Published online Dec 28, 2014. doi: 10.3748/wjg.v20.i48.18284
Revised: August 28, 2014
Accepted: October 15, 2014
Published online: December 28, 2014
Processing time: 341 Days and 11.2 Hours
AIM: To construct a tricistronic hepatitis C virus (HCV) replicon with double internal ribosome entry sites (IRESes) of only 22 nucleotides for each, substituting the encephalomyocarditis virus (EMCV) IRESes, which are most often used as the translation initiation element to form HCV replicons.
METHODS: The alternative 22-nucleotide IRES, RNA-binding motif protein 3 IRES (Rbm3 IRES), was used to form a tricistronic HCV replicon, to facilitate constructing HCV-harboring stable cell lines and successive antiviral screening using a luciferase marker. Briefly, two sequential Rbm3 IRESes were inserted into bicistronic pUC19-HCV plasmid, consequently forming a tricistronic HCV replicon (pHCV-rep-NeoR-hRluc), initiating the translation of humanized Renilla luciferase and HCV non-structural gene, along with HCV authentic IRES initiating the translation of neomycin resistance gene. The sH7 cell lines, in which the novel replicon RNA stably replicated, were constructed by neomycin and luciferase activity screening. The intracellular HCV replicon RNA, expression of inserted foreign genes and HCV non-structural gene, as well as response to anti-HCV agents, were measured in sH7 cells and cells transiently transfected with tricistronic replicon RNA.
RESULTS: The intracellular HCV replicon RNA and expression of inserted foreign genes and HCV non-structural gene in sH7 cells and cells transiently transfected with tricistronic replicon RNA were comparable to those in cells stably or transiently transfected with traditional bicistronic HCV replicons. The average relative light unit in pHCV-rep-NeoR-hRluc group was approximately 2-fold of those in the pUC19-HCV-hRLuc and Tri-JFH1 groups (1.049 × 108± 2.747 × 107vs 5.368 × 107± 1.016 × 107, P < 0.05; 1.049 × 108± 2.747 × 107vs 5.243 × 107± 1.194 × 107, P < 0.05), suggesting that the translation initiation efficiency of the first Rbm3 IRES in the two sequential IRESes was stronger than the HCV authentic IRES and EMCV IRES. The fold changes of 72 h/4 h relative light units in the pHCV-rep-NeoR-hRluc and pUC19-HCV-hRLuc groups were similar (159.619 ± 9.083 vs 163.536 ± 24.031, P = 0.7707), and were both higher than the fold change in the Tri-JFH1 group 159.619± 9.083 vs 140.811 ± 9.882, P < 0.05; 163.536 ± 24.031 vs 140.811 ± 9.882, P < 0.05), suggesting that the replication potency of the Rbm3 IRES tricistronic replicon matched the replication of bicistronic replicon and exceeded the potency of EMCV IRES replicon. Replication of tricistronic replicons was suppressed by ribavirin, simvastatin, atorvastatin, telaprevir and boceprevir. Interferon-alpha 2b could not block replication of the novel replicon RNA in sH7 cells. After interferon stimulation, MxA mRNA and protein levels were lower in sH7 than in parental cells.
CONCLUSION: Tricistronic HCV replicon with double Rbm3 IRESes could be applied to evaluate the replication inhibition efficacy of anti-HCV agents.
Core tip: Two sequential RNA-binding motif protein 3 internal ribosome entry sites (IRESs) of 22 nucleotides were used to construct a tricistronic hepatitis C virus (HCV) replicon, initiating translation of humanized Renilla luciferase and HCV non-structural gene, along with HCV authentic IRES initiating translation of neomycin resistance gene. Intracellular HCV replicon RNA and expression of inserted foreign genes and HCV non-structural gene in cells transiently and stably transfected with tricistronic replicon RNA were comparable to those in cells transfected with traditional bicistronic HCV replicons. The novel replicon could be applied to evaluate replication inhibition efficacy of anti-HCV agents, except for interferon-α2b, which might be attributed to suppressed interferon response pathway.
- Citation: Cheng X, Gao XC, Wang JP, Yang XY, Wang Y, Li BS, Kang FB, Li HJ, Nan YM, Sun DX. Tricistronic hepatitis C virus subgenomic replicon expressing double transgenes. World J Gastroenterol 2014; 20(48): 18284-18295
- URL: https://www.wjgnet.com/1007-9327/full/v20/i48/18284.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i48.18284
Hepatitis C virus (HCV) is a major cause of chronic liver disease, with 170 million people chronically infected worldwide, which are nearly 3% of the world’s population[1]. Efforts in developing new satisfactory therapeutic agents against HCV have been hampered until recently by difficulties in replicating the virus in cultured cells[2]. This barrier was eventually circumvented by a pivotal breakthrough; subgenomic replicons without structural genes[3,4], which were established in human hepatocellular carcinoma cell line Huh-7, were highly permissive for HCV replication[5], allowing for the screening of new antiviral compounds.
In the first in vitro culture system, HCV subgenomic RNA replicated and maintained itself as long as HCV gene was efficiently reproduced[4]. In HCV subgenomic replicon RNA, the structural protein region of HCV is replaced with the coding sequence of neomycin phosphotransferase (NeoR) that detoxifies neomycin harboring cytotoxicity. The HCV internal ribosome entry site (IRES) is preserved to direct translation of inserted NeoR, whereas the IRES derived from encephalomyocarditis virus (EMCV) is inserted at the downstream region of the NeoR gene to initiate translation of successive non-structural HCV proteins, which are located downstream within replicon RNA and involved in HCV gene replication.
For high-throughput compound screening, however, replicons containing both reporter gene and selectable marker have been proven to be the most useful. In that case, a sequence (e.g., 2A, ubiquitin) that mediates proteolytic cleavage is needed to link the reporter gene and selectable marker[6-8], forming fusion proteins that are structurally and functionally incompatible. The incompatibility might introduce impaired protein activity and solubility owing to unnatural folding processes of fusion proteins during translation and post-translational modification, as well as uncontrolled and insufficient cleavage efficiency of intracellular endogenous protease. Thus the tricistronic replicon is needed to simultaneously express selection marker and reporter gene to construct stable cell lines with high replication of HCV RNA, as well as to ensure sufficient production of reporter proteins. A tricistronic replicon, termed Tri-JFH1[9], was used to generate stable cell lines that constitutively synthesized HCV subgenomic RNA harboring the NeoR for drug selection and a luciferase reporter enzyme for assessing the levels of HCV RNA synthesis and the effect of small interfering RNA on HCV RNA replication.
Nevertheless, the major caveats with the current vector systems utilizing EMCV IRES are that the translation efficiency is significantly weaker than that of HCV IRES, and that the 450-nucleotide (nt) sequence might be too long for a replication-competent vector, which would occupy a substantial fraction of the limited space on the recombinant vector genome; the same holds for the majority of the other well-established viral IRES elements[10]. Thus we considered a shorter IRES of only 22 nt, which was truncated from the 720-nt 5’ leader of the RNA-binding motif protein 3 (Rbm3) mRNA by deletion and mutation analysis[11]. The Rbm3 sequence functions as an IRES when tested in isolation, and additionally, the Rbm3 IRES module binds specifically to 40 S ribosomal subunits as well. Therefore, we introduced two inserts of the short IRES into HCV replicons (with HCV IRES directing NeoR) to respectively initiate translation of humanized Renilla luciferase (hRLuc) and HCV non-structural proteins, forming a novel tricistronic HCV replicon independently and simultaneously expressing NeoR, luciferase and HCV non-structural proteins.
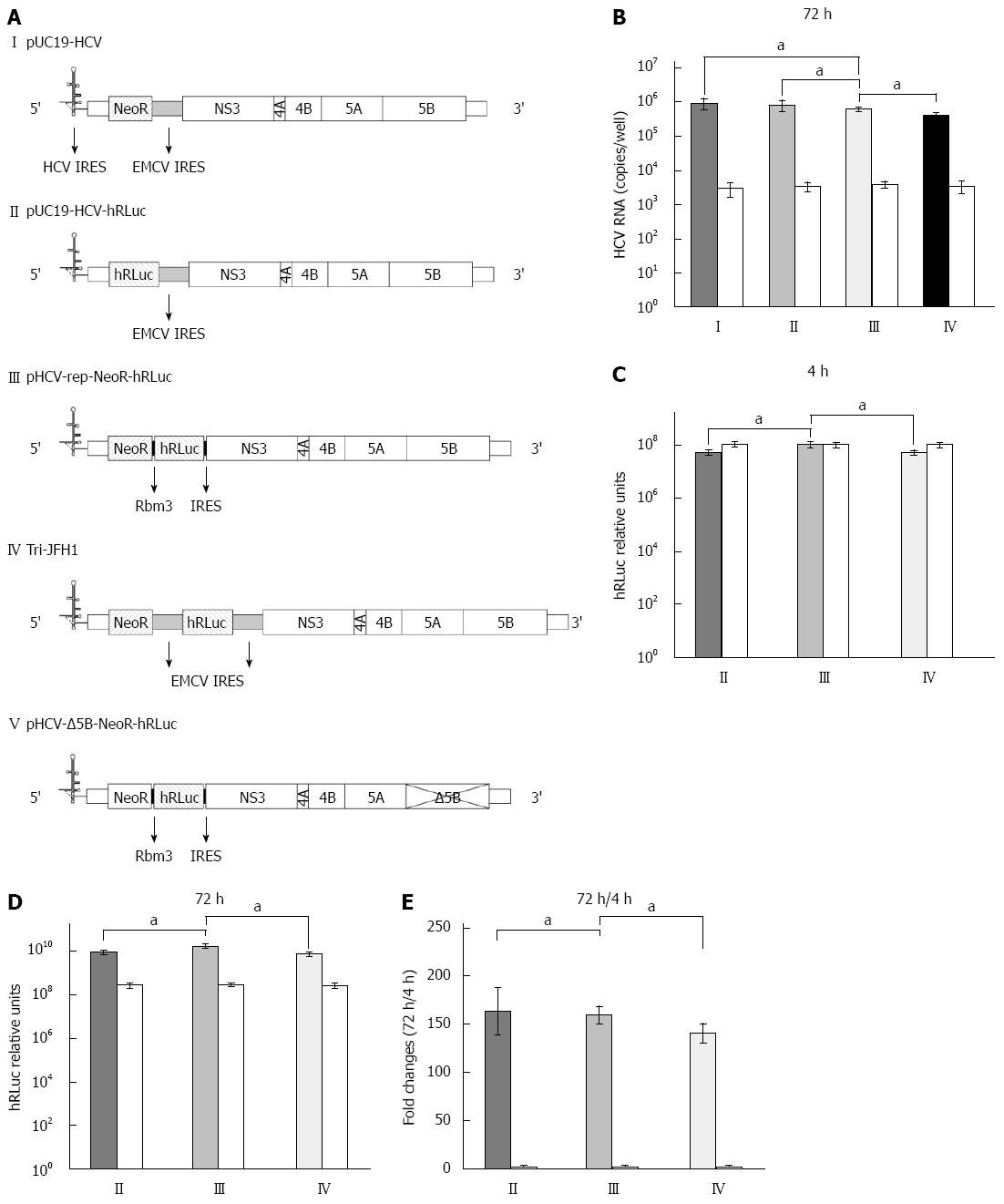
The bicistronic pUC19-HCV replicon of genotype 1b (Genbank accession number: AJ242652.1)[4] was gifted by Professor TS Benedict Yen at University of California, San Francisco. In this replicon, HCV structural genes were replaced by NeoR and EMCV IRES (Figure 1A, plasmid I). Subsequently, the plasmid was re-constructed into a similar one by replacing the NeoR with a segment of humanized Renilla luciferase (Figure 1A, plasmid II). The Rbm3 22-nt IRES sequence (along with space sequence and ATG) was: TTT ATA ATT TCT TCT TCC AGA AGA ATT TGT TGG TAA AGC CAC CAT G[11]. The IRES was synthesized by Sangon Biotech (Shanghai, China). The segment of Rbm3IRES-hRLuc-Rbm3IRES was amplified by PCR and subcloned into pUC19-HCV replicon, replacing the sequence of EMCV IRES, located downstream of the NeoR region and prior to HCV non-structural gene (Figure 1A, plasmid III). The newly constructed HCV replicon was named pHCV-rep-NeoR-hRluc and used as a template for cell-free transcription of HCV replicon RNA. A replication-defective vector (pHCV-ΔNS5B-NeoR- hRluc, Figure 1A, plasmid V) as a negative control was produced by digesting pHCV-rep-NeoR-hRluc with BglII, which liberates a 1000 bp region containing the active site of NS5B, followed by religation[12].
Tricistronic HCV replicon pHCV-rep-NeoR-hRluc was linearized by endonuclease ScaI (Takara, Otsu, Japan) and used as a template for cell-free in vitro transcription of HCV replicon RNA with RiboMAX Large Scale RNA Production Systems (Promega, Madison, WI, United States). Huh7 cells were planted in 6-well plates and were cultured in high-glucose DMEM (Gibco, New York, United States) without antibiotics, which was supplemented with 10% (v/v) fetal bovine serum (Gibco) and non-essential amino acids (Invitrogen, Carlsbad, CA, United States). RNA transcribed from HCV replicons was transfected into the cells with Lipofectamine 2000 (Invitrogen), following the manufacturer’s instructions. Neomycin G418 (Gibco) at a dose of 800 μg/mL was supplemented 48 h after transfection and the cells were screened with G418 for the following 4-8 wk. The cell clone with the highest luciferase expression was selected and proliferated for the subsequent HCV antiviral assay.
Total cellular RNA was extracted from Huh7 and HCV replicon transfected cells using an SV Total RNA Isolation System (Promega) according to the manufacturer’s instructions and quantified by measuring optical density at 260 nm with a BioPhotometer (Eppendorf, Hamburg, Germany). Northern blot for HCV replicon RNA was performed as previously described[13]. After ultraviolet irradiation cross-linking, the membrane was incubated with 32P-labeled negative-sense riboprobe that was complementary to nucleotides 8374-9440 of NS5B and the 3’ UTR[14]. Hybridization with negative-sense riboprobe for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served to normalize the total RNA amount loaded in each lane of the gel. Signals were detected by autoradiography.
Total cellular RNA extracted from Huh7 and HCV replicon transfected cells as described above was reverse transcribed and quantified for HCV RNA using a TaqMan EZ RT-PCR Kit (Applied Biosystems, Foster City, CA, United States) and SLAN Real-Time PCR system (Hongshi, Shanghai, China). The primers and probe for the HCV replicon containing NeoR have been previously described by Ali et al[15] and Dhanak et al[16], with 5’-CCGGCTACCTGCCCATTC-3’ as the forward primer, 5’-CCAGATCATCC TGATCGACAAG-3’ as the reverse primer and 5’-(FAM)-ACATCGCATCGA GCGAGCACG TAC-(TAMRA)-3’ as the probe, all of which were targeted to the NeoR sequence within the HCV replicon. The primers and probe for HCV replicon containing hRluc were 5’-GGGCGAGAAAATGGTGCTTG-3’ (forward primer), 5’-GAATGGCTCCAGGTAGGCAG-3’ (reverse primer), and 5’-TCGAGACCATGCTCCCAAGCAAG-3’ (probe), targeting the hRLuc sequence. The Myxovirus resistance gene A (MxA) mRNA level in Huh7 and the HCV replicon transfected cells was determined by real-time PCR and subsequent analysis via 2-∆∆Ct method. The primers for MxA mRNA were as follows: forward, 5’-CACCACAGAGGCTCTCAGCATGGCC-3’, and reverse, 5’-CTTCCTCCCCTACAGTTTCCTCTCC-3’. GAPDH was used as a reference with forward primer 5’-ACCACAGTCCATGCCATCAC-3’ and reverse primer 5’-TCCACCACCCTGTTGCTGTA-3’.
Cells transfected with tricistronic HCV replicon were lysed and subjected to an hRLuc activity assay using a GloMax 20/20 Luminometer following the manufacturer’s instructions (Promega). The relative light units were recorded for each cell strain. All luciferase assays were performed at least in quadruplicates.
A standard procedure was used for Western blot to detect HCV NS5B and MxA, with actin as a control[17]. Anti-NS5B, MxA and actin antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, United States). Signals were revealed with Western Blotting Luminol Reagent (Santa Cruz).
All of the data are presented as mean ± SD unless otherwise stated. Statistical analyses were performed by Student’s t-test and analysis of variance where appropriate. P < 0.05 was considered statistically significant.
HCV replicon RNA was in vitro transcribed using pUC19-HCV, pUC19-HCV-hRLuc, pHCV-rep-NeoR-hRluc, Tri-JFH1 and pHCV-Δ5B-NeoR- hRluc (Figure 1A) as templates, respectively. Seventy-two hours after the purified RNA derived from vectors of pUC19-HCV, pUC19-HCV-hRLuc, pHCV-rep-NeoR-hRluc and Tri-JFH1 was co-transfected into Huh-7 cells along with RNA from pHCV-Δ5B-NeoR-hRluc as a control, the HCV RNA was quantified by real-time PCR. Although slightly lower, the replication of tricistronic HCV replicon with Rbm3 IRESes was comparable to that of pUC19-HCV and pUC19-HCV-hRLuc (Figure 1B, III vs I, P = 0.1287, III vs II, P = 0.2206). The replication of replicon derived from pHCV-rep-NeoR-hRluc was significantly more potent than that from Tri-JFH1 (Figure 1B, III vs IV, P = 0.0136). The average replicon RNA copy numbers of pUC19-HCV, pUC19-HCV-hRLuc, pHCV-rep-NeoR-hRluc and Tri-JFH1 group in quadruplicated experiments were 8.93 × 105± 3.01 × 105, 8.08 × 105± 2.65 × 105, 6.17 × 105± 8.83 × 104, and 4.01 × 105± 8.87 × 104, respectively. The RNA levels of replication-defective “replicon” derived from pHCV-Δ5B-NeoR-hRluc were nearly equal when it was co-transfected with four different replication-competent replicons (Figure 1B, white columns). Although the total length was increased in the tricistronic replicon, the replication of replicon RNA was largely intact when compared with that of shorter bicistronic replicons. Moreover, the shorter Rbm3 IRESes showed a slighter impairment of replication potency of tricistronic replicon than the EMCV IRESes.
After replicon RNA transcription, three types of replication-competent replicons were co-transfected into Huh7 cells, respectively, along with the replication-defective RNA derived from pHCV-Δ5B-NeoR-hRluc as a control. Subsequently, cell lysates were harvested at 4 and 72 h after co-transfection and subjected to hRLuc activity assay. Four hours after transfection, expression of hRLuc was significantly higher in cells transfected with Rbm3 IRES tricistronic replicon than in cells transfected with replicons transcribed from pUC19-HCV-hRLuc and Tri-JFH1 (Figure 1C, III vs II, P = 0.0129, III vs IV, P = 0.0128). In the quadruplicated experiments, the average relative light unit in the pHCV-rep-NeoR-hRluc group was approximately 2-fold of those in the pUC19-HCV-hRLuc and Tri-JFH1groups (1.049 × 108± 2.747 × 107vs 5.368 × 107± 1.016 × 107, P < 0.05), suggesting that the translation initiation efficiency of the first Rbm3 IRES in the two sequential IRESes was stronger than the HCV authentic IRES and EMCV IRES. The hRLuc expression derived from pHCV-Δ5B-NeoR-hRluc in each group was similar to the hRLuc expression from pHCV-rep-NeoR-hRluc (Figure 1C, white columns), implying that the hRLuc activity at the time point of 4 h after RNA transfection could be regarded as a sensitive marker to compare the translation initiation efficiency of Rbm3 IRES with efficiency of HCV IRES and EMCV IRES without influence of replicon replication. Seventy-two hours after transfection, the hRLuc expression was also significantly higher in cells transfected with Rbm3 IRES tricistronic replicon than in cells transfected with replicons transcribed from pUC19-HCV-hRLuc and Tri-JFH1 (Figure 1D, III vs II, P = 0.0125, III vs IV, P = 0.0045). Subsequently, the hRLuc activity at 72 h after transfection was normalized to that at the 4 h time point to compare the replication potency of each replicon by diminishing the influence of basal difference of hRLuc activity between replicons. We found that the fold changes in 72 h/4 h relative light unit in the pHCV-rep-NeoR-hRluc and pUC19-HCV-hRLuc groups were similar (Figure 1E, III vs II, 159.619 ± 9.083 vs 163.536 ± 24.031, P = 0.7707, n = 4), and were both higher than the fold change in the Tri-JFH1 group (159.619 ± 9.083 vs 140.811 ± 9.882, P < 0.05, n = 4 and 163.536 ± 24.031 vs 140.811 ± 9.882, P < 0.05, n = 4), suggesting that the replication potency of Rbm3 IRES tricistronic replicon matched the replication of bicistronic replicon and exceeded the potency of EMCV IRES replicon. This finding was consistent with the results of replicon RNA quantification shown in Figure 1B.
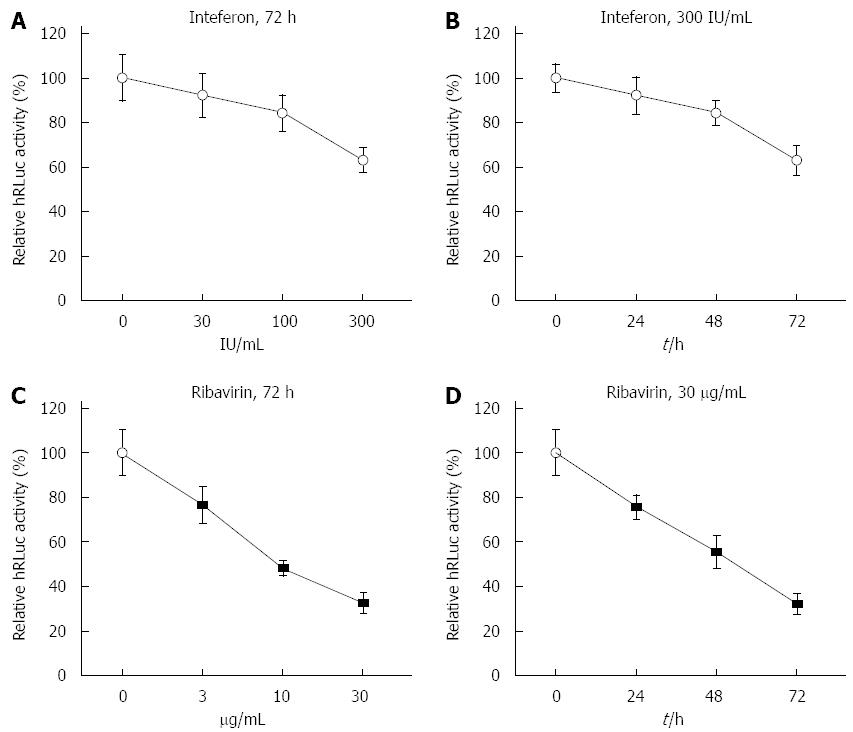
Prior to construction of stable transfection cell lines with tricistronic replicon, the effects of interferon-α2b (IFNα2b) and ribavirin on HCV replicon were investigated in Huh7 cells transiently transfected with the tricistronic replicon. Four hours after transfection, an increasing dose of IFNα2b up to 300 IU/mL was inoculated into cells for 72 h. Accordingly, 4 h after transfection, cells were successively treated with 300 IU/mL IFNα2b for 24, 48 and 72 h. Similarly, Huh7 cells were treated with ribavirin at increasing doses (0, 3, 10 and 30 μg/mL) and with a high dose for 0, 24, 48 and 72 h. The luciferase activity was substantially reduced in the Huh7 cells that were transiently transfected with tricistronic replicon in a dose- and time-dependent manner (Figure 2A and B). Correspondingly, the luciferase activity was markedly impeded by ribavirin in Huh-7 cells transiently transfected with replicon (Figure 2C and D), indicating that HCV RNA replication could be prevented by anti-HCV agents, including IFNα2b, in the novel replicon system when it was tested in a transient transfection setting.
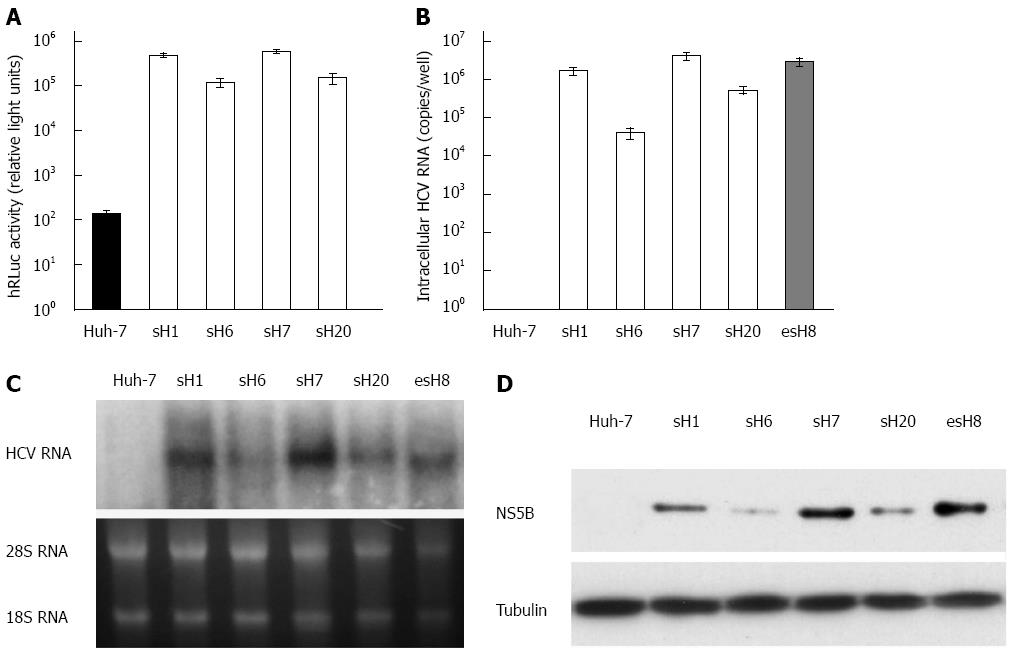
Fifty stable cell clones were screened with G418 from Huh-7 cells transfected with the purified tricistronic HCV replicon RNA. Four of the 50 clones with the highest hRLuc expression were selected and designated sH1, sH6, sH7, and sH20 (Figure 3A). The hRLuc activity was highest in the sH7 cells among all the surviving cell clones (Figure 3A). Huh-7 cells were also transfected with pUC19-HCV replicon and screened with G418 to form stable clones as a reference. Accordingly, the stable clone with the highest replicon RNA replication was selected and designated as esH8 to compare with tricistronic clones.
HCV RNA could be amplified from all the tricistronic replicon strains, especially in sH7 clones, in which HCV RNA copy number was higher than in any of the other strains, parental Huh-7 cells or esH8 cells (Figure 3B).
The RNA replication and NS5B expression could be determined by Northern and Western blot in all the tricistronic replicon strains, especially in the sH7 clones, in which the HCV RNA and NS5B levels were higher than those in parental Huh-7 cells and esH8 cells (Figure 3C and D). This finding implied that replicon RNA replication in stable clones selected by antibiotic and hRLuc activity assay was similar to the replication in clones selected traditionally by antibiotics alone. The consistency of the NS5B, RNA level and hRLuc activity in tricistronic replicon clones suggested that hRLuc activity could be regarded as a reliable marker of HCV replicon replication.
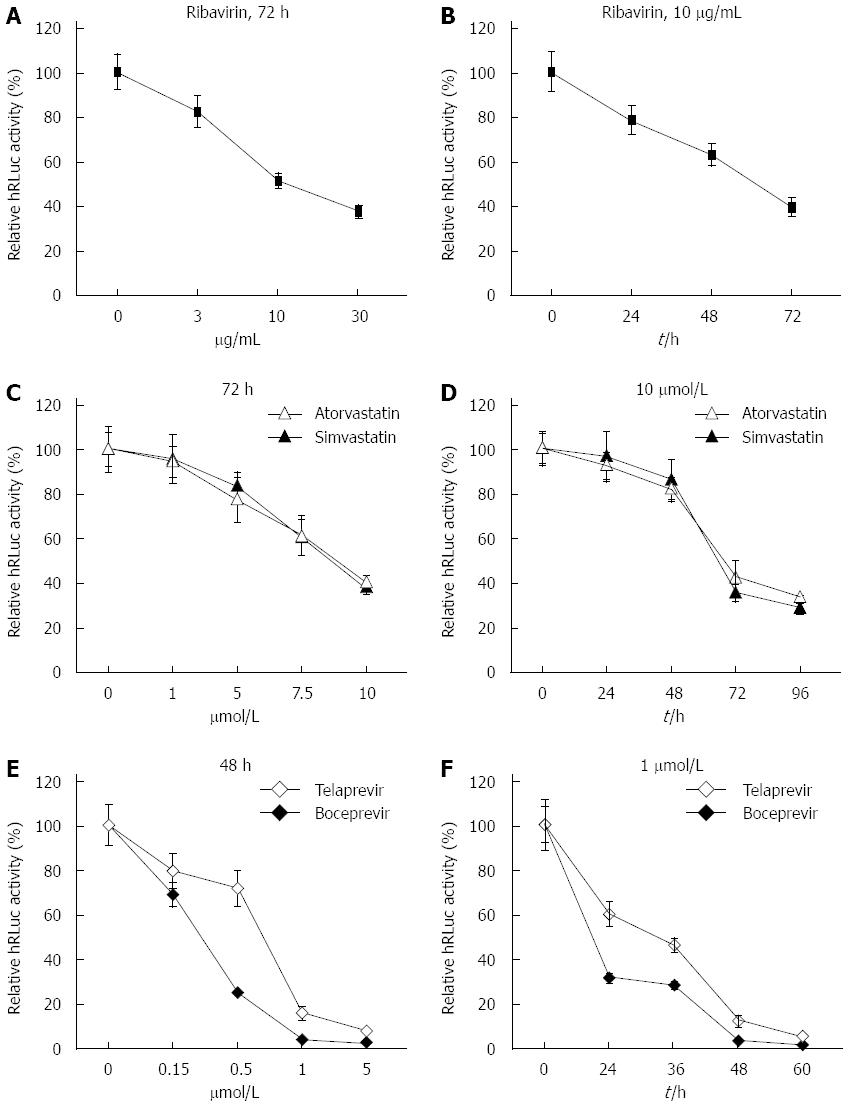
To test the practicability of our tricistronic HCV replicons in anti-HCV drug screening, ribavirin, two types of hydroxy methylglutaryl coenzyme A (HMG CoA) reductase inhibitors (simvastatin and atorvastatin) and two types of NS3/4A serine protease inhibitors (telaprevir and boceprevir) were incubated with sH7 cells, in which HCV replicon RNA was stably replicated along with hRLuc expression. Gradient doses of ribavirin, simvastatin, atorvastatin, telaprevir or boceprevir were added to the culture medium, as shown in Figure 4. The culture medium with ribavirin, simvastatin and atorvastatin was replaced after incubation for 72 h. The medium with telaprevir and boceprevir was replaced after 48 h. Accordingly, sH7 cells were treated with 10 μg/mL ribavirin, 10 μmol/L simvastatin, 10 μmol/L atorvastatin, 1 μmol/L telaprevir or 1 μmol/L boceprevir for different durations, as indicated in Figure 4. After sH7 cells were treated with ribavirin at increasing doses for 72 h, the luciferase activity sharply declined in a dose-dependent manner; especially in cells treated with 10 and 30 μg/mL ribavirin (Figure 4A). When sH7 cells were treated with 10 μg/mL ribavirin for increasing durations, the luciferase activity decreased in a time-dependent manner (Figure 4B), suggesting that the replication of HCV RNA in the sH7 cells incorporated with our tricistronic HCV replicon was impaired by ribavirin, which is the most widely used antiviral drug in patients with hepatitis C. Similar dose- and time-dependent patterns of inhibition were detected in sH7 cells treated with simvastatin, atorvastatin (Figure 4C and D), telaprevir or boceprevir (Figure 4E and F). The sensitivity of the tricistronic HCV replicon to antivirals contributed to application of this novel replicon and derived cell lines for anti-HCV agent screening, including novel HMG CoA reductase inhibitors and NS3/4A serine protease inhibitors.
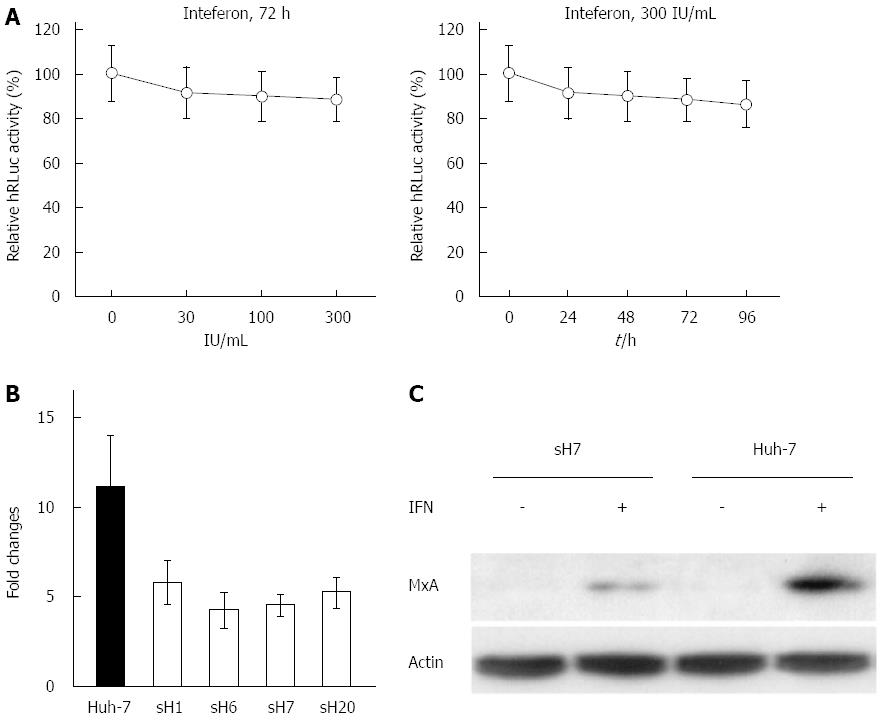
After sH7 cells were treated with increasing doses of IFNα2b for 72 h, there was no significant decrease in the luciferase activity, which was clearly inconsistent with the significant inhibitory effects of ribavirin, simvastatin and atorvastatin. Even in sH7 cells treated with 300 IU/mL IFNα2b, the luciferase activity was maintained at approximately 85% of that in untreated sH7 cells (Figure 5A, left panel), implying that IFNα2b up to 300 IU/mL was not sufficient to suppress HCV RNA replication in sH7 cells. Accordingly, the luciferase activity in sH7 cells treated with 300 IU/mL IFNα2b for as long as 96 h was largely intact compared with that in untreated sH7 cells (Figure 5A, right panel), suggesting that prolonged treatment with abundant IFNα2b does not inhibit HCV RNA replication in cells stably transfected with the newly formed tricistronic HCV replicon.
Given that the human MxA gene encodes a type I IFN-inducible protein that exhibits activity against a variety of RNA viruses, including HCV[18,19], MxA expression in IFNα2b-treated Huh-7 and sH7 cells was investigated to elucidate the mechanism of IFNα2b resistance in sH7 cells by real-time PCR. The fold changes were calculated to determine the ratio of MxA mRNA in IFN-treated cells to MxA mRNA in untreated cells. The fold change of MxA mRNA in Huh-7 cells after IFN treatment was higher than that in HCV replicon stably transfected cell lines after IFN treatment (Figure 5B). It showed that after IFNα2b stimulation, the MxA mRNA level was higher in Huh-7 cells than in sH7 cells in which HCV replicon was stably transfected.
Additionally, the MxA protein level was elevated after IFNα2b treatment in both Huh-7 and sH7 cells, whereas no MxA was detected in these two types of cells without IFNα2b stimulation. The MxA protein level was higher in parental Huh-7 cells than in sH7 cells (Figure 5C), indicating that the absence of responding MxA might contribute to the failure of IFNα2b to block HCV replicon RNA replication in sH7 cells.
As the activity of most IRESes is cell-type dependent, the lack of information concerning the activity of the Rbm3 IRES in hepatocyte-derived cell lines led us to test its activity in Huh7 cells. Moreover, a comparison of Rbm3 IRES and EMCV IRES in a HCV replicon context has not been reported thus far. We found that Rbm3 IRES activity was approximately 2-fold of the activity of EMCV IRES in Huh7 cells. The 22-nt IRES works well on an RNA driven by a heterologous cytomegalovirus promoter and by an authentic HCV promoter. It even works on a tricistronic RNA that mimics HCV RNA with two transgenes inserted between the HCV IRES and HCV non-structural gene. Furthermore, we found that the sequential double Rbm3 IRESes could efficiently direct translation of inserted transgene and authentic polymerase gene simultaneously in a replication-competent hepatitis B virus vector. These findings have recently been reported by our group[20]. In regard to the influence of IRES on replication potency of HCV replicons, we found that the sequential double Rbm3 IRESes did not significantly impair the replicon replication and showed an advantage over double EMCV IRESes, which might be attributed to the shorter length of Rbm3 IRES than EMCV IRES. The relationship between the length of inserted IRES into HCV replicon and the replication potency of replicon needs to be further addressed.
The introduction of the concise Rbm3 IRES facilitated the construction of tricistronic replicons and endowed the novel tricistronic replicons with an advantage over their bicistronic counterparts, which could be summarized as follows: the avoidance of potential structural and functional incompatibility in fusion proteins; the convenience of screening cell strains successively with antibiotic and fluorescence markers.
In sH7 cells stably harboring the HCV replicon, the HCV replication was efficiently suppressed by ribavirin, simvastatin, atorvastatin, telaprevir and boceprevir. In contrast, IFNα2b did not block HCV replicon RNA replication in sH7 cells. This observation was inconsistent with our expectation and with previous reports from other researchers[3,21]. Thus, we considered that the acquired IFN resistance of stably transfected cells during screening might be involved to explain this inconsistency. Endogenous IFN plays a crucial role in antiviral defense, including exerting a potent and direct replication-suppressive effect when host cells were infected by HCV. When the cells were transfected with an HCV replicon carrying the antibiotic-resistance gene NeoR, and were cultured in medium with abundant neomycin, only a small portion of cells, in which HCV replicon replicated vigorously and steadily, produced a sufficient amount of NeoR, and consequently survived the successive antibiotic screening for several months. To sustain the high level of replication in cells, two mechanisms may function during the screening process. The first mechanism is the adaptive mutation of the HCV replicon genome that boosted the efficiency of RNA replication. The second interpretation is the gradual adaptation of host factors that contributed to the “permissive” status of the host cells for HCV RNA replication, especially host factors that could restrict the antiviral function of IFN. In fact, several cell lines that are resistant to IFNα have been constructed. Using conditions that mimic the in vivo selection of IFN-resistant cells, the RST2 cell line has been isolated from the highly IFN-sensitive Daudi human Burkitt lymphoma cell line. The RST2 cell line is resistant to the antiviral, antiproliferative, and gene-induction activity of IFNα[22]. It has also been reported that IFN-resistant cells can be generated by selection with progressively increasing concentrations of IFNα[23]. Thus, the emergence of IFN-resistant strains from tricistronic replicon transfected cells was reasonable during antibiotic screening.
It has been demonstrated that HCV infection can suppress the IFN response in hepatocytes[24,25]. In particular, HCV NS5A inhibits IFNα signaling by suppressing the signal transducer and activator of transcription 1 phosphorylation in hepatocyte-derived cell lines. Furthermore, NS5A inhibits activation of IFN-stimulated gene factor 3 and IFN-stimulated response element-driven gene expression[26]. Thus, the accumulated NS5A in sH7 cells after several months of antibiotic screening might be a crucial factor conveying IFN resistance. In contrast, the factors in parental Huh-7 cells or HCV replicon transiently transfected cells, which impair the IFN response pathway, might not be sufficient to block the antiviral activity of IFN.
The HCV non-structural genes in our tricistronic replicon were directed by the novel Rbm3 IRES. The translation initiation efficiency of the 22-nt IRES was higher than that of the EMCV IRES, which is widely used to initiate translation of HCV non-structural genes in bicistronic replicons, therefore, NS5A and other HCV non-structural proteins in sH7 cells were present in higher quantities than in esH8 cells. Thus, the IFN response was largely suppressed in sH7 cells but predominantly preserved in esH8 cells. The resistance to IFN observed in our tricistronic HCV replicon system merits further detailed investigation focusing on the host factors as well as the adaptive mutation analysis of the HCV replicon genome.
MxA is a key mediator of the IFN-induced response against a wide range of viruses[27]. MxA expression is widely used as a surrogate marker for IFN activity in various experimental and clinical settings[28-30]. Therefore, we compared the IFN-induced MxA expression in HCV replicon stably transfected sH7 cells and transiently transfected Huh-7 cells to elucidate IFN resistance in sH7 cells. The impaired MxA mRNA and protein levels in sH7 cells, in contrast to those in Huh7 cells, revealed that the IFN pathway was retarded in sH7 cells, which contributed to the IFN resistance in sH7 cells. MxA might be just one among several IFN-stimulated genes, which were involved in the co-adaptation of the cells and replicon. The passive modification of cellular host factors during antibiotic selection might be addressed by proteomic research in the future. This instinct feature of sH7 cells, in which the HCV replicon potently and sustainably replicated with resistance to IFN, makes it a unique model by which to probe the mechanism underlying the unsatisfactory therapeutic effects of IFN in nearly 50% of patients with hepatitis C.
In conclusion, we have constructed a novel tricistronic HCV replicon with the aid of double Rbm3 IRESes, simultaneously expressing NeoR and luciferase along with HCV non-structural genes, which could be used in antiviral screening employing stable and transient transfection protocols.
Hepatitis C virus (HCV) replicons are widely used to evaluate the efficacy of anti-HCV agents. For high-throughput compound screening, however, replicons containing both a reporter gene and a selectable marker have been proven to be the most useful. A tricistronic replicon, termed Tri-JFH1, was used to generate stable cell lines that constitutively synthesized HCV subgenomic RNA harboring the neomycin phosphotransferase (NeoR) for drug selection and a luciferase reporter enzyme for assessing HCV RNA synthesis levels. Nevertheless, the major caveat with current vector systems utilizing the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) is that the translation efficiency of EMCV IRES is significantly weaker than that of HCV IRES, and that the 450-nt sequence might be too long for a replication-competent vector, which would occupy a substantial fraction of the limited space in the recombinant vector genome.
A shorter IRES of only 22 nucleotides (nt) was truncated from the 720-nt 5’ leader of the RNA-binding motif protein 3 (Rbm3) mRNA by deletion and mutation analysis. The Rbm3 sequence functions as an IRES when tested in isolation, and the Rbm3 IRES module binds specifically to 40 S ribosomal subunits. Therefore, we introduced two inserts of the short IRES into HCV replicons (with HCV IRES directing NeoR) to initiate translation of humanized Renilla luciferase (hRLuc) and HCV non-structural proteins, respectively, forming a novel tricistronic HCV replicon independently and simultaneously expressing NeoR, luciferase and HCV non-structural proteins.
The authors constructed a novel tricistronic HCV replicon with the aid of double Rbm3 IRESes, expressing NeoR and luciferase simultaneously along with HCV non-structural genes, which could be used in antiviral screening using stable and transient transfection protocols. As the activity of most IRESes is cell-type dependent, the lack of information concerning the activity of the Rbm3 IRES in hepatocyte-derived cell lines led us to test its activity in Huh7 cells. Moreover, a comparison of RMBP3 IRES and EMCV IRES in an HCV replicon context has not been reported thus far. Authors found that the activity of the Rbm3 IRES was approximately 2-fold of the activity of EMCV IRES in Huh7 cells. The 22-nt IRES works well on an RNA driven by a heterologous cytomegalovirus promoter and an authentic HCV promoter. It even works on a tricistronic RNA replicon that mimics HCV RNA with two transgenes inserted between the HCV IRES and the HCV non-structural gene. Furthermore, they found that the sequential double Rbm3 IRESes could efficiently direct translation of inserted transgene and authentic polymerase gene simultaneously in a replication-competent hepatitis B virus vector. With regard to the influence of IRES on replication potency of HCV replicons, authors found that the sequential double Rbm3 IRESes did not significantly impair the replicon replication and showed an advantage over double EMCV IRESes, which might be attributed to the shorter length of Rbm3 IRES than EMCV IRES. In sH7 cells stably harboring the HCV replicon, HCV replication was efficiently suppressed by ribavirin, simvastatin, atorvastatin, telaprevir and boceprevir. In contrast, interferon-α2b (IFNα2b) did not block HCV replicon RNA replication in sH7 cells. Authors considered that the acquired IFN resistance of stably transfected cells during screening might be involved to explain this inconsistency.
The newly-formed tricistronic HCV replicon could be used to construct cell lines stably or transiently harboring potent replication of HCV replicon and applied in antiviral screening.
The authors reporting the tricistronic HCV subgenomic replicon with Rbm3 IRES expressing double transgenes in different cell lines and studying the replicative potentials and suppression of replication by different types of antivirals. They concluded that the tricistronic replicon had best replicative potentials in four of the tested strains, namely sH7. The inhibitory activity was demonstrated for direct-acting antivirals but not for IFN which, as concluded by the authors, might be attributed to suppressed IFN response pathway. The study is very interesting and well designed. The results are well presented and discussed.
P- Reviewer: Borg BB, Sira MM S- Editor: Gou SX L- Editor: Wang TQ E- Editor: Ma S
| 1. | Negro F, Alberti A. The global health burden of hepatitis C virus infection. Liver Int. 2011;31 Suppl 2:1-3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 107] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 2. | Bukh J. A critical role for the chimpanzee model in the study of hepatitis C. Hepatology. 2004;39:1469-1475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 131] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 3. | Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972-1974. [PubMed] |
| 4. | Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110-113. [PubMed] |
| 5. | Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001-13014. [PubMed] |
| 6. | Murray EM, Grobler JA, Markel EJ, Pagnoni MF, Paonessa G, Simon AJ, Flores OA. Persistent replication of hepatitis C virus replicons expressing the beta-lactamase reporter in subpopulations of highly permissive Huh7 cells. J Virol. 2003;77:2928-2935. [PubMed] |
| 7. | Vrolijk JM, Kaul A, Hansen BE, Lohmann V, Haagmans BL, Schalm SW, Bartenschlager R. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J Virol Methods. 2003;110:201-209. [PubMed] |
| 8. | Yi M, Bodola F, Lemon SM. Subgenomic hepatitis C virus replicons inducing expression of a secreted enzymatic reporter protein. Virology. 2002;304:197-210. [PubMed] |
| 9. | Jones DM, Domingues P, Targett-Adams P, McLauchlan J. Comparison of U2OS and Huh-7 cells for identifying host factors that affect hepatitis C virus RNA replication. J Gen Virol. 2010;91:2238-2248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 10. | Walsh D, Mohr I. Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol. 2011;9:860-875. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 323] [Cited by in RCA: 391] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 11. | Chappell SA, Mauro VP. The internal ribosome entry site (IRES) contained within the RNA-binding motif protein 3 (Rbm3) mRNA is composed of functionally distinct elements. J Biol Chem. 2003;278:33793-33800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 59] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 12. | Sumpter R, Wang C, Foy E, Loo YM, Gale M. Viral evolution and interferon resistance of hepatitis C virus RNA replication in a cell culture model. J Virol. 2004;78:11591-11604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 68] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 13. | Hwang DR, Tsai YC, Lee JC, Huang KK, Lin RK, Ho CH, Chiou JM, Lin YT, Hsu JT, Yeh CT. Inhibition of hepatitis C virus replication by arsenic trioxide. Antimicrob Agents Chemother. 2004;48:2876-2882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | Bukh J, Pietschmann T, Lohmann V, Krieger N, Faulk K, Engle RE, Govindarajan S, Shapiro M, St Claire M, Bartenschlager R. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc Natl Acad Sci USA. 2002;99:14416-14421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 200] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 15. | Ali S, Pellerin C, Lamarre D, Kukolj G. Hepatitis C virus subgenomic replicons in the human embryonic kidney 293 cell line. J Virol. 2004;78:491-501. [PubMed] |
| 16. | Dhanak D, Duffy KJ, Johnston VK, Lin-Goerke J, Darcy M, Shaw AN, Gu B, Silverman C, Gates AT, Nonnemacher MR. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J Biol Chem. 2002;277:38322-38327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 163] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 17. | Brohm C, Steinmann E, Friesland M, Lorenz IC, Patel A, Penin F, Bartenschlager R, Pietschmann T. Characterization of determinants important for hepatitis C virus p7 function in morphogenesis by using trans-complementation. J Virol. 2009;83:11682-11693. [PubMed] |
| 18. | Mekky RY, Hamdi N, El-Akel W, Esmat G, Abdelaziz AI. Estrogen-related MxA transcriptional variation in hepatitis C virus-infected patients. Transl Res. 2012;159:190-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 19. | Numajiri Haruki A, Naito T, Nishie T, Saito S, Nagata K. Interferon-inducible antiviral protein MxA enhances cell death triggered by endoplasmic reticulum stress. J Interferon Cytokine Res. 2011;31:847-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Wang Z, Wu L, Cheng X, Liu S, Li B, Li H, Kang F, Wang J, Xia H, Ping C. Replication-competent infectious hepatitis B virus vectors carrying substantially sized transgenes by redesigned viral polymerase translation. PLoS One. 2013;8:e60306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 21. | Frese M, Pietschmann T, Moradpour D, Haller O, Bartenschlager R. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J Gen Virol. 2001;82:723-733. [PubMed] |
| 22. | Du Z, Fan M, Kim JG, Eckerle D, Lothstein L, Wei L, Pfeffer LM. Interferon-resistant Daudi cell line with a Stat2 defect is resistant to apoptosis induced by chemotherapeutic agents. J Biol Chem. 2009;284:27808-27815. [PubMed] |
| 23. | Blomberg J, Höglund A, Eriksson D, Ruuth K, Jacobsson M, Lundgren E, Nilsson JA. Inhibition of cellular FLICE-like inhibitory protein abolishes insensitivity to interferon-α and death receptor stimulation in resistant variants of the human U937 cell line. Apoptosis. 2011;16:783-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 24. | Hiraga N, Imamura M, Tsuge M, Noguchi C, Takahashi S, Iwao E, Fujimoto Y, Abe H, Maekawa T, Ochi H. Infection of human hepatocyte chimeric mouse with genetically engineered hepatitis C virus and its susceptibility to interferon. FEBS Lett. 2007;581:1983-1987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 25. | Tsuge M, Fujimoto Y, Hiraga N, Zhang Y, Ohnishi M, Kohno T, Abe H, Miki D, Imamura M, Takahashi S. Hepatitis C virus infection suppresses the interferon response in the liver of the human hepatocyte chimeric mouse. PLoS One. 2011;6:e23856. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 26. | Lan KH, Lan KL, Lee WP, Sheu ML, Chen MY, Lee YL, Yen SH, Chang FY, Lee SD. HCV NS5A inhibits interferon-alpha signaling through suppression of STAT1 phosphorylation in hepatocyte-derived cell lines. J Hepatol. 2007;46:759-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 27. | Haller O, Kochs G. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J Interferon Cytokine Res. 2011;31:79-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 279] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 28. | MacQuillan GC, de Boer WB, Allan JE, Platten MA, Reed WD, Jeffrey GP. Hepatocellular MxA protein expression supports the differentiation of recurrent hepatitis C disease from acute cellular rejection after liver transplantation. Clin Transplant. 2010;24:252-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 29. | Roers A, Hochkeppel HK, Horisberger MA, Hovanessian A, Haller O. MxA gene expression after live virus vaccination: a sensitive marker for endogenous type I interferon. J Infect Dis. 1994;169:807-813. [PubMed] |
| 30. | Shaker O, Ahmed A, Doss W, Abdel-Hamid M. MxA expression as marker for assessing the therapeutic response in HCV genotype 4 Egyptian patients. J Viral Hepat. 2010;17:794-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |