Published online Jan 7, 2015. doi: 10.3748/wjg.v21.i1.196
Peer-review started: July 3, 2014
First decision: July 21, 2014
Revised: August 19, 2014
Accepted: October 15, 2014
Article in press: October 15, 2014
Published online: January 7, 2015
Processing time: 188 Days and 13.5 Hours
AIM: To investigate the expression of forkhead box protein M1 (FoxM1) in the process of epithelial mesenchymal transition in hepatocellular carcinoma (HCC) and its role in metastasis.
METHODS: FoxM1 and E-cadherin expression in HCC tissue microarray specimens was evaluated by immunohistochemical staining, and statistical methods were applied to analyze the correlation between FoxM1 and epithelial-mesenchymal transition (EMT). Kaplan-Meier analysis of the correlation between the FoxM1 expression level and recurrence or overall survival of HCC patients was performed. The expression of FoxM1, E-cadherin and snail homologue 1 (SNAI1) in HCC cell lines was evaluated by real-time reverse transcription-polymerase chain reaction and Western blot. Hepatocyte growth factor (HGF) was used to induce EMT and stimulate cell migration in HCC cells. The expression of FoxM1 and SNAI1 was regulated by transfection with plasmids pcDNA3.1 and siRNAs in vitro. The occurrence of EMT was evaluated by Transwell assay, morphologic analysis and detection of the expression of EMT markers (E-cadherin and vimentin). Luciferase and chromatin immunoprecipitation assays were used to evaluate whether SNAI1 is a direct transcriptional target of FoxM1.
RESULTS: FoxM1 expression was increased significantly in HCC compared with para-carcinoma (10.7 ± 0.9 vs 8.2 ± 0.7, P < 0.05) and normal hepatic (10.7 ± 0.9 vs 2.7 ± 0.4, P < 0.05) tissues. Overexpression of FoxM1 was correlated with HCC tumor size, tumor number, macrovascular invasion and higher TNM stage, but was negatively correlated with E-cadherin expression in microarray specimens and in cell lines. FoxM1 overexpression was correlated significantly with HCC metastasis and EMT. In vitro, we found that FoxM1 plays a key role in HGF-induced EMT, and overexpression of FoxM1 could suppress E-cadherin expression and induce EMT changes, which were associated with increased HCC cell invasiveness. Next, we confirmed that FOXM1 directly binds to and activates the SNAI1 promoter, and we identified SNAI1 as a direct transcriptional target of FOXM1. Moreover, inhibiting the expression of SNAI1 significantly inhibited FoxM1-mediated EMT.
CONCLUSION: FoxM1 overexpression promotes EMT and metastasis of HCC, and SNAI1 plays a critical role in FoxM1-mediated EMT.
Core tip: Epithelial-mesenchymal transition (EMT) has emerged as a pivotal event affecting cancer invasion and metastasis, and forkhead box protein M1 (FoxM1) may regulate the EMT phenotype of pancreatic cancer cells by activation of mesenchymal cell markers. The present study demonstrated that FoxM1 plays a pivotal role in EMT and metastasis of hepatocellular carcinoma (HCC). Immunohistochemical analysis indicated that FoxM1 overexpression correlated significantly with HCC metastasis and EMT. In vitro, we found that FoxM1 plays a key role in HGF-induced EMT, and overexpression of FoxM1 could suppress E-cadherin expression and induce EMT changes by increasing snail homologue 1 expression.
- Citation: Meng FD, Wei JC, Qu K, Wang ZX, Wu QF, Tai MH, Liu HC, Zhang RY, Liu C. FoxM1 overexpression promotes epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma. World J Gastroenterol 2015; 21(1): 196-213
- URL: https://www.wjgnet.com/1007-9327/full/v21/i1/196.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i1.196
Hepatocellular carcinoma (HCC) accounts for 90% of cases of primary liver cancer and is the fifth most common cancer worldwide. It ranks third in mortality after gastric cancer and esophageal cancer, and half of the deaths from HCC occur in China[1,2]. Due to the high rate of recurrence or intrahepatic metastasis after curative resection, the overall prognosis of HCC patients remains poor despite obvious improvements in surgical techniques and perioperative management[3,4]. Therefore, effective therapy is imperative. The most common causes of death among patients with HCC are recurrence, metastasis, and the deterioration of primary tumors[5]. Most reports in the literature show that intrahepatic and extrahepatic metastases occur in > 50% of patients after resection of HCC with a high incidence of intrahepatic metastasis[6]. The common sites of extrahepatic metastasis include the lung, bone, peritoneum, spleen, and lymph nodes. HCC invasiveness is related to the ability of tumor cells to invade the capsule and portal vein[7,8].
Epithelial cell layers lose polarity and cell-cell contacts after epithelial-mesenchymal transition (EMT), a key event in the tumor invasion process, resulting in dramatic remodeling of the cytoskeleton[9]. E-cadherin is a central component of cell-cell adhesion junctions in the maintenance of cell polarity and environment[10,11], and the loss of E-cadherin expression is a hallmark of EMT and is associated with tumor invasiveness, metastasis, and poor prognosis[12]. The activation of various ligands, including fibroblast growth factor, transforming growth factor-β (TGF-β), bone morphogenetic protein, Wnt, epidermal growth factor (EGF), vascular endothelial growth factor (VEGF), and hepatocyte growth factor (HGF) and its receptor can upregulate the expression of EMT-regulating transcription factors, including snail homolog 1 (SNAI1), snail homolog 2 (SNAI2), zinc-finger E-box binding homeobox 1 (ZEB1), zinc-finger E-box binding homeobox 2 (ZEB2), and Twist[13]. Recent studies have demonstrated that EMT features can be induced by HGF in HCC, and E-cadherin transcription is repressed by several EMT-inducing regulators, of which Snail-related zinc finger transcription factors (Snail and Slug) are the most prominent via interaction with specific E-boxes of the proximal E-cadherin promoter[9,14-16]. To date, numerous clinicopathologic studies have shown positive correlations between the expression of the transcription factors SNAI1 and SNAI2, key inducible factors of EMT, and poor clinical outcomes in breast, ovarian, colorectal, and lung cancer, squamous cell carcinoma, melanoma, and HCC[13].
FoxM1, which consists of more than 50 amino acid residues and is characterized by a conserved 100 amino acid DNA binding domain, is a member of the FoxM family; FoxM1 is also a transcription factor that plays important roles in cell proliferation, organogenesis, aging and cancer[17-19]. FoxM1 has been clearly suggested to be an oncogenic protein complex, playing important roles in angiogenesis, invasion, and metastasis[20,21]. Interestingly, FoxM1 may regulate the EMT phenotype of pancreatic cancer cells by activation of mesenchymal cell markers[22], whereas the underlying mechanisms are unknown.
In the present study, we sought to determine the role of FoxM1 in EMT and metastasis of HCC as well as the regulatory role of FoxM1 in SNAIL expression and function. We discovered that the novel FoxM1-SNAIL signaling pathway critically regulates EMT, invasion, and metastasis of HCC.
Tissue specimens for tissue microarray (TMA) were obtained from 172 patients who underwent hepatectomy for HCC and 12 normal hepatic tissues were obtained from patients with hepatic hemangioma from 2006 to 2010 at The First Affiliated Hospital of Xi’an Jiaotong University, Shaanxi, China. Ethical approval was obtained from the research ethics committee of The First Affiliated Hospital of Xi’an Jiaotong University, and written informed consent was obtained from each patient. The preoperative clinical diagnosis of HCC met the diagnostic criteria of the American Association for the Study of Liver Diseases.
All tissue samples of the HCC TMA were embedded in paraffin for array studies and were freshly sectioned and stained with hematoxylin and eosin (HE). The representative regions of the lesion were reviewed carefully and defined by two pathologists. Based on the clinicopathologic information, specimens were grouped in tissue cylinders, and a diameter of 1 mm was taken from the selected regions of the donor block and then punched precisely into a recipient paraffin block using a tissue array instrument (Beecher Instruments, Silver Spring, MD). Consecutive 5 μm sections of the microarray blocks were made using a microtome. Finally, a TMA section with 172 HCC and 12 normal liver samples was constructed.
Formalin-fixed and paraffin-embedded sections with a thickness of 4 μm were dewaxed in xylene and graded alcohols, hydrated, and washed in PBS. After pretreatment in a microwave oven [12 min in sodium citrate buffer (pH 6)], the endogenous peroxidase was inhibited with 0.3% H2O2 for 15 min, and the sections were incubated with 10% normal goat serum for 15 min. Primary antibodies were applied overnight in a moist chamber at 4 °C. A standard avidin-biotin peroxidase technique (DAKO, Carpinteria, CA) was applied. Briefly, biotinylated goat anti-rabbit immunoglobulin and avidin-biotin peroxidase complex were applied for 30 min each, with 15-min washes in PBS. The reaction was finally developed using the Dako Liquid DAB+ substrate chromogen system (DAKO).
The HCC cell lines HepG2, HUH-7, SK-Hep1 and MHCC-97H used in this study were obtained from The Chinese Academy of Sciences (Shanghai, China), and were maintained in DMEM containing 10% fetal bovine serum (Gibco, Grand Island, NY, United States) at 37 °C with 5% CO2.
The plasmid pcDNA3.1-FoxM1 and control vector pcDNA3.1 were purchased from GenePharma China. The siRNA sequences targeting FoxM1 and SNAI1 were as follows: FoxM1-1, 5′-GGA CCA CUU UCC CUA CUU U-3′; FoxM1-2, 5′-CUC UUC UCC CUC AGA UAU A-3′; SNAI1-1, 5′-AGC UCA CAU CGC AUA CCG UA-3′; SNAI1-2, 5′-ACU CAG AUG UCA AGA AGU AUU-3′; Control, 5′-GCA AGC UGA CCC UGA AGU UCA U-3′.
The plasmids and/or oligonucleotides were transfected into cells using Lipofectamine 2000 (Invitrogen) or Lipofectamine RNAiMAX (Invitrogen) and were incubated for 48 h according to the manufacturer’s instructions. Stable cell lines were selected using the appropriate antibiotics for at least 48 h after transfection.
Cells samples were lysed in ice-cold RIPA lysis buffer (1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 150 mmol/L NaCl and 10 mmol/L Tris-HCl) containing a protease inhibitor cocktail. The concentration of total protein was calculated using the Bio-Rad Protein Assay (Bio-Rad, Hercules, CA, United States). Equivalent amounts of proteins (30 μg) were then separated by 10% SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA, United States). After being blocked in Tris-buffered saline containing 5% BSA, the membranes were incubated with primary antibodies [rabbit polyclonal anti-FoxM1 (1:500, Santa Cruz Biotechnology); rabbit polyclonal anti-SNAI1 (1:1000, Cell Signaling Technology); rabbit polyclonal anti-E-cadherin (1:1000, Cell Signaling Technology); rabbit polyclonal anti-vimentin (1:1000, Cell Signaling Technology); mouse monoclonal anti-β-actin (1:2000, Santa Cruz Biotechnology)] at 4 °C for 12 h and then with a horseradish peroxidase conjugated anti-rabbit or anti-mouse secondary antibody (Zhongshan, Beijing, China) at a dilution of 1:8000 at room temperature for 1 h. Signals were detected on X-ray film using an electrochemiluminescence detection system (Pierce, Rockford, IL, United States). Equal protein loading was assessed by the expression of β-actin.
Total RNA was isolated using Trizol reagent according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA). cDNA was synthesized using reverse transcriptase (Bio-Rad). Real-time RT-PCR amplification was performed using a Thermal Cycler Dice (Takara) in accordance with the manufacturer’s instructions under the following conditions: 95 °C for 6 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. Glyceraldehyde 3-phosphate dehydrogenase was used to normalize the expression levels in the subsequent quantitative analyses. To amplify the target genes, the following primers were purchased from TaKaRa: FoxM1, forward 5′-TGC AGC AGG GAT GTG AAT CTT C-3′ and reverse 5′-GGA GCC CAG TCC ATC AGA ACT-3′; SNAI1, forward 5′-TCT AGG CCC TGG CTG CTA CAA-3′ and reverse 5′-ACA TCT GAG TGG GTC TGG AGG TG-3′; E-cadherin, forward 5′-TAA CCG ATC AGA ATG AC-3′ and reverse 5′-TTT GTC AGG GAG CTC AGG AT-3′; and GAPD, forward 5′-GCA CCG TCA AGG CTG AGA AC-3′ and reverse 5′-ATG GTG GTG AAG ACG CCA GT-3′.
Cells were cultivated for 24 h and then transferred on the top of Matrigel-coated chambers (24-well insert, 8-μm pore size, BD Biosciences, San Jose, United States) in serum-free DMEM. DMEM containing 20% fetal calf serum was added to the lower chamber as a chemoattractant. After incubation for 48 h, non-invaded cells were removed from the upper well with cotton swabs, while the invaded cells were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, and photographed (× 200) in five independent fields for each well. Each test was repeated in triplicate.
HepG2 and Huh7 cells (2 × 104 cells/well, respectively) were seeded in 6-well tissue culture dishes. After 24 h of incubation, the cells were analyzed using a light microscope. The experiment was performed in triplicate.
The SNAI1 promoter was PCR amplified and cloned into the pGL2-basic luciferase vector using the following primers: 5′-TCT TAC CCC GGG CCT TTC CCC TCG-3′ and 5′-CCG CTC GAG TGG CCA GAG CGA CCT AG-3′. Dual luciferase assay (Promega, Madison, Wisconsin) was performed 24 h after transfection. Cells were crosslinked by the addition of formaldehyde and sonicated to create DNA fragments between 500 and 1000 bp. Protein/DNA complexes were used for immunoprecipitation with Foxm1 antibodies (C20, Santa Cruz Biotechnology) or control rabbit serum. Reverse crosslinked chromatin immunoprecipitation (ChIP) DNA samples were subjected to real-time PCR using primers specific to human SNAI1 (5′-TTC AAC GAA ACT CTA ACC AGG TCC-3′ and 5′-TGA GGG AGA CAG ACG AAG TAA ACA G-3′).
All graphical values are represented by mean ± SE from three independent experiments with each measured in triplicate. The differences between two groups were analyzed using unpaired two-tailed Student’s t-test. Categorical data were analyzed by Fisher’s exact test. The cumulative recurrence and overall survival rates were calculated by the Kaplan-Meier method and the log-rank test. P-values less than 0.05 were considered statistically significant and are indicated with asterisks as described in the figure legends.
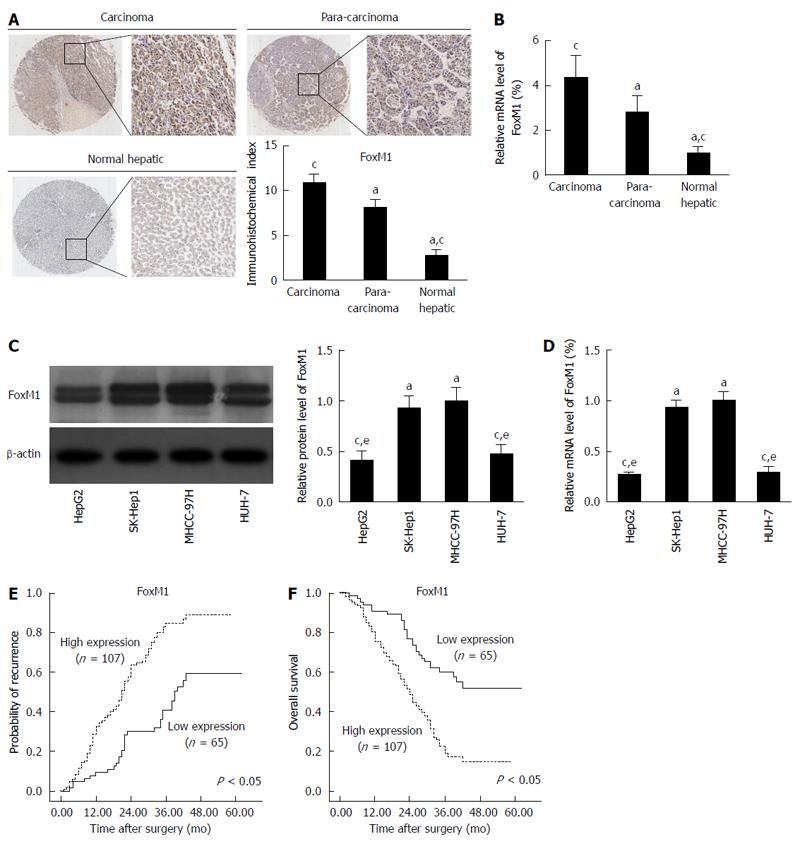
The clinicopathologic characteristics of the patients are summarized in Table 1. The overall follow-up durations ranged from 1 to 62 mo (median, 31 mo). To determine the role of FoxM1 in HCC metastasis, we evaluated FoxM1 expression in TMA specimens by immunostaining. In 172 carcinoma tissue specimens from Chinese HCC patients, their matched para-carcinoma tissues and 12 normal hepatic tissues (from the patients with hepatic hemangioma), the immunohistochemical indexes of FoxM1 were 10.7 ± 0.9, 8.2 ± 0.7 and 2.7 ± 0.4, respectively (Figure 1A). FoxM1 expression increased significantly in carcinoma compare with para-carcinoma (P < 0.05) and normal hepatic tissues (P < 0.05). Additionally, we used qRT-PCR to gauge the mRNA expression of FoxM1 in carcinoma, para-carcinoma and normal hepatic tissues, and the results were consistent with the immunohistochemical data (Figure 1B). Combined with clinical data analysis, abnormal FoxM1 expression was interrelated with tumor size (P = 0.002), tumor number (P = 0.010), macrovascular invasion (P = 0.002) and higher TNM stage (P = 0.008), whereas no substantial differences were found regarding patient age, gender, underlying liver disease, liver function, preoperative AFP level, cirrhotic background and tumor differentiation between high and low levels of FoxM1 (Table 2). In vitro, we compared FoxM1 expression in various HCC cell lines with different metastatic potentials by Western blot and qRT-PCR. A significant increase in FoxM1 at both the protein and mRNA levels was observed in metastatic MHCC-97H and SK-Hep1 compared with nonmetastatic HepG2 and Huh-7 cell lines (Figure 1C and D). To further confirm that FOXM1 was associated with metastasis, we used Kaplan-Meier analysis to reveal that patients with high expression of FoxM1 had a higher recurrence risk and shorter overall survival than those with low expression of FoxM1 (Figure 1E and F). Using a multivariate Cox proportional hazard model, we confirmed that the FoxM1 expression level was an independent and significant factor for disease-free survival in HCC patients after surgical resection (Table 3). Collectively, FoxM1 overexpression correlates significantly with HCC metastasis.
| Characteristic | Patients |
| Gender | |
| Male | 116 (67) |
| Female | 56 (33) |
| Age (yr) | |
| ≤ 50 | 73 (42) |
| > 50 | 99 (58) |
| Underlying liver disease | |
| HBV | 133 (77) |
| Others | 39 (23) |
| Liver function (Child-Pugh classification) | |
| Grade A | 157 (91) |
| Grade B | 11 (6) |
| Grade C | 4 (3) |
| Preoperative AFP level | |
| ≤ 200 ng/mL | 66 (38) |
| > 200 ng/mL | 90 (52) |
| Missing data | 16 (10) |
| Tumor size | |
| ≤ 8 cm | 116 (67) |
| > 8 cm | 51 (30) |
| Undetermined | 5 (3) |
| Tumor number | |
| ≤ 3 | 157 (91) |
| > 3 | 15 (9) |
| Cirrhotic background | |
| No | 39 (23) |
| Yes | 133 (77) |
| Macrovascular invasion | |
| No | 149 (87) |
| Yes | 23 (13) |
| Tumor differentiation | |
| G1/G2 | 154 (90) |
| G3 | 10 (6) |
| Undetermined | 8 (4) |
| TNM stage | |
| Stage I + II | 135 (78) |
| Stage III (IIIA+IIIB+IIIC) | 37 (22) |
| Variable | Patients | FOXM1 expression | P-value | E-cadherin expression | P-value | ||
| Low | High | Low | High | ||||
| Gender | |||||||
| Male | 116 (67) | 48 (41) | 68 (59) | 0.185 | 73 (63) | 43 (37) | 0.187 |
| Female | 56 (33) | 17 (30) | 39 (70) | 29 (52) | 27 (48) | ||
| Age (yr) | |||||||
| ≤ 50 | 73 (42) | 24 (33) | 49 (67) | 0.270 | 42 (58) | 31 (42) | 0.754 |
| > 50 | 99 (58) | 41 (41) | 58 (59) | 60 (61) | 39 (39) | ||
| Underlying liver disease | |||||||
| HBV | 133 (69) | 51 (38) | 82 (62) | 0.852 | 80 (60) | 53 (40) | 0.713 |
| Others | 39 (31) | 14 (36) | 25 (64) | 22 (56) | 17 (44) | ||
| Liver function (Child-Pugh classification) | |||||||
| Grade A | 157 (91) | 59 (38) | 98 (62) | 0.720 | 93 (59) | 64 (41) | 0.913 |
| Grade B | 11 (6) | 4 (36) | 7 (64) | 7 (64) | 4 (36) | ||
| Grade C | 4 (3) | 2 (50) | 2 (50) | 2 (50) | 2 (50) | ||
| Preoperative AFP level | |||||||
| ≤ 200 ng/mL | 66 (42) | 19 (29) | 47 (71) | 0.095 | 40 (61) | 26 (39) | 0.330 |
| > 200 ng/mL | 90 (58) | 38 (42) | 52 (58) | 47 (52) | 43 (48) | ||
| Tumor size | |||||||
| ≤ 8 cm | 116 (69) | 52 (45) | 64 (55) | 0.002a | 71 (61) | 45 (39) | 0.611 |
| > 8 cm | 51 (31) | 10 (20) | 41 (80) | 29 (57) | 22 (43) | ||
| Tumor number | |||||||
| ≤ 3 | 157 (91) | 64 (40) | 93 (60) | 0.010a | 95 (61) | 62 (39) | 0.410 |
| > 3 | 15 (9) | 1 (7) | 14 (93) | 7 (47) | 8 (53) | ||
| Cirrhotic background | |||||||
| No | 39 (23) | 11 (28) | 28 (72) | 0.191 | 18 (46) | 21 (54) | 0.066 |
| Yes | 133 (77) | 54 (41) | 79 (59) | 84 (63) | 49 (37) | ||
| Macrovascular invasion | |||||||
| No | 149 (87) | 63 (42) | 86 (58) | 0.002a | 82 (55) | 67 (45) | 0.003a |
| Yes | 23 (13) | 2 (9) | 21 (91) | 20 (87) | 3 (13) | ||
| Tumor differentiation | |||||||
| G1/G2 | 154 (94) | 56 (34) | 98 (66) | 0.502 | 87 (56) | 67 (44) | 0.047a |
| G3 | 10 (6) | 5 (80) | 5 (20) | 9 (90) | 1 (10) | ||
| TNM stage | |||||||
| Stage I + II | 135 (78) | 58 (43) | 77 (57) | 0.008a | 73 (53) | 62 (47) | 0.008a |
| Stage III (IIIA+IIIB+IIIC) | 37 (22) | 7 (19) | 30 (81) | 29 (78) | 8 (22) | ||
| Variable | P-value | |||
| Univariate analysis | Multivariate analysis | |||
| RFS P-value | OS | RFS P-value | OS P-value | |
| Gender (male vs female) | 0.152 | 0.331 | - | - |
| Age ( ≤ 50 yr vs > 50 yr) | 0.478 | 0.663 | - | - |
| Underlying disease (HBV vs others) | 0.461 | 0.859 | - | - |
| Liver function (A/B vs C) | 0.764 | 0.493 | - | - |
| Preoperative AFP level ( ≤ 200 ng/mL vs > 200 ng/mL) | 0.179 | 0.228 | - | - |
| Tumor size ( ≤ 8 cm vs > 8 cm) | < 0.002a | 0.012a | 0.174 | 0.391 |
| Tumor number ( ≤ 3 vs > 3) | 0.002a | 0.015a | 0.228 | 0.348 |
| Cirrhotic background (no vs yes) | 0.172 | 0.373 | - | - |
| Macrovascular invasion (no vs yes) | < 0.001a | < 0.001a | < 0.001a | 0.003a |
| Tumor differentiation (G1/G2 vs G3) | 0.131 | 0.179 | - | - |
| TNM stage (I/II vs III) | < 0.001a | 0.006a | 0.013a | 0.011a |
| FOXM1 expression (low vs high) | 0.004a | 0.007a | 0.012a | 0.008a |
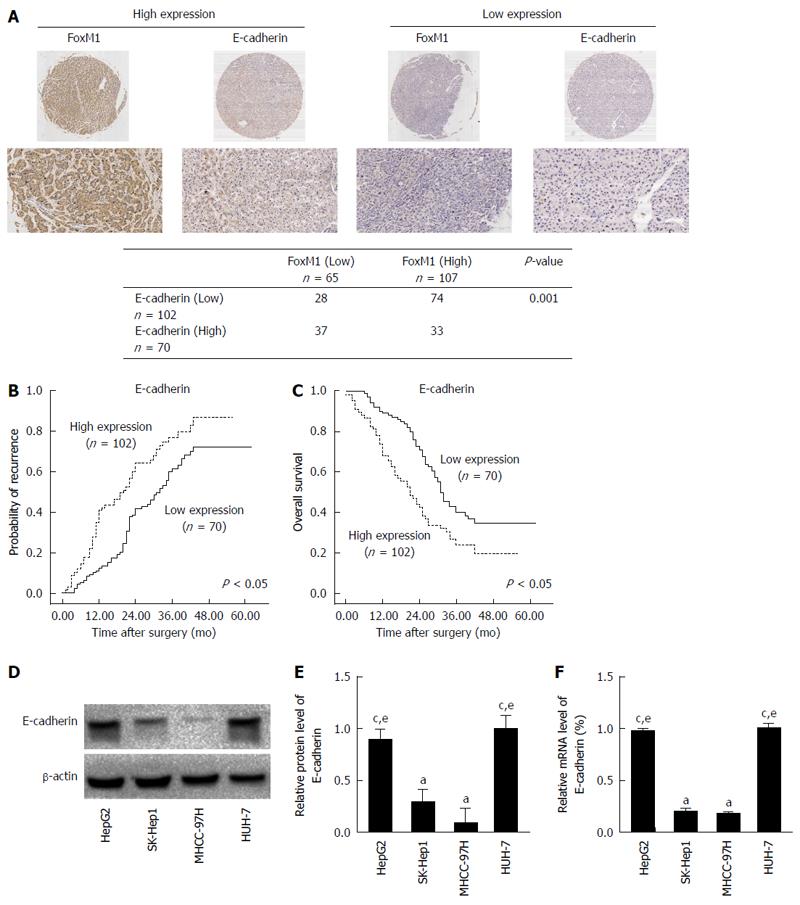
To further estimate the possible correlation of FoxM1 expression with metastasis and EMT, we evaluated the protein expression of E-cadherin in another set of TMA specimens. The E-cadherin expression was correlated with FoxM1 in TMA specimens, and FoxM1 expression was negatively correlated with E-cadherin expression in HCC tissues (Figure 2A). Statistical analysis showed that E-cadherin overexpression was negatively correlated with microvascular invasion, malignant differentiation, and TNM stage (Table 2). Kaplan-Meier analysis revealed that patients with low expression of E-cadherin had a higher recurrence rate and shorter overall survival than those with high expression of E-cadherin (Figure 2B and C). Additionally, we investigated the correlation of FoxM1 with E-cadherin in HCC by examination of E-cadherin mRNA and protein levels in the above HCC cell lines with different metastatic potentials by Western blot and qRT-PCR. A significant decrease in E-cadherin at the protein and mRNA levels was revealed in metastatic MHCC-97H and SK-Hep1 compared with nonmetastatic HepG2 and Huh-7 cell lines (Figure 2D and E). The above evidence indicates that FoxM1 overexpression was negatively correlated with E-cadherin expression in HCC, and may be correlated with EMT.
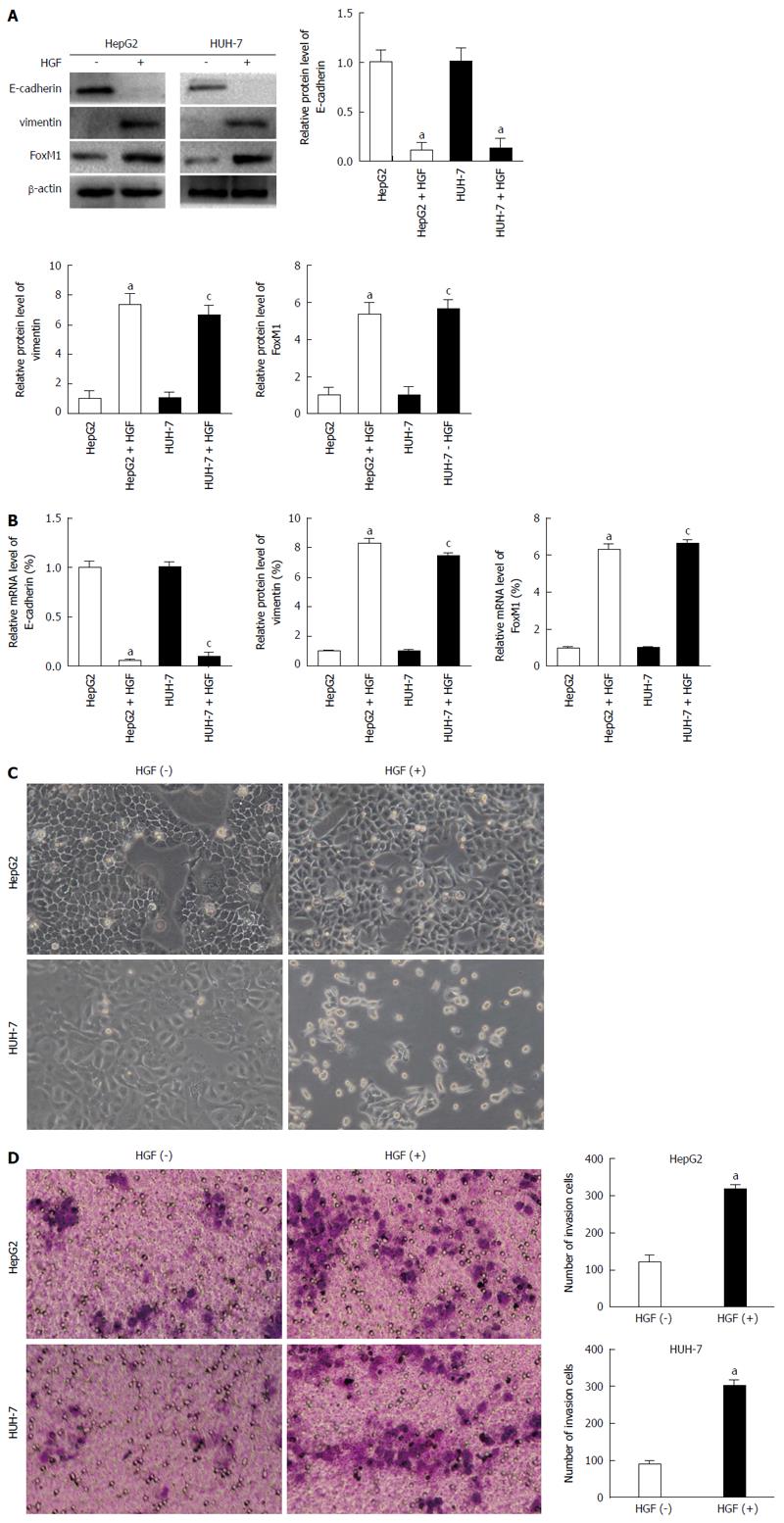
In previous studies, it had been demonstrated that HGF induces EMT and stimulates cell migration in HCC cells. In HepG2 and HUH-7 cells, we observed that both the protein and mRNA levels of FoxM1 and vimentin were significantly increased by HGF, in contrast to those of E-cadherin (Figure 3A and B). Meanwhile, we examined cellular morphology after HGF stimulation to evaluate whether HGF mediates the morphologic changes. HGF clearly mediated both cell scattering and the elongation of the cell shape, and resulted in morphologic changes from tightly packed colonies to scattered growth structure in HepG2 and HUH-7 cell lines, which are consistent with mesenchymal morphology (Figure 3C). These data indicate that HGF mediates the morphologic changes that are compatible with the induction of EMT in the HCC cell lines. In addition, HGF stimulation resulted in enhanced cell invasion (Figure 3D).
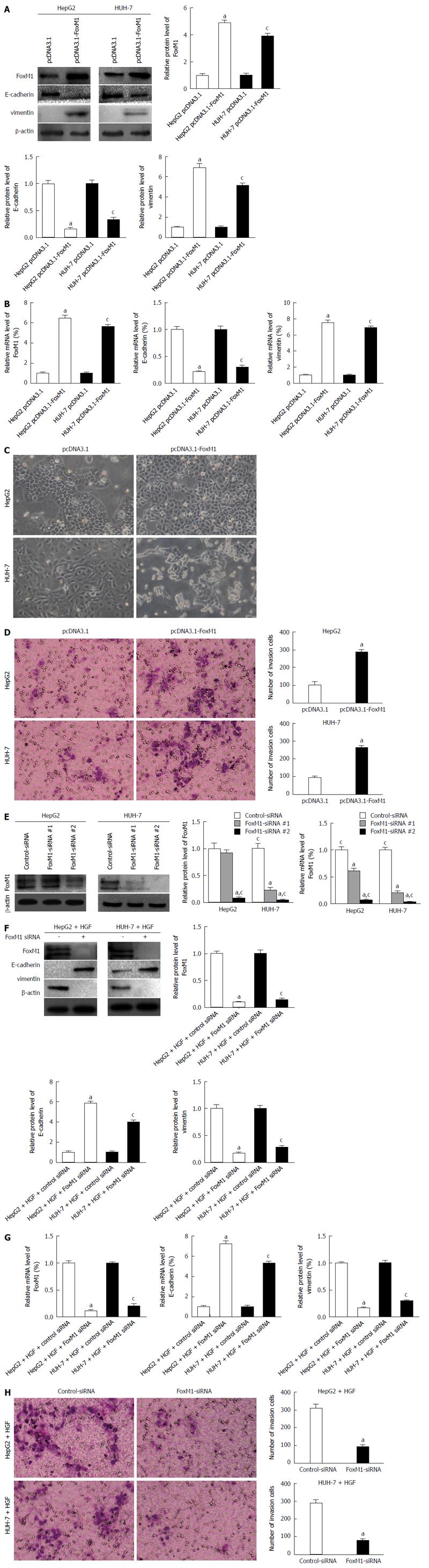
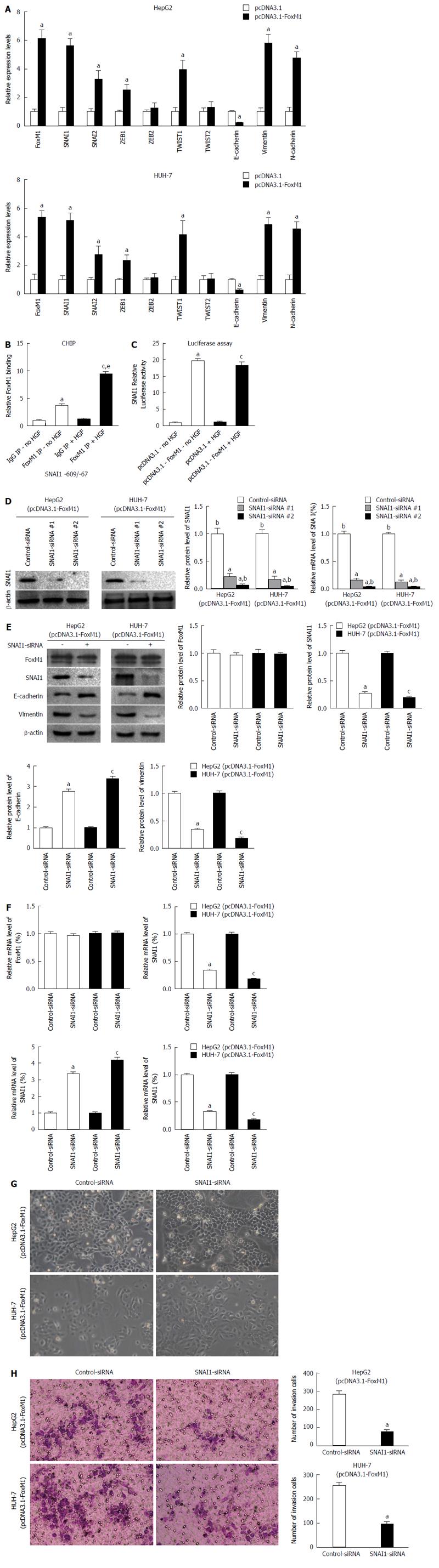
To confirm the effect of altered FoxM1 expression on the epithelial or mesenchymal phenotype of HCCs, we transfected the FoxM1 expression vector pcDNA3.1-FoxM1 or the control vector pcDNA3.1 into HepG2 and HUH-7 cells, which typically express low levels of FoxM1 and display an epithelial phenotype. We observed that both the protein and mRNA levels of vimentin were significantly increased by elevated expression of FoxM1 in HepG2 and HUH-7 cells, in contrast to those of E-cadherin (Figure 4A and B), and caused both typical morphology and invasion changes of EMT (Figure 4C and D). To further confirm the effect of FoxM1 in HGF-induced EMT, we transfected two sequences of FoxM1-siRNA (1 and 2) or control-siRNA (consists of a scrambled sequence that will not lead to the specific degradation of any cellular message) into HepG2 and HUH-7 cells. Western blot and qRT-PCR showed that the expression of FoxM1 was more suppressed by FoxM1-siRNA-2 in the HepG2 and HUH-7 cells (Figure 4E); thus, mixed siRNA was used in subsequent experiments to avoid unexpected off-target effects. FoxM1 knockdown led to a decrease in vimentin but an increase in E-cadherin expression (Figure 4F and G), and caused invasion changes in both cell lines (Figure 4H). The above results indicated that FoxM1 plays a key role in HGF-induced EMT. Moreover, altered FoxM1 expression may contribute to the epithelial or mesenchymal phenotype of HCC cells.
Several transcription factors have the potential to repress the expression of E-cadherin and induce EMT, including SNAI1, SNAI2, ZEB1, ZEB2, Twist1 and Twist2. We observed that the expression of EMT-related molecules was altered under the effect of FoxM1, and the expression of SNAIL1 had a greater magnitude change compared to other molecules (Figure 5A). Considering that FoxM1 induces the expression of SNAI1, we investigated whether SNAI1 is a direct transcriptional target of FoxM1. We identified a potential FoxM1-binding site within the - 1.0 kb promoter regions of human SNAI1 genes. Next, we determined whether FoxM1 binds to the promoter region of the SNAI1 gene by ChIP assay in HepG2 cells. After HGF treatment, the specific binding of FoxM1 protein to the SNAI1 promoter DNA was increased (Figure 5B). Because we transfected pcDNA3.1-FoxM1 into HepG2 cells, the activity of the SNAI1 promoter region was significantly increased in a luciferase reporter assay (Figure 5C). To further confirm the effect of SNAI1 in FoxM1-mediated EMT, we used two sequences of SNAI1-siRNA (1 and 2). Western blot and qRT-PCR showed that the FoxM1-mediated upregulation of SNAI1 was suppressed by both SNAI1-siRNA-1 and SNAI1-siRNA-2 in the HepG2 and HUH-7 cells (Figure 5D); thus, mixed siRNA was used in subsequent experiments to avoid unexpected off-target effects. SNAI1-siRNA strongly reversed the FoxM1- mediated downregulation of E-cadherin and upregulation of vimentin in both HepG2 and Huh7 cells (Figure 5E and F). Similarly, we found that the FoxM1-mediated morphologic changes were prevented by siRNA-mediated knockdown of SNAI1 in both HepG2 and HUH-7 cells, whereas control-siRNA did not show an effect (Figure 5G). Finally, we confirmed that the siRNA-mediated knockdown of SNAI1 inhibited FoxM1-mediated cellular invasion (Figure 5H). Collectively, these results indicate that SNAI1 is required to induce the FoxM1-mediated EMT in HCC cells.
HCC invasiveness indicates poor prognosis as a key step leading to metastasis[23]. Recently, growing evidence has shown that tumor progression, including local invasion, spreading through the circulation and metastasis, is mediated by EMT, which is a process first identified in embryogenesis[24]. In the present study, we assessed the expression levels of FoxM1 by immunostaining in a cohort of 172 Chinese HCC patients and demonstrated that elevated expression of FoxM1 was associated with several clinicopathological factors and predicted a poor prognosis in HCC patients. Next, we found that the abnormal increase in FoxM1 expression was significantly correlated with low expression of E-cadherin, representing EMT and higher metastatic abilities. In vitro, we found that FoxM1 plays a pivotal role in HGF-induced EMT and increased expression of FoxM1 can induce EMT in epithelioid-phenotype HCC cells. Additionally, we have confirmed that FOXM1 directly binds to and activates the SNAI1 promoter, constituting a novel signaling pathway that affects EMT, invasion, and metastasis of HCC cells, and controlling the clinicopathologic behavior of HCC.
Metastasis is a complicated multi-step process[25] that includes decreased adhesion and increased motility, cell attachment, matrix dissolution and migration[26]. In the process of tumor progression, cancer cells undergo significant changes in cytoskeletal organization to adopt an invasive phenotype, ultimately metastasizing to other organs[27]. Emerging evidence has suggested a key role of EMT in metastasis through its significant influence on the migratory activity of tumor cells[28,29]. Morphology changes in epithelial cells toward the acquisition of a spindle-like shape, epithelial marker loss (such as E-cadherin), and mesenchymal marker emergence (typically, N-cadherin and vimentin) indicate typical EMT features[30]. In various tumor cell lines, HGF has been shown to cause EMT and stimulate cell invasion[31]. In addition, EMT could also be induced by certain other factors, such as TGF-β, basic fibroblast growth factor, EGF, VEGF, and Wnt ligands[30]. In the present study, HGF was used to induce a mesenchymal phenotype with EMT features. We found that the expression level of E-cadherin decreased significantly in the presence of HGF, whereas the expression level of vimentin was increased. In addition, HGF increased the expression level of FoxM1, and reduced expression of FoxM1 blunts HGF-induced EMT. The latter result indicates that HGF-mediated FoxM1 up-regulation is responsible for the epithelial-mesenchymal transition.
Many solid tumors show elevated expression of FoxM1, including but not limited to HCC, breast carcinoma, non-small cell lung cancer, and colon carcinoma, and FoxM1 has been demonstrated as one of the most commonly up-regulated genes by microarray analysis of human solid tumors[32]. Recently, research has shown that the increased FoxM1 expression could induce the occurrence of EMT in pancreatic, lung and breast cancer cells[33,34]; however, a similar effect has not yet been found in HCC. Our studies have shown that high expression of FoxM1 was correlated with low expression of E-cadherin and poor prognosis in HCC, preliminarily indicating that the correlation between FoxM1 and poor prognosis of liver cancer might be associated with EMT. Furthermore, we have confirmed that FOXM1 directly binds to and activates the SNAI1 promoter, and identified SNAI1 as a direct transcriptional target of FOXM1. This finding provides a mechanism by which FOXM1 induces EMT and potentially contributes to the metastasis of HCC. Other mechanisms have been suggested in previous studies to explain how FOXM1 regulates EMT in cancer. For example, FOXM1 can modulate the migration of breast cancer cells through regulation of the expression of extracellular matrix degradation factors such as urokinase-type plasminogen activator (uPA), uPA receptor, matrix metalloproteinase-2, and matrix metalloproteinase-9[35]. Additionally, overexpression of FOXM1 can induce the EMT process indirectly by up-regulating ZEB1 and ZEB2 and down-regulating microRNA-200b[22]. Evidence has also shown enhancement of Snail expression during radiation-induced pulmonary fibrosis by EMT induction by FOXM1[36]. Whether FOXM1 directly or indirectly affects the expression of other EMT key factors requires further investigation.
The mechanisms underlying the SNAI1-induced metastatic and aggressive phenotypes of cancer cells in both basic and clinical research studies have recently been intensively investigated. It has been suggested that SNAI1 may act as a switch to promote the EMT program in various cells because SNAI1 was sufficient to induce EMT and the expression of EMT-associated genes. Consistent with the critical role of SNAI1 in EMT, the present study confirmed that SNAI1 was induced during FoxM1-mediated EMT. We found that the expression of SNAI1 was significantly increased in the presence of FoxM1, a finding that may be due to the transcriptional activity of SNAI1 promoter region being able to be induced by FoxM1 in co-transfection experiments. Moreover, inhibiting the expression of SNAI1 can significantly inhibit FoxM1-mediated EMT. Overall, these results indicate that SNAI1 is required to induce the FoxM1-mediated EMT in HCC cells.
In summary, the present study conclusively presents evidence of the following: the role of FOXM1 in promoting EMT and metastasis of hepatocellular carcinoma; the critical role of the upstream FoxM1 transcription factor in HGF-induced EMT; and the critical role of SNAI1 in FoxM1-mediated EMT. Collectively, a novel molecular mechanism of HCC EMT could be proposed, based on which a potentially effective therapeutic approach could be developed for the inhibition of migration, invasion, and metastasis of HCC.
Hepatocellular carcinoma (HCC) is a rapidly growing tumor associated with a high propensity for vascular invasion and metastasis. The epithelial-mesenchymal transition (EMT) has emerged as a pivotal event in the development of the invasive and metastatic potentials, although the cause of this process is largely unknown. Progression of this disease is associated with a proliferation-associated transcription factor, forkhead box M1 (FoxM1), which is a crucial molecule involved in cell proliferation, organogenesis, aging and cancer.
FoxM1 overexpression is common in most malignant tumors of the digestive system, and it has been clearly suggested to be an oncogenic protein complex, playing important roles in angiogenesis, invasion, and metastasis. However, the actual role of FoxM1 in the epithelial-mesenchymal transition of hepatocellular carcinoma remains unclear. In the present study, the authors demonstrate that the overexpression of FoxM1 could be a potential mechanism for mediating the epithelial-mesenchymal transition, and snail homolog 1 (SNAI1) plays a critical role in FoxM1-mediated epithelial-mesenchymal transition.
Recent reports have highlighted the importance of various transcription factors, including FoxM1, in the development of the EMT. In this study, authors demonstrated that elevated expression of FoxM1 was associated with several clinicopathological factors and predicted a poor prognosis in HCC patients. Moreover, they found that the abnormal increase of FoxM1 expression was significantly correlated with low expression of E-cadherin, which represents the epithelial- mesenchymal transition and higher metastatic abilities. In vitro, they first found that FoxM1 plays a pivotal role in HGF-induced EMT. Additionally, they have confirmed that FoxM1 directly binds to and activates the SNAI1 promoter, constituting a novel signaling pathway that affects the EMT, invasion, and metastasis of hepatocellular carcinoma cells and suggesting that FoxM1 is a potential therapy target.
By understanding how FoxM1 is functionally involved in the epithelial-mesenchymal transition in hepatocellular carcinoma and by blocking its expression, this study proposes a future strategy for therapeutic intervention in the treatment of patients with hepatocellular carcinoma.
Morphology changes of epithelial cells toward the acquisition of a spindle-like shape, epithelial marker loss (such as E-cadherin), and mesenchymal marker emergence (typically, N-cadherin and vimentin) indicate typical EMT features. Such a mechanism is thought to be crucial in the invasion and metastasis of cancer. Not surprisingly, the progression of EMT and metastasis is promoted by FoxM1 overexpression in hepatocellular carcinoma, and SNAI1 plays a critical role in FoxM1-mediated EMT.
The authors described the roles of FOXM1 in the EMT in hepatocellular carcinoma. They discovered that the novel FoxM1-SNAIL signaling pathway critically regulates hepatocellular carcinoma EMT, invasion, and metastasis. This research is well organized and of relevant interest and scientific innovation.
P- Reviewer: Hong J, Miyazaki T S- Editor: Gou SX L- Editor: Wang TQ E- Editor: Wang CH
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25524] [Article Influence: 1823.1] [Reference Citation Analysis (7)] |
| 2. | Nordenstedt H, White DL, El-Serag HB. The changing pattern of epidemiology in hepatocellular carcinoma. Dig Liver Dis. 2010;42 Suppl 3:S206-S214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 404] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 3. | Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 929] [Cited by in RCA: 1102] [Article Influence: 100.2] [Reference Citation Analysis (1)] |
| 4. | Tang ZY, Ye SL, Liu YK, Qin LX, Sun HC, Ye QH, Wang L, Zhou J, Qiu SJ, Li Y. A decade’s studies on metastasis of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2004;130:187-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 336] [Cited by in RCA: 361] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 5. | Lee SC, Tan HT, Chung MC. Prognostic biomarkers for prediction of recurrence of hepatocellular carcinoma: current status and future prospects. World J Gastroenterol. 2014;20:3112-3124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 63] [Cited by in RCA: 69] [Article Influence: 6.3] [Reference Citation Analysis (2)] |
| 6. | Tung-Ping Poon R, Fan ST, Wong J. Risk factors, prevention, and management of postoperative recurrence after resection of hepatocellular carcinoma. Ann Surg. 2000;232:10-24. [PubMed] |
| 7. | Rodríguez-Perálvarez M, Luong TV, Andreana L, Meyer T, Dhillon AP, Burroughs AK. A systematic review of microvascular invasion in hepatocellular carcinoma: diagnostic and prognostic variability. Ann Surg Oncol. 2013;20:325-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 504] [Cited by in RCA: 493] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 8. | Toyosaka A, Okamoto E, Mitsunobu M, Oriyama T, Nakao N, Miura K. Intrahepatic metastases in hepatocellular carcinoma: evidence for spread via the portal vein as an efferent vessel. Am J Gastroenterol. 1996;91:1610-1615. [PubMed] |
| 9. | Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2783] [Cited by in RCA: 2975] [Article Influence: 141.7] [Reference Citation Analysis (0)] |
| 10. | Hirohashi S, Kanai Y. Cell adhesion system and human cancer morphogenesis. Cancer Sci. 2003;94:575-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 11. | Sugimachi K, Taguchi K, Aishima S, Tanaka S, Shimada M, Kajiyama K, Sugimachi K, Tsuneyoshi M. Altered expression of beta-catenin without genetic mutation in intrahepatic cholangiocarcinoma. Mod Pathol. 2001;14:900-905. [PubMed] |
| 12. | Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1400] [Cited by in RCA: 1657] [Article Influence: 127.5] [Reference Citation Analysis (0)] |
| 13. | Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2365] [Cited by in RCA: 2490] [Article Influence: 138.3] [Reference Citation Analysis (0)] |
| 14. | Nagai T, Arao T, Furuta K, Sakai K, Kudo K, Kaneda H, Tamura D, Aomatsu K, Kimura H, Fujita Y. Sorafenib inhibits the hepatocyte growth factor-mediated epithelial mesenchymal transition in hepatocellular carcinoma. Mol Cancer Ther. 2011;10:169-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 195] [Reference Citation Analysis (0)] |
| 15. | Peinado H, Portillo F, Cano A. Transcriptional regulation of cadherins during development and carcinogenesis. Int J Dev Biol. 2004;48:365-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 434] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 16. | Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1272] [Cited by in RCA: 1328] [Article Influence: 57.7] [Reference Citation Analysis (0)] |
| 17. | Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. 2002;250:1-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 624] [Cited by in RCA: 674] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 18. | Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 484] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 19. | Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 211] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 20. | Gartel AL. FoxM1 inhibitors as potential anticancer drugs. Expert Opin Ther Targets. 2008;12:663-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 21. | Park HJ, Gusarova G, Wang Z, Carr JR, Li J, Kim KH, Qiu J, Park YD, Williamson PR, Hay N. Deregulation of FoxM1b leads to tumour metastasis. EMBO Mol Med. 2011;3:21-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 22. | Bao B, Wang Z, Ali S, Kong D, Banerjee S, Ahmad A, Li Y, Azmi AS, Miele L, Sarkar FH. Over-expression of FoxM1 leads to epithelial-mesenchymal transition and cancer stem cell phenotype in pancreatic cancer cells. J Cell Biochem. 2011;112:2296-2306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 175] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 23. | Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10002] [Cited by in RCA: 10447] [Article Influence: 696.5] [Reference Citation Analysis (0)] |
| 24. | Franco-Chuaire ML, Magda Carolina SC, Chuaire-Noack L. Epithelial-mesenchymal transition (EMT): principles and clinical impact in cancer therapy. Invest Clin. 2013;54:186-205. [PubMed] |
| 25. | Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559-1564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3059] [Cited by in RCA: 3604] [Article Influence: 257.4] [Reference Citation Analysis (0)] |
| 26. | Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423-1437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4060] [Cited by in RCA: 5713] [Article Influence: 519.4] [Reference Citation Analysis (0)] |
| 27. | Choi C, Helfman DM. The Ras-ERK pathway modulates cytoskeleton organization, cell motility and lung metastasis signature genes in MDA-MB-231 LM2. Oncogene. 2014;33:3668-3676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 28. | Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, Thompson EW. Epithelial--mesenchymal and mesenchymal--epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213:374-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 783] [Cited by in RCA: 827] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 29. | Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548-558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1362] [Cited by in RCA: 1447] [Article Influence: 72.4] [Reference Citation Analysis (0)] |
| 30. | Zheng H, Kang Y. Multilayer control of the EMT master regulators. Oncogene. 2014;33:1755-1763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 254] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 31. | Cañadas I, Rojo F, Taus Á, Arpí O, Arumí-Uría M, Pijuan L, Menéndez S, Zazo S, Dómine M, Salido M. Targeting epithelial-to-mesenchymal transition with Met inhibitors reverts chemoresistance in small cell lung cancer. Clin Cancer Res. 2014;20:938-950. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 109] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 32. | Wierstra I. FOXM1 (Forkhead box M1) in tumorigenesis: overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Adv Cancer Res. 2013;119:191-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 151] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 33. | Balli D, Ustiyan V, Zhang Y, Wang IC, Masino AJ, Ren X, Whitsett JA, Kalinichenko VV, Kalin TV. Foxm1 transcription factor is required for lung fibrosis and epithelial-to-mesenchymal transition. EMBO J. 2013;32:231-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 149] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 34. | Yang C, Chen H, Tan G, Gao W, Cheng L, Jiang X, Yu L, Tan Y. FOXM1 promotes the epithelial to mesenchymal transition by stimulating the transcription of Slug in human breast cancer. Cancer Lett. 2013;340:104-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 86] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 35. | Ahmad A, Wang Z, Kong D, Ali S, Li Y, Banerjee S, Ali R, Sarkar FH. FoxM1 down-regulation leads to inhibition of proliferation, migration and invasion of breast cancer cells through the modulation of extra-cellular matrix degrading factors. Breast Cancer Res Treat. 2010;122:337-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 114] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 36. | Wang X, Quail E, Hung NJ, Tan Y, Ye H, Costa RH. Increased levels of forkhead box M1B transcription factor in transgenic mouse hepatocytes prevent age-related proliferation defects in regenerating liver. Proc Natl Acad Sci USA. 2001;98:11468-11473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 168] [Article Influence: 7.0] [Reference Citation Analysis (0)] |