pathway and colon and rectal cancer. Mol Carcinog. 2013;52:155-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 152] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 14. | Gao B. Cytokines, STATs and liver disease. Cell Mol Immunol. 2005;2:92-100. [PubMed] |
| 15. | Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1278] [Cited by in RCA: 1300] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 16. | Hong F, Jaruga B, Kim WH, Radaeva S, El-Assal ON, Tian Z, Nguyen VA, Gao B. Opposing roles of STAT1 and STAT3 in T cell-mediated hepatitis: regulation by SOCS. J Clin Invest. 2002;110:1503-1513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 202] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 17. | Siebler J, Wirtz S, Klein S, Protschka M, Blessing M, Galle PR, Neurath MF. A key pathogenic role for the STAT1/T-bet signaling pathway in T-cell-mediated liver inflammation. Hepatology. 2003;38:1573-1580. [PubMed] |
| 18. | Kim WH, Hong F, Radaeva S, Jaruga B, Fan S, Gao B. STAT1 plays an essential role in LPS/D-galactosamine-induced liver apoptosis and injury. Am J Physiol Gastrointest Liver Physiol. 2003;285:G761-G768. [PubMed] |
| 19. | Chen G, Wang H, Xie S, Ma J, Wang G. STAT1 negatively regulates hepatocellular carcinoma cell proliferation. Oncol Rep. 2013;29:2303-2310. [PubMed] |
| 20. | Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13:795-804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 289] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 21. | Carter-Su C, Smit LS. Signaling via JAK tyrosine kinases: growth hormone receptor as a model system. Recent Prog Horm Res. 1998;53:61-82; discussion 82-83. [PubMed] |
| 22. | Pankov YA. Growth hormone and a partial mediator of its biological action, insulin-like growth factor I. Biochemistry (Mosc). 1999;64:1-7. [PubMed] |
| 23. | Bromberg J, Darnell JE. The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468-2473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 927] [Cited by in RCA: 968] [Article Influence: 38.7] [Reference Citation Analysis (0)] |
| 24. | Levy DE, Darnell JE. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2306] [Cited by in RCA: 2456] [Article Influence: 106.8] [Reference Citation Analysis (0)] |
| 25. | Good SR, Thieu VT, Mathur AN, Yu Q, Stritesky GL, Yeh N, O’Malley JT, Perumal NB, Kaplan MH. Temporal induction pattern of STAT4 target genes defines potential for Th1 lineage-specific programming. J Immunol. 2009;183:3839-3847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 60] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 26. | Kaplan MH. STAT4: a critical regulator of inflammation in vivo. Immunol Res. 2005;31:231-242. [PubMed] |
| 27. | Jiang DK, Sun J, Cao G, Liu Y, Lin D, Gao YZ, Ren WH, Long XD, Zhang H, Ma XP. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet. 2013;45:72-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 244] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 28. | Kudo M. Hepatocellular carcinoma 2009 and beyond: from the surveillance to molecular targeted therapy. Oncology. 2008;75 Suppl 1:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 72] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 29. | Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1723] [Cited by in RCA: 1813] [Article Influence: 86.3] [Reference Citation Analysis (0)] |
| 30. | Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778-809, table of contents. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2029] [Cited by in RCA: 2003] [Article Influence: 83.5] [Reference Citation Analysis (0)] |
| 31. | Wubetu GY, Utsunomiya T, Ishikawa D, Yamada S, Ikemoto T, Morine Y, Iwahashi S, Saito Y, Arakawa Y, Imura S. High STAT4 expression is a better prognostic indicator in patients with hepatocellular carcinoma after hepatectomy. Ann Surg Oncol. 2014;21 Suppl 4:S721-S728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 32. | Calvisi DF, Ladu S, Gorden A, Farina M, Conner EA, Lee JS, Factor VM, Thorgeirsson SS. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117-1128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 538] [Cited by in RCA: 553] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 33. | Liu S, Li L, Zhang Y, Zhang Y, Zhao Y, You X, Lin Z, Zhang X, Ye L. The oncoprotein HBXIP uses two pathways to up-regulate S100A4 in promotion of growth and migration of breast cancer cells. J Biol Chem. 2012;287:30228-30239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 34. | Ni Z, Lou W, Lee SO, Dhir R, DeMiguel F, Grandis JR, Gao AC. Selective activation of members of the signal transducers and activators of transcription family in prostate carcinoma. J Urol. 2002;167:1859-1862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 52] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 35. | Silver DL, Naora H, Liu J, Cheng W, Montell DJ. Activated signal transducer and activator of transcription (STAT) 3: localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res. 2004;64:3550-3558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 217] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 36. | Wang Y, Feng D, Wang H, Xu MJ, Park O, Li Y, Gao B. STAT4 knockout mice are more susceptible to concanavalin A-induced T-cell hepatitis. Am J Pathol. 2014;184:1785-1794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 37. | Shen XD, Ke B, Zhai Y, Gao F, Anselmo D, Lassman CR, Busuttil RW, Kupiec-Weglinski JW. Stat4 and Stat6 signaling in hepatic ischemia/reperfusion injury in mice: HO-1 dependence of Stat4 disruption-mediated cytoprotection. Hepatology. 2003;37:296-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 86] [Article Influence: 3.9] [Reference Citation Analysis (0)] |