Published online Mar 14, 2015. doi: 10.3748/wjg.v21.i10.2926
Peer-review started: September 13, 2014
First decision: October 14, 2014
Revised: October 22, 2014
Accepted: December 1, 2014
Article in press: December 1, 2014
Published online: March 14, 2015
Processing time: 184 Days and 6.1 Hours
AIM: To study the role of Twist gene in gastric cancer by gene silencing, including the potential of induction of apoptosis, cell cycle arrest, and proliferation inhibition in human malignant gastric SGC7901 cells.
METHODS: The expression level of Twist in gastric cancer samples was measured by immunohistochemistry. The effects of Twist gene silencing were detected at both mRNA and protein levels by RT-PCR and Western blot. We also evaluated the cell proliferation and apoptosis by CCK-8 assay and flow cytometry. We determined the activity of caspase-3 and caspase-9 with a caspase activity assay kit. Cell cycle distribution was analyzed by flow cytometry. Cell migration and invasion ability was evaluated by wound scratch assay and Boyden chamber assay.
RESULTS: Twist protein was highly expressed in gastric cancer samples. Twist gene silencing significantly induced apoptosis, cell cycle arrest at G0/G1 phase, proliferation inhibition, and reduced the ability of migration and invasion in human gastric cancer SGC7901 cells. Meanwhile, both caspase-3 and caspase-9 were activated.
CONCLUSION: The Twist gene could serve as a potential molecular target for gene therapy of gastric cancer with targeted small interfering RNA.
Core tip: Twist is a highly conserved transcription factor gene in gastric cancer. Silencing of the Twist gene could induce apoptosis of human gastric cancer SGC7901 cells, induce cell cycle arrest at the G0/G1 phase, inhibit cell proliferation, and reduce the ability of cell migration and invasion. The current research provides a potential gene therapy target for gastric cancer.
- Citation: Zhang H, Gong J, Kong D, Liu HY. Anti-proliferation effects of Twist gene silencing in gastric cancer SGC7901 cells. World J Gastroenterol 2015; 21(10): 2926-2936
- URL: https://www.wjgnet.com/1007-9327/full/v21/i10/2926.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i10.2926
Twist, a highly conserved transcription factor with helix-loop-helix structures, is considered to regulate essential oncogenic properties in many kinds of cancer cells[1-5]. Twist was firstly identified in Drosophila as an essential gene for the dorsoventral polarity[6]. Previous work suggests that Twist promotes the colony formation of mouse embryonic fibroblasts in soft agar and antagonizes the growth inhibition induced by p53. Recently, Twist was found involved in the metastasis of breast cancer by promoting the process of epithelial to mesenchymal transition in vivo[7]. It was also reported that Twist expression is significantly enhanced in several different types of carcinomas, such as prostate cancer, sarcomas, lymphomas and melanoma[1,5,8,9]. The inhibition against Twist could induce cell cycle arrest at the G0/G1 phase. Additionally, Twist also plays an important role in numerous physiological processes in metastasis, like angiogenesis, invadopodia, extravasation, and chromosomal instability[5]. These findings suggest that Twist is a novel oncogene associated with tumorigenesis as well as tumor progression[1,2,7].
Gastric cancer is the second most common cause of death associated with carcinomas, and has low survival rates in patients[10,11]. Due to the difficulties in diagnosis at early stage, a majority of patients bearing gastric cancer have a poor clinical outcome[12]. Despite the evidence suggesting the oncogene role for Twist, there are little reports regarding the expression of Twist in gastric cancer.
In the present study, we examined the expression level of Twist in both normal and carcinomatous human gastric tissues. In addition, we quantified the expression of Twist at both mRNA and protein levels in human gastric cancer SGC7901 cells and human gastric epithelial GES-1 cells. To determine the role of Twist in gastric cancer, we studied the effects of Twist gene silencing on proliferation, migration and invasion of SGC7901 cells using RNA interference (RNAi) technique. In addition, the effects of Twist gene silencing on cell cycle distribution and apoptosis induction were also investigated.
We collected a total of 127 patients who were diagnosed with gastric cancer, and these patients accepted resection at No. 1 Hospital of Jilin University from December 2006 to January 2008. Among them, there were 71 males and 56 females with the mean age of 55 years (range, 33-73 years). As to the progression of cancer, there were 15 cases of stage I, 33 cases of stage II, 43 cases of stage III and 36 cases of stage IV. No patients were treated with chemotherapy or radiotherapy prior to the surgery. Another 19 normal gastric tissues were collected to compare Twist expression, and all samples were confirmed by two experienced pathological experts.
Since gastric epithelial GES-1 cells and normal gastric epithelial cells have similar biological properties, GES-1 cells were used in this study. Human gastric cancer cell line SGC7901[13] and human gastric epithelium GES-1 cell line were kindly provided by No. 3 Hospital of Jilin University, all cell lines have been identified by Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China), and cell lines were grown in RPMI 1640 medium (Hyclone, Logan, UT, United States) supplemented with 10% fetal calf serum at 37 °C in a humidified atmosphere of 5% CO2. The antibodies against human Twist, mouse/rat Bax and Bcl-2 were purchased from Santa Cruz Corporation (Santa Cruz, CA, United States). Caspase 3 and caspase 9 activity assay kits were purchased from BiYunTian Biotechnology Research Institute (Shanghai, China). The immunoglobulin G (IgG) labelled with horseradish peroxidase was purchased from Zhongshan Company (Zhongshan, China). pGenesil-2 vector carrying Twist siRNA was synthesized by Wuhan Cell marker Biotechnology Co., Ltd. (Wuhan, China). The target sequence of the Twist siRNA was AAGCTGAGCAAGATTCAGACCCTCAAGCT.
The total RNA was isolated using Trizol reagent (Invitrogen, United States) and transcribed into cDNA using a reverse transcription kit (Toyobo, Japan). RT-PCR was carried out using a MX3000 machine (Eppendorf, Germany) using the following condition: 95 °C 30 s, 55 °C 30 s, 72 °C 30 s for 35 cycles. Each sample was determined in triplicate. For Twist gene amplification, the forward primer was 5’ GGG AGT CCG CAG TCT TAC GA 3’, and the reverse primer was: 5’ CCA GAC CGA GAA GGC GTA GC 3’. GAPDH expression was used as an internal control, and the forward primer was: 5’ AGC CAC ATC GCT CAG ACAC 3’, and the reverse primer was 5’ GAG GCA TTG CTG ATG ATC TTG 3’. The 1.5% agarose gel was used to view the PCR product with a Kodak UV detector (Kodak, United States). For the total protein extraction, cells were harvested and lysed with mammalian cell lysis/extraction reagent (Sigma-Aldrich). Western blot analysis was conducted according to the standard procedure. Briefly, harvested cells were washed twice with phosphate buffered saline (PBS) and lysed in 100 μL radioimmunoprecipitation assay lysis buffer. The protein concentration was determined with Bradford reagent (Sigma−Aldrich, Saint Louis, MO, United States) according to the manufacturer’s instructions. Proteins were resolved by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Roche Diagnostics GmbH, Mannheim, Germany). After non-specific reactivity was blocked with 5% non-fat milk in Tris-buffered saline for 3 h, the membranes were incubated overnight with primary antibodies. The secondary antibodies used were horseradish peroxidase-conjugated goat anti-rabbit or -mouse IgG from Tiangen (Beijing, China). Membranes were incubated with the secondary antibodies for 1 h at room temperature, and densitometric scans of the autoradiographs were digitized on a ScanMaker 3860 (Microtek, Shanghai, China) with Scanwizard 5 software (Microtek).
Tissue immunohistochemical staining was completed with peroxidase-labeled streptavidin in the immunohistochemistry kit according to the instructions. Briefly, samples were embedded in paraffin, and the sections were deparaffinized. After the depletion of endogenous peroxidase, nonspecific binding sites were saturated with bovine serum albumin. Human Twist antibody (dilution, 1:1000) was added and incubated overnight at 4 °C. After washing with PBS, biotinylated goat anti-mouse IgG was added to the tissue sections, and incubated at room temperature for 30 min. After washing with PBS and incubation with avidin DH-biotinylated peroxidase, these tissues were detected with diaminobenzidine color development reagent.
The cell viability was measured using Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Rockville, MD, United States) according to the instructions. Cells were seeded in 96-well flat-bottom plates, and 10 μL of CCK-8 reagent was added at different time points and then incubated for another 1 h. The absorbance was measured with a microplate reader (Bio-Rad, United States) at 450 nm. The CCK8 assay was conducted at daily intervals from the next day to the seventh day after seeding, and the average OD value at each time point from different groups was plotted with Excel (Microsoft, United States).
In the soft-agar assay[14], cells (7 × 103) were suspended in 1 mL of 0.3% agarose and then added to each well of a 24-well plate with a foundation layer of 0.5% agarose. Fifteen days later, the colonies were observed and counted.
Matrigel Plate Coating was prepared according to previous reports[15]. Cells (2 × 105) were plated in 96-well Matrigel-coated plates and incubated at 37 °C for 1 h. After three times of wash using PBS to remove suspended cells, the adherent cells were observed under a microscope and quantified using CCK-8 assay. The adherent rate for the three different cell populations were calculated. Specifically, the stained SGC-7901 cells with rhodamine 123 were seeded into the Matrigel labeled Boyden chamber (2 × 105/well). After induction by NIH/3T3, SGC-7901 cells migrated into the Matrigel 3 h later, and the invasion depth was studied with a laser scanning confocal microscope.
The effects of RNAi on the invasion were also evaluated in vivo. Twist RNAi transfected SGC-7901 cells, together with control cells, were injected under the kidney capsule. After cell transplantation for 6 d, we sacrificed the animals and studied the invasion depth as well as the effects on the xenograft under the kidney. The effects were observed under a microscope after HE staining.
Matrix metalloproteinases (MMPs) play an important role in the process of migration, invasion and metastasis of tumor infiltration. The expression of MMP-2 was measured by gelatin zymography.
The decrease of mitochondrial transmembrane potential (MTP, ΔΨm) is generally regarded as one of the earliest events in apoptotic cascade reaction process. Once the MTP is destroyed, irreversible apoptosis would be started[16,17]. We measured the MTP by rhodamine-123 staining.
As the important checkpoint in the commonly apoptotic signaling pathway, the ratio of pro-apoptotic factors (Bax) to anti-apoptotic ones (Bcl-2) critically regulates the activity of caspase-3 and -9[18], and Bax and Bcl-2 regulate apoptosis by controlling MTP as well as membrane permeability[18,19]. Therefore, the effects of Twist RNAi on the activity of caspase-3 and -9 were tested; the expression of Bax and Bcl-2 was measured by fluorescent activated cell sorting (FACS).
Tumor suppressor protein p53 also plays an important role in the regulation of apoptosis[20]. A previous study has suggested that the loss of p53 expression or function is highly associated with an increased risk of carcinogenesis[21]. Multiple signaling pathways mediate p53-induced apoptosis, one of which involves both Bcl-2 and Bax proteins. Actually, the pro-apoptotic protein Bax is a downstream target of p53 while the anti-apoptotic protein Bcl-2 could be suppressed by p53 at the transcriptional level[18,20,22]. On these basis, we examined the expression of p53 in Twist RNAi SGC-7901 cells by FACS.
Cell cycle analysis was performed by FACS as described previously[23]. Briefly, the cells in logarithmic growth phase were harvested, fixed in 70% ethanol at 4 °C for 12 h, and stained with propidium iodide (PI) in a phosphate buffered saline solution containing RNase. Data were analyzed with Cellfil software.
In the current study, the cells were seeded on the coverslips in 6-well plates (1.5 × 105/well). Apoptotic cells were detected by TUNEL assay using the in situ cell death assay kit according to the instructions. The apoptosis index was calculated as the percentage of cells with definite positive terminal deoxynucleotidyl transferase-mediated digoxigenin-dUTP nick-end labeling (TUNEL) staining and was obtained by counting five randomly chosen visual fields for each slide under a microscope.
All the data were statistically analyzed using t-test or χ2 test, and are expressed as mean ± SD based on three independent experiments. All the analyses were carried out with SPSS 12.0 statistical software. P values less than 0.05 were considered statistically significant.
Comparison of surgically resected samples demonstrated that little expression of Twist was only observed in 3 of 19 normal gastric samples. In contrast, we found that Twist was largely expressed in the neoplastic gastric samples with statistical significance compared with normal tissues (P < 0.01) (Table 1).
| Stage | No. of samples | Twist expression | Twist positive rate | |||
| - | + | ++ | +++ | |||
| Normal | 19 | 16 | 3 | 0 | 0 | 15.8% |
| I | 15 | 7 | 7 | 1 | 0 | 53.3% |
| II | 33 | 8 | 12 | 12 | 1 | 75.8% |
| III | 43 | 7 | 20 | 14 | 2 | 83.7% |
| IV | 36 | 3 | 6 | 15 | 12 | 91.7% |
| Total | 127 | 25 | 45 | 42 | 15 | 80.3% |
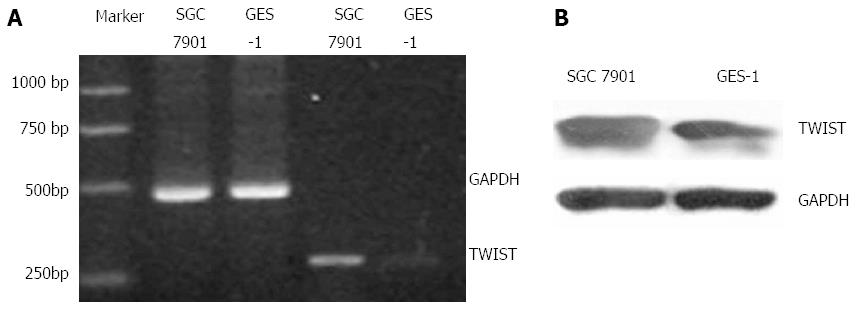
On the other hand, RT-PCR and Western blot results showed the expression of Twist in gastric cancer SGC7901 cells and human gastric epithelial GES-1 cells. The results showed increased expression in SGC7901 cells compared to GES-1 cells (Figure 1).
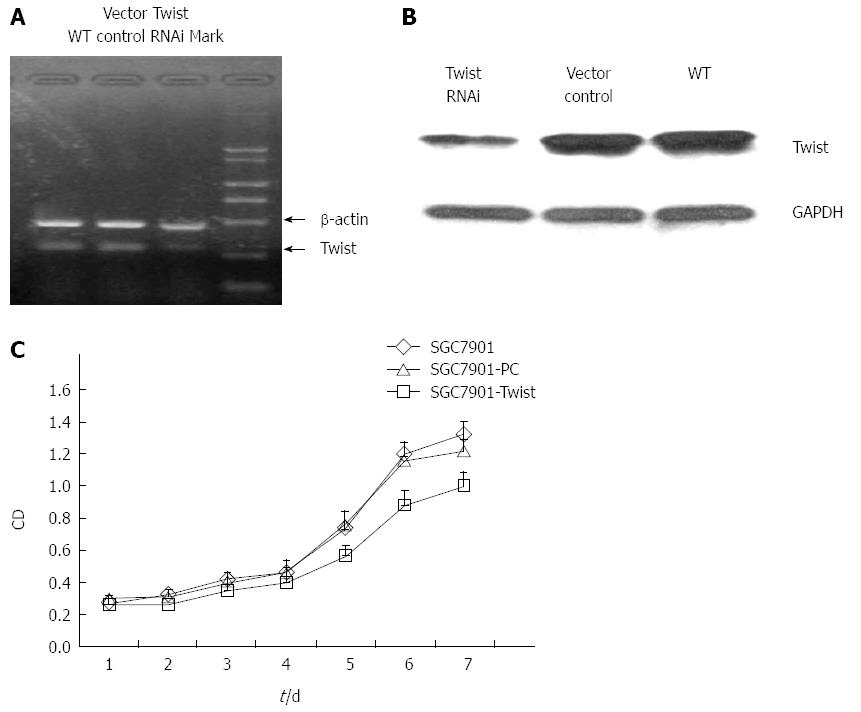
After transfecting of SGC7901 cells with the pGensil-2/Twist or a control vector (pGensil-2), we detected the expression of Twist. The expression level of Twist decreased approximately by 70% in cells transfected with the pGensil-2/Twist (Figure 2A and B). We measured the proliferation of both Twist RNAi cells and vector control cells (Figure 2C), and found that the growth of Twist siRNA transfected cells was significantly inhibited at each time point, which demonstrated the decreased cell division and increased apoptosis.
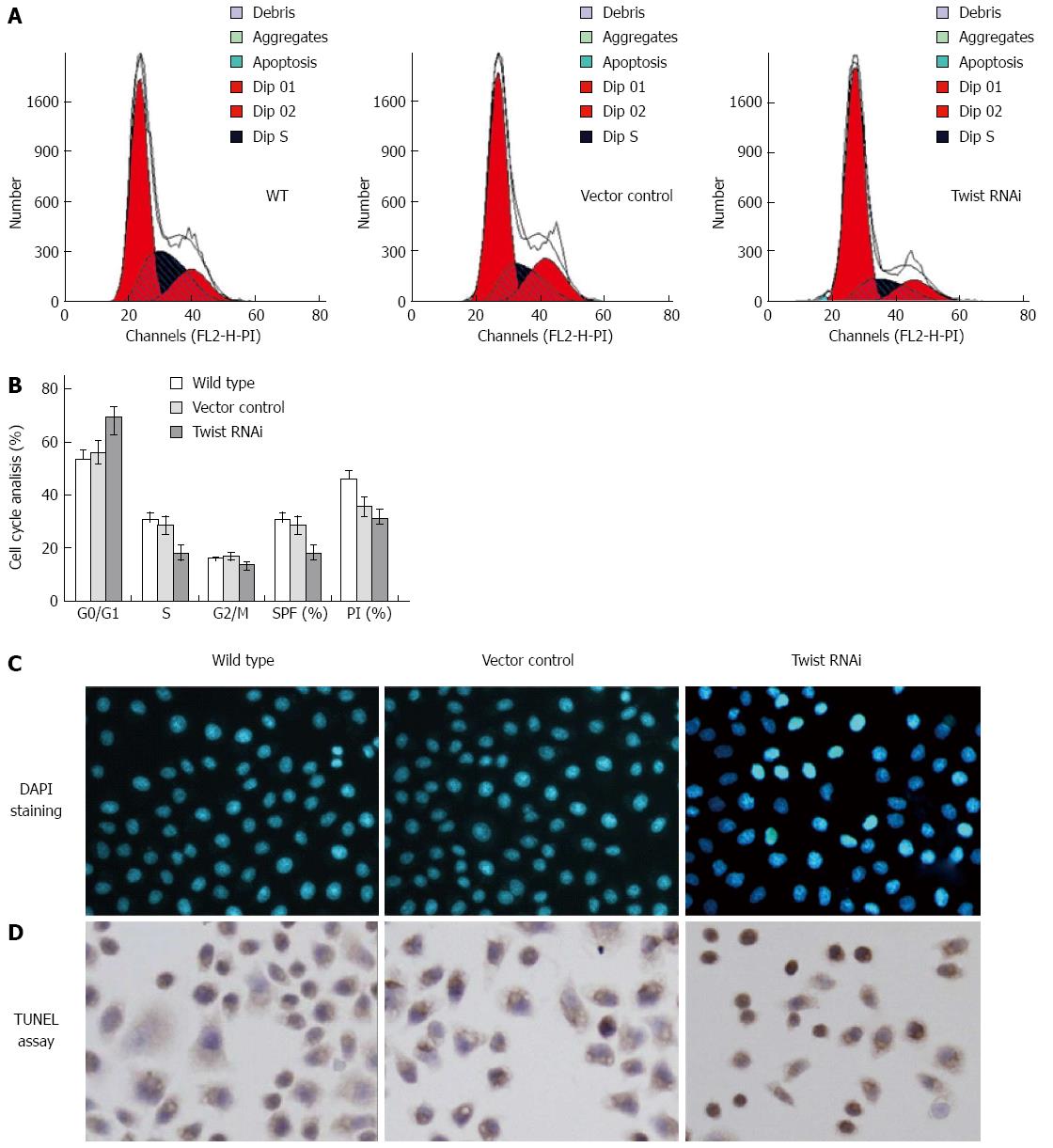
The changes of cell cycle distribution and apoptosis were further examined by PI staining, 4’,6-diamidino-2-phenylindole (DAPI) staining and TUNEL assays, respectively. For the cell cycle analysis, we found significant accumulation of cells at the G0/G1 phase in Twist siRNA treated cells, with reduction at S phase, compared to the vector control and wild type (Figure 3A and 3B). The proliferation index[24] for Twist siRNA treated cells decreased from 35.7% to 31.0%. The SPF also decreased from 28.7% to 17.9%. The results suggested that Twist RNAi could induce cell cycle arrest at G0/G1 phase.
In the DAPI staining assay, we found the round and clear-edged nuclear morphology in vector control cells, which is similar to that observed in the wild type cells. In the Twist RNAi cells, the apoptosis (characterized by nuclear pyknosis and heavier coloring) increased significantly compared to the vector control cells (Figure 3C). Similar results were also observed in TUNEL assay. The apoptotic cells with brown granules in the nucleus were much more in Twist RNAi SGC-7901 cells (35.1 ± 8.2/field) than vector control cells (15.2 ± 2.1/field) (Figure 3D) with statistical significance (P < 0.001).
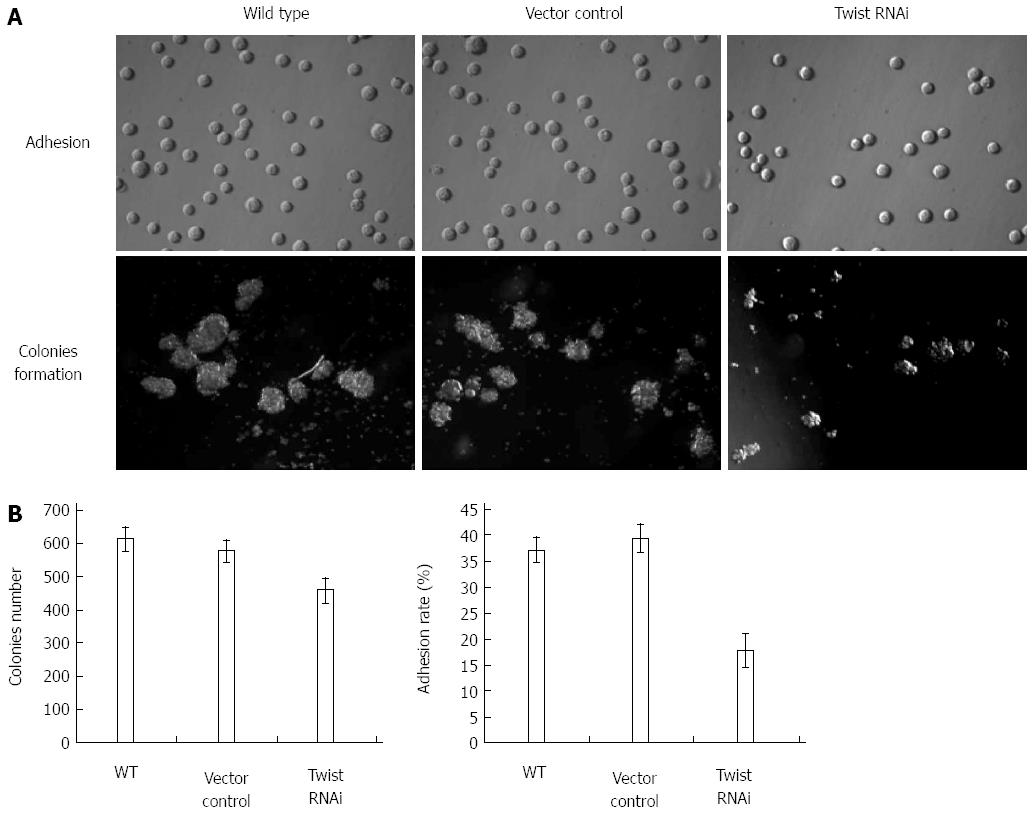
As to the adhesion, result showed there was a significant difference in the adherent rate between Twist siRNA treated cells (17.8%) and the vector control cells (39.5%) (P < 0.001). In contrast, there was no significant difference between vector cells and wild type cells (Figure 4).
Figure 4 demonstrates the anchorage-independent growth results. As we can see, Twist RNAi resulted in a significant decrease (about 19.5%) in colony formation in SGC-7901 cells compared with vector control (P < 0.001). However, no significant difference between vector control and wild type cells was found. These results suggest that the decreased expression of Twist could lead to the difficulties in forming colonies in soft agar for gastric cancer cells.
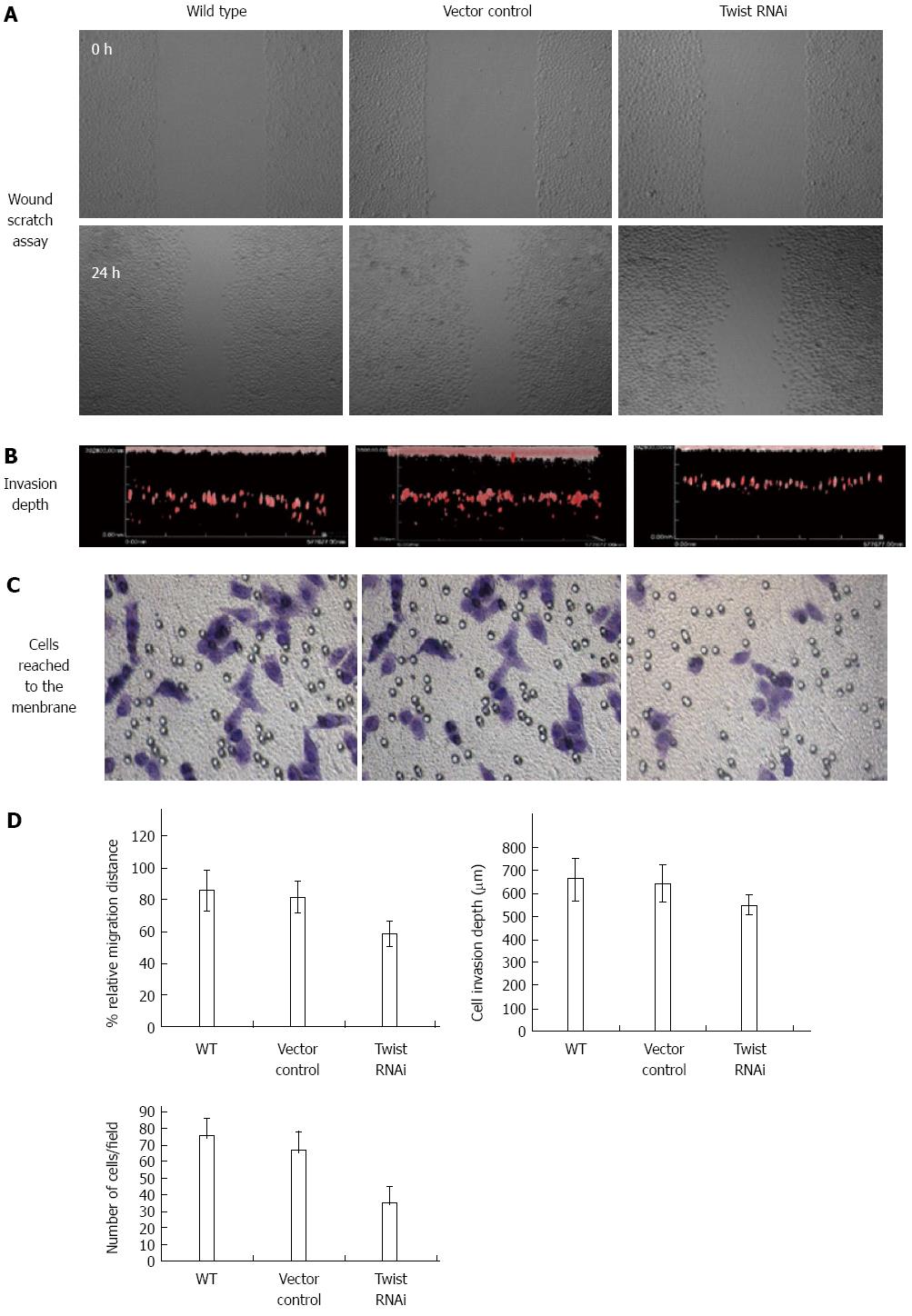
We measured the relative migrating distance after 18 h with the result of 58.5% in Twist siRNA treated cells, which was much lower than that in control vector treated cells (81.8%) (P < 0.05). In contrast, there was no significant difference between vector and wild type cells (Figure 5).
For the effects on invasion (Figure 5), the invasion depth was determined to be 640.1 ± 77.6 and 660.0 ± 89.4 μm for vector control cells and wild type cells, respectively. In contrast, the invasion depth of Twist RNAi cells decreased to 550.5 ± 36.5 μm. In addition, we observed the cells reaching the membrane of Boyden chamber by crystal violet staining. As shown in Figure 5, compared to the vector control (66.3 ± 11.2/field) and wild type cells (75.5 ± 10.1/field), the number of cells in the Twist RNAi group decreased largely (35.2 ± 9.6/field). These results suggest that knockdown of Twist could significantly inhibit the migration and invasion ability of SGC-7901 cells.
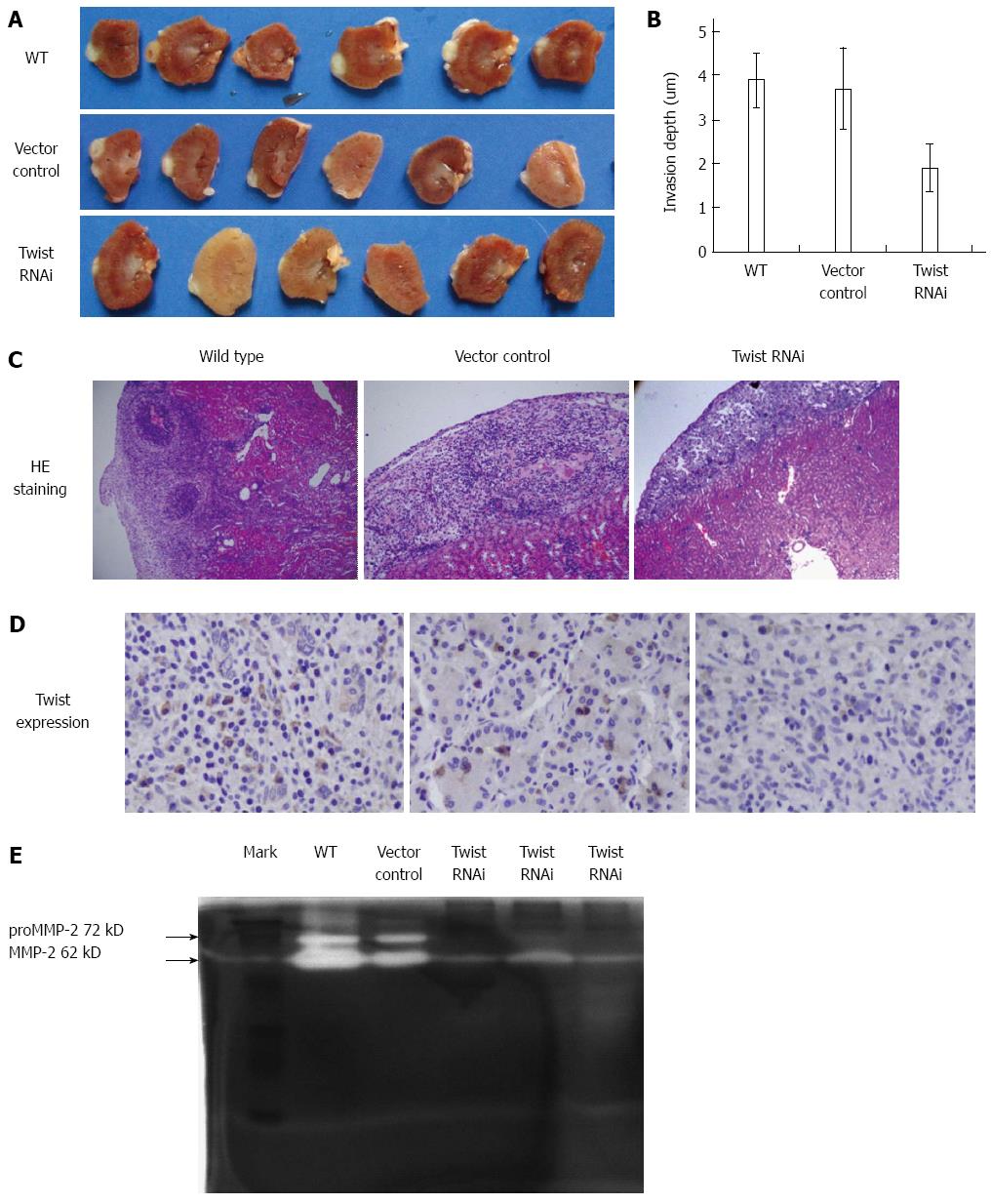
As shown in Figure 6A and B, the invasion depth in wild type and vector control was determined as 3.9 ± 0.6 mm, 3.7 ± 0.9 mm, respectively. In contrast, the invasion depth of Twist RNAi SGC-7901 cells decreased to 1.9 ± 0.5 mm. As shown in Figure 6C, cancer cells infiltrated into and oppressed the renal cortex in xenografts derived from wild type and vector control cells, which led to the damage of renal structure. In contrast, much alleviated effects were observed in the Twist RNAi group. We measured the expression of Twist by immunohistochemistry. As shown in Figure 6D, high expression of Twist was found in cytoplasm for both the wild type and control vector groups. However, Twist expression was decreased in the Twist RNAi group. These results suggest that the invasion was more aggressive in xenografts derived from control vector transfected and wild type cells than from Twist siRNA transfected cells. What’s more, the invasion ability was consistent with the expression level of Twist.
As shown in Figure 6E, the amounts of secreted pro-MMP-2 (72 kD) and activated MMP-2 (62 kD) largely decreased in the Twist RNAi group compared to the vector control and wild type groups, which suggests that Twist gene silencing in SGC7901 cells led to the inhibition of secretion and activation of MMP-2.
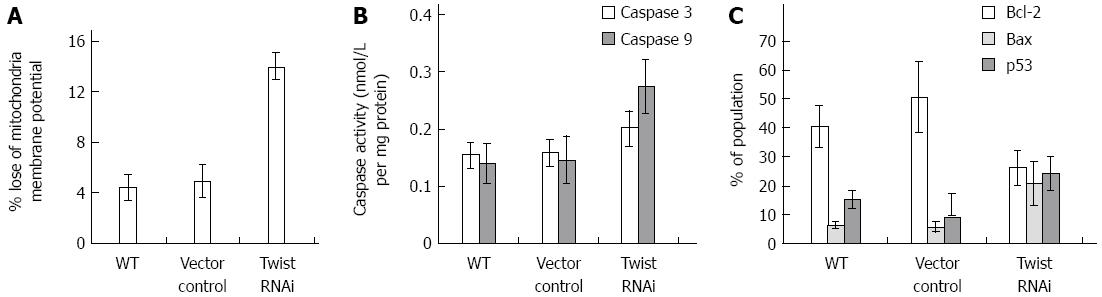
MTP testing results showed that compared with the empty vector transfected control cells, the number of rhodamine-123 negatively stained cells was increased significantly for Twist RNAi cells (P < 0.001) (Figure 7A). This suggests that knockdown of Twist could reduce the MTP.
As we can see from Figure 7B, the activity of caspase-3 and -9 increased in Twist RNAi SGC-7901 cells, compared to the vector control and wild type cells. In addition, the expression of Bax in the Twist RNAi cells increased by 15.16%, while the expression of Bcl-2 decreased by 24.35% compared to the vector control (Figure 7C).
The expression of p53 also increased significantly in the Twist RNAi cells compared to the vector and wild type control cells. This suggests Twist RNAi can induce apoptosis by means of enhancing the expression the p53 gene (Figure 7C).
In the present study, we studied the expression of Twist in carcinomatous samples by immunohistochemistry, which were collected from 127 patients with gastric cancer. According to the previous reports, Twist is widely expressed in most of human cancers, including lung[7,25], colon[2], pancreas[1], prostate[3,8], breast[26-29], and gastric cancers[30,31]. We found that Twist was expressed in different stages of neoplastic gastric samples, and the expression level in cancer was much higher than that in normal tissues. In addition, compared to human gastric epithelial GES-1 cells, both mRNA and protein expression levels of Twist consistently increased significantly in the cultured gastric cancer SGC7901 cells.
To clarify the roles of Twist in gastric cancer cells, we measured the effect of Twist knockdown on the neoplastic characteristics of gastric cancer SGC7901 cells. We found that vector control cells showed much similar properties with wild type SGC7901 cells. In contrast, Twist RNAi inhibited the proliferation of SGC7901 cells, demonstrating that Twist gene silencing could induce apoptosis and cell cycle arrest in SGC7901 cells.
Moreover, we observed that other important oncogenic characteristics were also affected in Twist RNAi SGC7901 cells, such as the adhesion, anchorage-independent growth, migration and invasion, all of these functions decreased considerably in Twist RNAi cells.
It is generally accepted that the disruption of mitochondrial function could induce apoptosis, which involves the abnormal expression of Bcl-2 and/or Bax, and release of cytochrome c from mitochondria into cytosol[22]. From our results, we found that Twist RNAi can lead to the decrease of MTP. Meanwhile, the ratio of Bax/Bcl-2 in Twist RNAi cells also decreased. In contrast, the expression of p53 increased. Previous reports have suggested that Twist could inhibit the expression of ARF (alternative reading frame)[2], which binds to and promotes the rapid degradation of MDM2 and leads to stabilization and accumulation of p53[32]. On these basis, it could be hypothesized that Twist exerts its functions by regulating the expression of p53 as well as Bax and Bcl-2 in the gastric cancer cells, which affects the MTP and the activity of caspases.
In conclusion, the current study demonstrates that Twist gene silencing could significantly inhibit the oncogenic properties of gastric cancer SGC7901cells. It holds promise as a potential molecular target in the gene therapy of gastric cancer.
Gastric cancer is one of the most common malignant tumors of the digestive system. Currently, the early diagnosis of gastric cancer is difficult due to lacking of effective preventive measures. Studies indicate that high expression of Twist gene is closely related with the occurrence, development and invasion of a variety of tumors including gastric cancer.
In this study, RNA interference (RNAi) technology was used to silence Twist gene expression, and then we observed its effect on the biological behavior of human malignant gastric cancer cell line SGC7901.
The expression of Twist in gastric cancer sample provides a meaningful indicator to judge the malignant degree and invasion of gastric cancer. Since silencing the expression of Twist in gastric cancer cells could inhibit the growth of tumor cells, Twist may be a potential target for gene therapy of gastric cancer.
This study demonstrated that transfection with the constructed Twist-siRNA plasmid inhibited the expression of Twist at both the mRNA and protein levels, and affected the proliferation, migration, invasion and apoptosis of SGC7901 cells.
RNAi is a biological process in which RNA molecules inhibit gene expression, typically by causing the destruction of specific mRNA molecules. RNAi is a powerful gene-silencing process that holds great promise in the field of cancer therapy.
This is an interesting study on RNAi targeting of molecules crucial for cancer therapy. Many gene products involved in carcinogenesis have already been explored as targets for RNAi. The authors take Twist as the target, and demonstrate that Twist targeted siRNA generated significant effects on biological behavior of gastric cancer cells. This study provides a new theoretical and experimental basis for Twist targeted gene therapy.
P- Reviewer: Chmiela M, Furka A, Zouiten-Mekki L S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Liu XM
| 1. | Ohuchida K, Mizumoto K, Ohhashi S, Yamaguchi H, Konomi H, Nagai E, Yamaguchi K, Tsuneyoshi M, Tanaka M. Twist, a novel oncogene, is upregulated in pancreatic cancer: clinical implication of Twist expression in pancreatic juice. Int J Cancer. 2007;120:1634-1640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 62] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 2. | Maestro R, Dei Tos AP, Hamamori Y, Krasnokutsky S, Sartorelli V, Kedes L, Doglioni C, Beach DH, Hannon GJ. Twist is a potential oncogene that inhibits apoptosis. Genes Dev. 1999;13:2207-2217. [PubMed] |
| 3. | Raatikainen S, Aaltomaa S, Palvimo JJ, Kärjä V, Soini Y. TWIST overexpression predicts biochemical recurrence-free survival in prostate cancer patients treated with radical prostatectomy. Scand J Urol. 2015;49:51-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 4. | Mironchik Y, Winnard PT, Vesuna F, Kato Y, Wildes F, Pathak AP, Kominsky S, Artemov D, Bhujwalla Z, Van Diest P. Twist overexpression induces in vivo angiogenesis and correlates with chromosomal instability in breast cancer. Cancer Res. 2005;65:10801-10809. [PubMed] |
| 5. | Khan MA, Chen HC, Zhang D, Fu J. Twist: a molecular target in cancer therapeutics. Tumour Biol. 2013;34:2497-2506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 161] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 6. | Thisse B, el Messal M, Perrin-Schmitt F. The twist gene: isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res. 1987;15:3439-3453. [PubMed] |
| 7. | Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2783] [Cited by in RCA: 2982] [Article Influence: 142.0] [Reference Citation Analysis (0)] |
| 8. | Kwok WK, Ling MT, Lee TW, Lau TC, Zhou C, Zhang X, Chua CW, Chan KW, Chan FL, Glackin C. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65:5153-5162. [PubMed] |
| 9. | Kyo S, Sakaguchi J, Ohno S, Mizumoto Y, Maida Y, Hashimoto M, Nakamura M, Takakura M, Nakajima M, Masutomi K. High Twist expression is involved in infiltrative endometrial cancer and affects patient survival. Hum Pathol. 2006;37:431-438. [PubMed] |
| 10. | Roder DM. The epidemiology of gastric cancer. Gastric Cancer. 2002;5 Suppl 1:5-11. [PubMed] |
| 11. | Jing JJ, Liu HY, Hao JK, Wang LN, Wang YP, Sun LH, Yuan Y. Gastric cancer incidence and mortality in Zhuanghe, China, between 2005 and 2010. World J Gastroenterol. 2012;18:1262-1269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 12. | Yasui W, Oue N, Aung PP, Matsumura S, Shutoh M, Nakayama H. Molecular-pathological prognostic factors of gastric cancer: a review. Gastric Cancer. 2005;8:86-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 212] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 13. | Lin CH, Fu ZM, Liu YL, Yang JL, Xu JF, Chen QS, Chen HM. Investigation of SGC-7901 cell line established from human gastric carcinoma cells. Chin Med J (Engl). 1984;97:831-834. [PubMed] |
| 14. | Wang H, Zhao LN, Li KZ, Ling R, Li XJ, Wang L. Overexpression of ribosomal protein L15 is associated with cell proliferation in gastric cancer. BMC Cancer. 2006;6:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 80] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 15. | Partridge J, Flaherty P. An in vitro FluoroBlok tumor invasion assay. J Vis Exp. 2009;pii: 1475. [PubMed] |
| 16. | Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35:495-516. [PubMed] |
| 17. | Chen WH, Xu XD, Luo GF, Jia HZ, Lei Q, Cheng SX, Zhuo RX, Zhang XZ. Dual-targeting pro-apoptotic peptide for programmed cancer cell death via specific mitochondria damage. Sci Rep. 2013;3:3468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 86] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 18. | Gross A, McDonnell JM, Korsmeyer SJ. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 1999;13:1899-1911. [PubMed] |
| 19. | Martinou JC, Youle RJ. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell. 2011;21:92-101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1103] [Cited by in RCA: 1117] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 20. | Ryan KM, Phillips AC, Vousden KH. Regulation and function of the p53 tumor suppressor protein. Curr Opin Cell Biol. 2001;13:332-337. [PubMed] |
| 21. | Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275-283. [PubMed] |
| 22. | Katiyar SK, Roy AM, Baliga MS. Silymarin induces apoptosis primarily through a p53-dependent pathway involving Bcl-2/Bax, cytochrome c release, and caspase activation. Mol Cancer Ther. 2005;4:207-216. [PubMed] |
| 23. | Wang H, Tan SS, Wang XY, Liu DH, Yu CS, Bai ZL, He DL, Zhao J. Silencing livin gene by siRNA leads to apoptosis induction, cell cycle arrest, and proliferation inhibition in malignant melanoma LiBr cells. Acta Pharmacol Sin. 2007;28:1968-1974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 24. | Fondrevelle ME, Kantelip B, Reiter RE, Chopin DK, Thiery JP, Monnien F, Bittard H, Wallerand H. The expression of Twist has an impact on survival in human bladder cancer and is influenced by the smoking status. Urol Oncol. 2009;27:268-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 53] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 25. | Hui L, Liu B, Zhao L, Zhang S, Zhou B, Qiu X, Cui Z. [High expression of twist is positively correlated with the differentiation of lung cancer.]. Zhongguo Fei Ai Zazhi. 2009;12:294-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 26. | Vesuna F, Winnard P, Glackin C, Raman V. Twist overexpression promotes chromosomal instability in the breast cancer cell line MCF-7. Cancer Genet Cytogenet. 2006;167:189-191. [PubMed] |
| 27. | Watanabe O, Imamura H, Shimizu T, Kinoshita J, Okabe T, Hirano A, Yoshimatsu K, Konno S, Aiba M, Ogawa K. Expression of twist and wnt in human breast cancer. Anticancer Res. 2004;24:3851-3856. [PubMed] |
| 28. | Martin TA, Goyal A, Watkins G, Jiang WG. Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann Surg Oncol. 2005;12:488-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 391] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 29. | Zhao M, Hu HG, Huang J, Zou Q, Wang J, Liu MQ, Zhao Y, Li GZ, Xue S, Wu ZS. Expression and correlation of Twist and gelatinases in breast cancer. Exp Ther Med. 2013;6:97-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 30. | Luo GQ, Li JH, Cao L, Zhou YH, Wen JF. Activator protein-1 involvement in proliferation inhibition by gene silencing of Twist in gastric cancer cells. Pathology. 2011;43:697-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 31. | Yan-Qi Z, Xue-Yan G, Shuang H, Yu C, Fu-Lin G, Fei-Hu B, Shi-Ren S, Xu-Feng W, Jie D, Dai-Ming F. Expression and significance of TWIST basic helix-loop-helix protein over-expression in gastric cancer. Pathology. 2007;39:470-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 47] [Article Influence: 2.6] [Reference Citation Analysis (0)] |