Published online Dec 28, 2014. doi: 10.3748/wjg.v20.i48.18249
Revised: July 4, 2014
Accepted: July 29, 2014
Published online: December 28, 2014
Processing time: 215 Days and 23.1 Hours
AIM: To investigate ethanol-induced hepatic steatosis after liver resection and the mechanisms behind it.
METHODS: First, the preliminary examination was performed on 6 sham-operated (Sham) and 30 partial hepatectomy (PH) male Wistar rats (8-wk-old) to evaluate the recovery of the liver weight and liver function after liver resection. PH rats were sacrificed at the indicated time points (4, 8, and 12 h; 1, 3, and 7 d) after PH. Second, the time point for the beginning of the chronic ethanol exposure (1 wk after sham- or PH-operation) was determined based on the results of the preliminary examination. Finally, pair-feeding was performed with a controlled diet or with a 5-g/dL ethanol liquid diet for 28 d in another 35 age-matched male Wistar rats with a one-week recovery after undergoing a sham- (n = 15) or PH-operation (n = 20) to evaluate the ethanol-induced liver injury after liver resection. Hepatic steatosis, liver function, fatty acid synthase (Fas) gene expression level, the expression of lipid metabolism-associated enzyme regulator genes [sterol regulatory element binding protein (Srebp)-1 and peroxisome proliferator-activated receptor (Ppar)-α], the mediators that alter lipid metabolism [plasminogen activator (Pai)-1 gene expression level and tumor necrosis factor (Tnf)-α production], and hepatic class-1 alcohol dehydrogenase (Adh1)-associated ethanol elimination were investigated in the 4 groups based on histological, immunohistochemical, biochemical, Western blotting, reverse transcriptase chain reaction, and blood ethanol concentration analyses. The relevant gene expression levels, liver weight, and liver function were assessed before and 1 wk after surgery to determine the subject’s recovery from the liver resection using the rats that had been subjected to the preliminary examination.
RESULTS: In the PH rats, ethanol induced marked hepatic steatosis with impaired liver functioning, as evidenced by the accumulation of fatty droplets within the hepatocytes, the higher increases in their hepatic triglyceride and blood alanine aminotransferase and blood aspartate aminotransferase levels after the 28-d pair-feeding period. The Sham-ethanol rats, not the PH-ethanol rats, demonstrated the up-regulation of Srebp-1 and the down-regulation of Ppar-α mRNA expression levels after the 28-d pair-feeding period. The 28-d ethanol administration induced the up-regulation of Pai-1 gene expression level and an overproduction of TNF-α in the Sham and the PH rats; however, the effect was more significant in the PH rats. The PH-ethanol rats (n = 4) showed higher residual blood ethanol concentrations than did the Sham-ethanol rats (n = 6) after a 5-h fast (0.66 ± 0.4 mg/mL vs 0.2 ± 0.1 mg/mL, P < 0.05); these effects manifested without up-regulation of Adh1 gene expression, which was present in the Sham-ethanol group after the 28-d pair-feeding period. One week after the liver resection, the liver weight, function, the gene expression levels of Fas, Srebp-1, Ppar-α, Pai-1 and Tnf-α recovered; however, the Adh1 gene expression did not recover in rats.
CONCLUSION: Desensitization to post-hepatectomy ethanol treatment and slow recovery from PH in Adh1 gene expression enhanced the susceptibility to ethanol-induced hepatic steatosis after PH in rats.
Core tip: Pair-feeding was performed with control or ethanol liquid diet for 28 d in sham-operated (Sham) and partial hepatectomy (PH) rats. In PH rats, ethanol induced hepatic steatosis with liver dysfunction and higher residual blood ethanol concentrations without up-regulation of hepatic class-1 alcohol dehydrogenase (Adh1) gene expression, which was present in the Sham-ethanol rats. One week after PH, liver weight, function, and lipid metabolism-related gene expressions recovered; but Adh1 gene expression did not. Desensitization to post-hepatectomy ethanol treatment and the slow recovery of Adh1 expression from PH enhanced the susceptibility to ethanol-induced hepatic steatosis in the rats post PH.
- Citation: Liu X, Hakucho A, Liu J, Fujimiya T. Delayed ethanol elimination and enhanced susceptibility to ethanol-induced hepatosteatosis after liver resection. World J Gastroenterol 2014; 20(48): 18249-18259
- URL: https://www.wjgnet.com/1007-9327/full/v20/i48/18249.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i48.18249
Hepatic surgery is the curative treatment for patients with primary or secondary malignant liver tumors[1-3], and the procedure is performed in living donor liver transplantations (LDLT) to overcome the shortage of cadaver organ donations, particularly in Asia[4,5]. The benefit of a LDLT for the recipient could not be achieved without exposing the living donor to some degree of risk. Few studies have investigated the effects of LDLT on the living donor. A global systematic review showed that living liver donor morbidity ranged from 0% to 100%, with a median of 16.1%; the living liver donor death rate was 0.1%-0.3%[5-7]. Despite intensive care treatment, extended liver resections induce a high risk of liver failure, particularly in patients with parenchymal liver disease such as hepatic steatosis[8].
Hepatic steatosis increases the risk of postoperative mortality and morbidity by affecting liver regeneration and recovery[9,10]. Patients with steatosis had up to a two-fold increased risk of postoperative complications[2]. The prevalence rates of alcohol abuse, obesity, diabetes mellitus, and metabolic syndrome are reaching epidemic proportions globally, and the effect of hepatic steatosis, which frequently accompanies these conditions, on postoperative outcomes is not well understood. Hepatic steatosis, particularly alcoholic fatty liver disease (AFLD), after hepatic surgery in patients and living donors is unclear.
The consumption of alcohol is extensive, and alcohol is a well-known hepatotoxin. Alcohol abstention is difficult for some individuals. One mechanism by which ethanol induces fatty liver is that hepatic class-1 alcohol dehydrogenase (ADH1)-related ethanol metabolism induces the reduction of NAD+ to NADH, which could inhibit the NAD+-requiring tricarboxylic acid cycle and the β-oxidation of fatty acids[11]. ADH1 is required for the oxidization of excess retinol to retinoic acid and is down-regulated to prevent apoptosis by increased retinol levels during the early stages of liver regeneration after a partial hepatectomy (PH)[12].
The PH procedure was first described by Higgins and Anderson and was developed as a useful model for studying liver regeneration[13]. In this study, PH rats were used in a liver resection animal model to assess the effects of liver resection and post-hepatectomy ethanol treatment on the gene expression of Adh1, the key ethanol-metabolizing enzyme, and to determine the contribution of the PH to the delayed ethanol elimination, the enhanced susceptibility to AFLD, and the impaired liver function found after hepatic surgery. The liver weight, liver function, and the relevant gene expression levels were evaluated before and 1 wk after the PH to describe the subject’s recovery from the liver resection in the rats.
In the 1st stage of the present study, 8-wk-old male Wistar rats from Charles River Japan, Inc. (Tokyo) were randomly assigned to either the PH- or the sham-operation (Sham) group for a better chance at detecting if the observed changes were due to chance or due to the PH.
The rats were anesthetized with 1%-2% isoflurane in oxygen, and the PH was performed according to the technique described by Higgins and Anderson[13], with some adaptations. The abdomen was opened through a midline laparotomy, and the left, middle, and caudate lobes were ligated and resected. The abdomen was closed with a silk running suture. With the exception of the liver excision, the identical surgical procedures were performed on the rats in the Sham group. The operations were performed between 9:00 am and 12:00 pm to minimize potential variability in the progression of liver regeneration associated with the surgery time and the circadian clock.
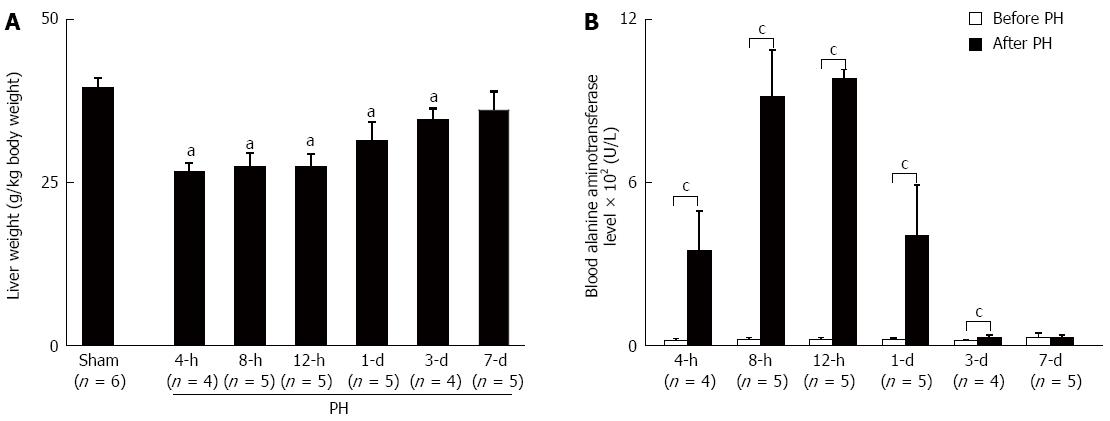
In the 2nd stage of the present study, the time point for the beginning of the chronic ethanol exposure (1 wk after sham- or PH-operation) was determined based on the results of the preliminary examination (Figure 1). Six Sham and 30 PH rats were used to determine the recovery of the liver weight and function. PH rats were sacrificed at the indicated time points (4, 8, and 12 h; 1, 3, and 7 d) after the PH; their livers were excised and weighed, and their liver tissues were snap-frozen in liquid nitrogen and stored at -80 °C until further analysis.
The blood samples collected from the rat tails before the PH (before PH) and at the indicated time points after the PH were used to measure the blood alanine aminotransferase levels (ALT, U/L) as the indicator of the liver function.
Two PH rats, died from PH operation, were discarded from the preliminary examination. The results showed that the liver weight recovered to the level of the Sham rats (Figure 1A), and the liver function recovered to the level of the before PH (Figure 1B) 7 d after the liver resection for all of the PH rats.
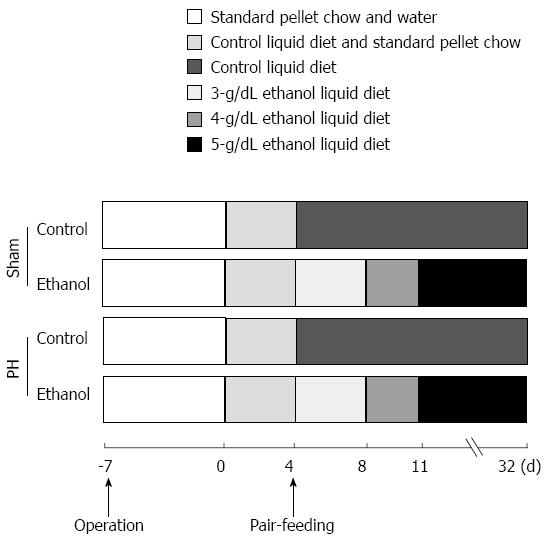
In the 3rd stage of the present study, another 35 age-matched male Wistar rats were housed individually with a 12-h light/12-h dark cycle and were fed a nutritionally adequate control diet or a 5-g/dL ethanol liquid diet (Oriental Yeast Co., Ltd., Tokyo, Japan) after a 1-wk recovery from the surgery at the Institute of Laboratory Animals, Yamaguchi University. The chronic ethanol exposure protocol was similar to the procedure used in our previous study[14]; there were 28 d of overall ethanol exposure (Figure 2). The groups were divided as follows: Sham rats given a controlled liquid diet (Sham-control, n = 7); Sham rats given a 5-g/dL ethanol liquid diet (Sham-ethanol, n = 8); PH rats given a controlled liquid diet (PH-control, n = 10); and PH rats given a 5-g/dL ethanol liquid diet (PH-ethanol, n = 10). At the end of the pair-feeding, the rats were anesthetized, and blood samples were collected from the tails of the rats. The liver samples were harvested, and the liver weights were measured; the samples were fixed in 4% paraformaldehyde in 0.1 M phosphate buffered saline (PBS) at 4 °C and/or snap-frozen in liquid nitrogen. The liver samples were stored at -80 °C until further analysis.
All of the experimental protocol on animal protection and welfare satisfied the guidelines of our Institutional Animal Care Committee and the guidelines of the National Institutes of Health. Anesthesia consisted of the spontaneous inhalation of 1%-2% isoflurane in oxygen was performed when necessary.
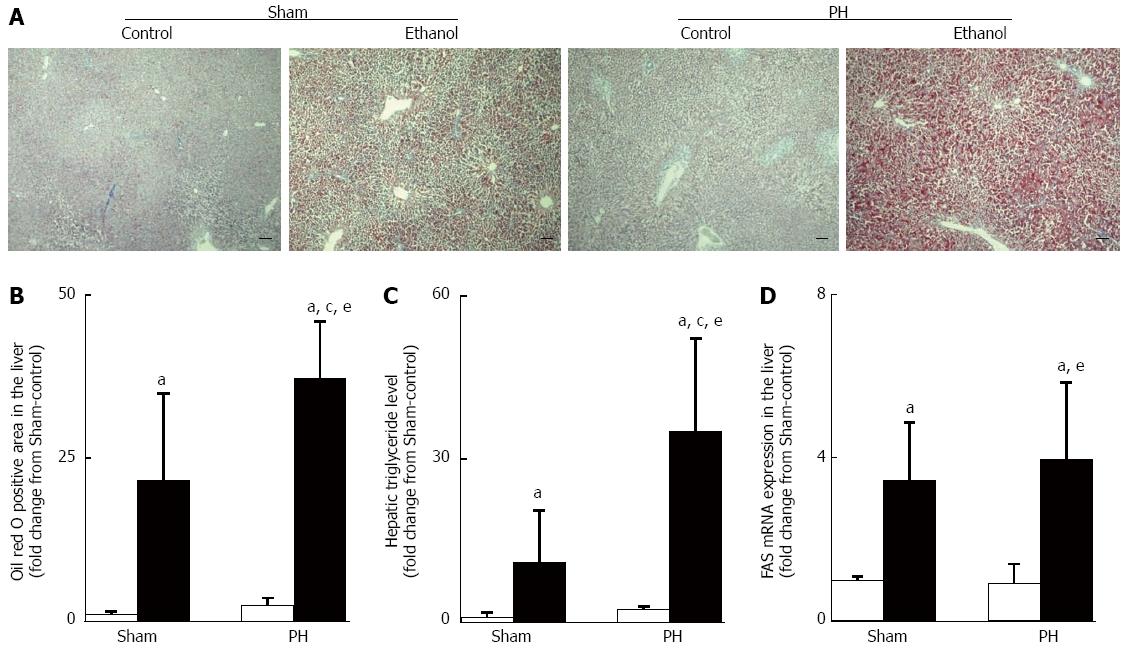
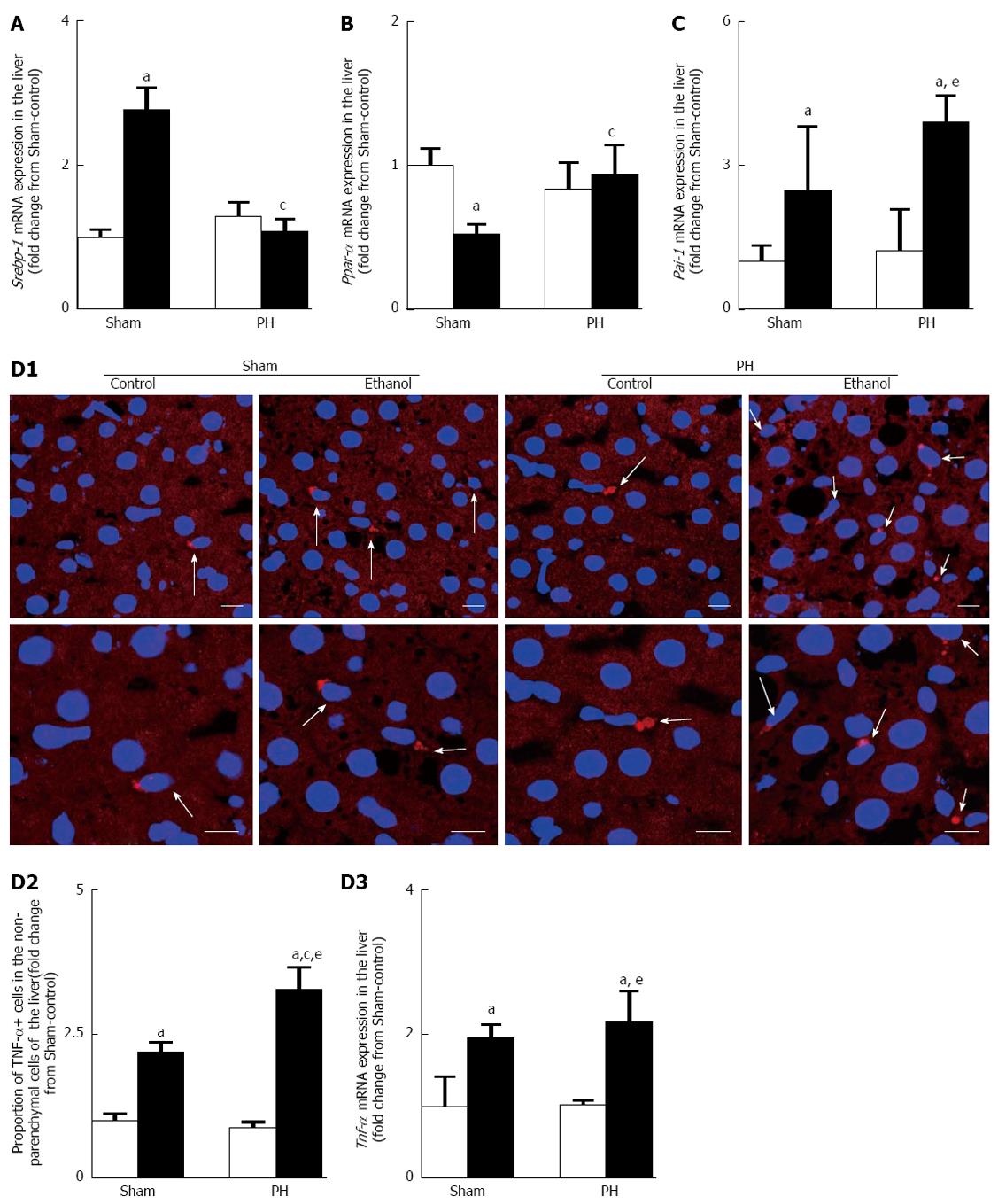
Sections (5-μm thick) of the paraformaldehyde/PBS-fixed rat livers harvested at the end of the pair-feeding period were frozen and sliced in a cryostat for Oil red O staining. Oil red O staining was used to quantitatively evaluate the hepatic steatosis using a computerized microscope system (BZ-II VIEWER and BZ-II ANALYZER 2007; KEYENCE Corporation, Osaka, Japan) at 10 × magnification. Three randomly selected areas in each specimen were analyzed. The ratio of each value was calculated as the percentage of Oil red O positive-stained areas and was expressed as the fold change from the Sham-control value.
For the immunofluorescence staining, 8-μm thick sections of the paraformaldehyde/PBS-fixed rat livers harvested at the end of the pair-feeding period were frozen and sliced in a cryostat (Leica CM1850). The sections were incubated with a tumor necrosis factor-α (TNF-α; N-19, goat polyclonal; Santa Cruz Biotechnology) primary antibody (1:100 dilution) and then incubated with a Cy3-labeled anti-goat IgG (Chemicon, Temecula, CA) diluted to 1:1000. The nuclei were stained with TO-PRO®-3 iodide (Molecular Probes, Carlsbad, CA), and the sections were observed under a confocal laser-scanning microscope (LSM5 Pascal/Version 3.2, Carl Zeiss Microlmaging Co. Ltd., Zeiss, Germany). The total number of non-parenchymal cells around the hepatocytes (the number of nuclei) and the total number of TNF-α positive cells in the non-parenchymal cells (immunofluorescence-expressing cells that had nuclei) were counted in 25 consecutive regions with a confocal microscope under a 63 × objective. The ratio of each value was calculated as the percentage of TNF-α positive cell and was expressed as the fold change from the Sham-control value.
The total RNA was extracted from the liver tissue using the RNeasy® Mini Kit (Qiagen, Tokyo, Japan) and was reverse-transcribed using the High Capacity RNA-to-cDNA Kit (Applied Biosystems, Tokyo, Japan). Reverse transcriptase polymerase chain reaction (RT-PCR) was performed for the quantitative assessment of mRNA expression using an Applied Biosystems StepOneTM system (Applied Biosystems). The following were purchased from Applied Biosystems: gene expression assays for fatty acid synthase (Fas; ID: Rn01645297_g1); sterol regulatory element binding protein-1 (Srebp-1; ID: Rn01446560_m1), which is a gene that plays an important role in the regulation of the transcription of genes involved in hepatic triglyceride synthesis, such as FAS[15]; peroxisome proliferator-activated receptor-α (Ppar-α, ID: Rn00566193_m1), which is a member of the nuclear hormone receptor superfamily that coordinates a number of metabolic pathways to dispose of excess fatty acids[16]; plasminogen activator-1 (Pai-1, ID: Rn 01481341_m1), which is a major inhibitor of tissue-type and urokinase-type plasminogen activators involved in fatty livers[16]; Tnf-α (ID: Rn99999017_m1); and Adh1 (ID: Rn00570670_m1), which is the key alcohol-metabolizing enzyme to detoxify incoming ethanol by converting it into acetaldehyde[17]. The relative expression of the target gene mRNA was normalized to the amount of the Gapdh (ID: Rn99999916_s1) mRNA in an identical cDNA sample, using the comparative quantitative method recommended by the manufacturer; the relative expression was expressed as the fold change from the Sham-control value or the fold change from before PH value.
The liver tissues harvested at the end of the pair-feeding period were homogenized with Extraction Buffer 1 of the ProteinExtract® Transmembrane Protein Extraction Kit (Novagen®, Merck KGaA, Darmstadt, Germany) and centrifuged at 1000 g for 5 min at 4 °C. The resultant supernatants were used for the total hepatic cytosolic proteins and subjected to Western blotting analysis and hepatic triglyceride concentration measurement. The protein concentrations were determined using Bradford’s method.
The total hepatic cytosolic protein was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis using a 4-20% (w/v) gel; the proteins were then transferred to a polyvinyl difluoride membrane. Each blot was incubated with an anti-beta-actin antibody (β-actin 1:5000; A2228, mouse monoclonal, Sigma-Aldrich, St. Louis, MO) or an ADH1 antibody (1:5000; rabbit, kindly provided by Dr. Takeshi Haseba, Tokyo, Japan); the blots were then incubated with the appropriate secondary horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG antibodies. Finally, the Western blotting bands were analyzed using Quantity One software (Bio-Rad Laboratories, Inc., CA). The ADH1 reaction products were normalized to β-actin and expressed as fold change from the Sham-control value.
The blood samples collected from the tails of the rats were centrifuged at 3000 g for 10 min, and the resultant supernatants were used to measure the ALT and blood aspartate aminotransferase (AST) levels (U/L). The total hepatic cytosolic proteins were used to measure the hepatic triglyceride concentrations (mg/g total liver protein, the fold change from the Sham-control value). The levels of blood ALT, AST, and the hepatic triglycerides were measured using an automatic analyzer (FDC4000; Fuji Medical Systems, Tokyo, Japan).
The blood samples (0.1 mL), collected at 08:30-09:00 and after a 5-h fast (13:30-14:00) at the end of the pair-feeding period, were mixed with 1.0 mL of 0.6-N cold perchloric acid and centrifuged at 3000 g for 5 min at 4 °C. The ethanol concentration in the supernatant was determined by headspace gas chromatography (GC-14B and HSS-2A, C-R6A; Shimadzu Co., Kyoto, Japan), as previously described[14].
Continuous data was expressed as the mean ± SD. Extreme value was excluded by Smirnov‐Grubbs test when necessary. The statistical significances of the continuous variables were assessed by 1-way analysis of variance (ANOVA) using Statcel2 for Windows software (OMS Publishing, Inc., Saitama, Japan). When an F-value was found to be significant by the ANOVA, a Bonferroni/Dunn post hoc test was used for the multiple comparisons. For the comparison of the means between two groups the 2-tailed Student t test was used. Values of P < 0.05 were considered statistically significant.
Body weight and liver function before and 1 wk after the sham- or PH-operation among the 4 groups were compared (Table 1). No significant differences were shown in the body weight and ALT among the 4 groups both before and 1 wk after the sham- or PH-operation. The data suggested that the body weight and liver function recovered from the PH operation at the beginning of the pair-feeding, which be performed for the ethanol treatment in the rats that had been subjected to PH.
| Sham | PH | |||
| Control | Ethanol | Control | Ethanol | |
| (n = 7) | (n = 8) | (n = 10) | (n = 10) | |
| Body weight (g) | ||||
| Before operation | 264 ± 7 | 259 ± 8 | 247 ± 20 | 246 ± 26 |
| 1 wk after operation | 274 ± 28 | 276 ± 15 | 257 ± 22 | 260 ± 25 |
| ALT (U/L) | ||||
| Before operation | 27 ± 2 | 29 ± 2 | 29 ± 5 | 29 ± 6 |
| 1 wk after operation | 26 ± 3 | 28 ± 3 | 28 ± 4 | 27 ± 5 |
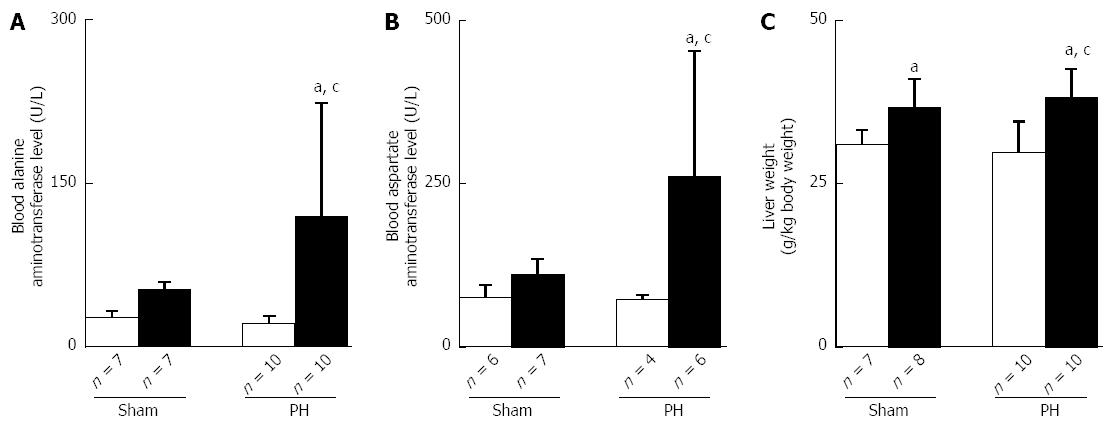
Similar amounts of total ethanol were consumed in the Sham-ethanol and the PH-ethanol group rats (85 ± 5 g vs 85 ± 6 g, P > 0.05) during the 28-d pair-feeding period. The rats that had been subjected to PH showed significant signs of hepatic steatosis and impaired liver function, as evidenced by the Oil red O staining analyses (Figure 3A and B), marked elevations in the hepatic triglyceride levels (Figure 3C) and their blood ALT and AST levels (Figure 4A and B). Moreover, the PH-ethanol rats showed the more significant upward intend (the statistical analysis found no differences from Sham-ethanol group) levels in their Fas gene expression (Figure 3D), liver function (Figure 4A and B), and liver weights (Figure 4C) after the 28-d pair-feeding period.
Up-regulation of Srebp-1 and down-regulation of Ppar-α gene expression levels have been shown to accompany ethanol-induced fatty liver[14]. In this study, the Sham-ethanol rats, and not the PH-ethanol rats, demonstrated up-regulation of Srebp-1 (Figure 5A) and the down-regulation of the Ppar-α mRNA expression levels (Figure 5B) after the 28-d pair-feeding period. Those discrepant results led us to assess the other mediators that alter lipid metabolism such as PAI-1 and TNF-α.
PAI-1, an acute-phase protein, showed an important role in liver dysfunction after PH in a rat model[18], and alcohol-induced hepatic steatosis was accompanied by a significant up-regulation of hepatic PAI-1 expression in mice[19]. TNF-α overproduction was responsible for the ethanol-induced hepatic steatosis[14,16]. Consistent with this previous report, we observed that the 28-d ethanol administration induced the up-regulation of Pai-1 gene expression and TNF-α overproduction in the Sham and the PH rats; however, the effect was shown the more significant upward intend (the statistical analysis found no differences) in the PH rats (Figure 5C and D).
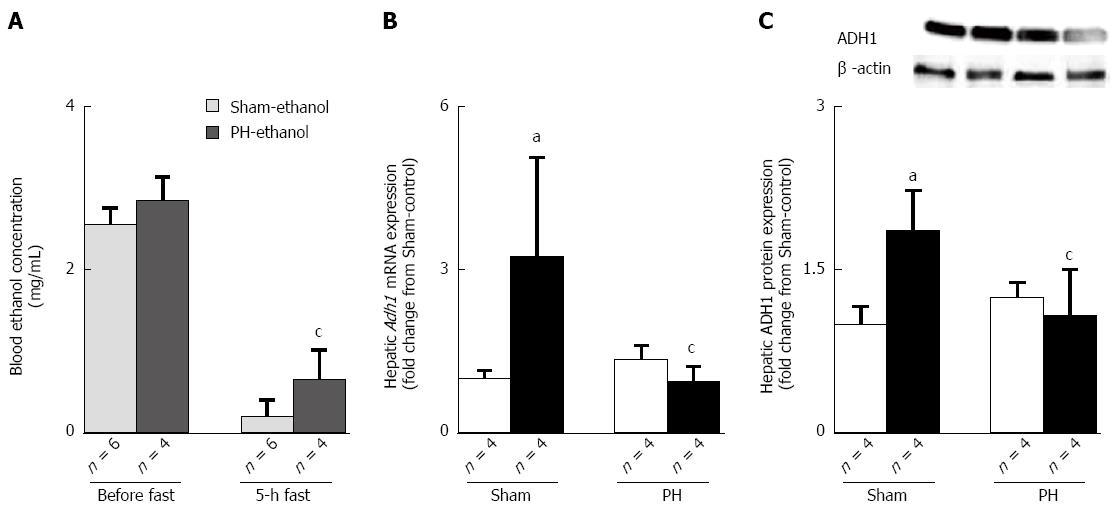
ADH1 is the major enzyme involved in ethanol oxidation[20]. Treatment of male Sprague-Dawley rats with a 5-g/dL ethanol liquid diet for 4 wk significantly increased the mRNA levels of Adh1 in the liver[21]. In this study, no significant difference was found in the blood ethanol concentrations before fasting (Figure 6A); however, the PH-ethanol rats showed a higher residual blood ethanol concentration compared with the Sham-ethanol rats after a 5-h fast at the end of the pair-feeding period (Figure 6A), suggesting delayed ethanol elimination after liver resection. In the Sham-ethanol group, the mRNA (Figure 6B) and protein expression levels (Figure 6C) of ADH1 were found to be up-regulated by the chronic ethanol treatment; the Adh1 gene (Figure 6B) and protein (Figure 6C) expression levels fully recovered at the end of the 28-d pair-feeding period in the PH-control rats. The PH-ethanol rats showed significantly reduced Adh1 mRNA and protein expression levels compared with those of Sham-ethanol rats (Figure 6B and C). These results suggest that the 28-d post-hepatectomy ethanol treatment induced the desensitization of the ADH1 expression in the rats that had been subjected to liver resection.
To clarify the roles of specific genes in the PH-ethanol rats, the expression levels of several key genes were assessed before and 1 wk after surgery to determine the subject’s recovery from the liver resection using the rats that had been subjected to the preliminary examination. The mRNA expression levels of Fas, the lipid metabolism-associated transcription factors (Srebp-1 and Ppar-α), the mediators that alter lipid metabolism (Pai-1 and Tnf-α) showed full recovery; however, the levels of Adh1 did not show full recovery (Table 2).
| Before PH | 1 wk after PH | |
| Fas (fold change from before PH) | 1.0 ± 0.34 | 0.6 ± 0.22 |
| Srebp-1 (fold change from before PH) | 1.0 ± 0.26 | 1.03 ± 0.20 |
| Ppar-α (fold change from before PH) | 1.0 ± 0.43 | 1.04 ± 0.29 |
| Pai-1 (fold change from before PH) | 1.0 ± 0.27 | 0.87 ± 0.33 |
| Tnf-α (fold change from before PH) | 1.0 ± 0.20 | 0.99 ± 0.11 |
| Adh1 (fold change from before PH) | 1.0 ± 0.30 | 0.40 ± 0.06c |
Using PH rats in a liver resection animal model, we identified enhanced susceptibility to ethanol-induced hepatic steatosis and liver function impairment after liver resection. The incomplete recovery of hepatic Adh1 gene expression at 1 wk after liver resection and the desensitization of Adh1 gene expression because of post-hepatectomy ethanol treatment might play roles in the enhanced susceptibility to ethanol hepatotoxicity after liver resection. Lower Adh1 gene expression levels after PH resulted in a delay in ethanol elimination and higher residual blood ethanol concentrations, leading to ethanol-induced hepatic steatosis and liver function impairment. These findings elucidate a basic pathogenic mechanism of ethanol-induced hepatic steatosis after liver resection in rats and might be valuable for individuals considering hepatic surgery and/or liver resection as living donors in LDLT.
Developments in surgical techniques and postoperative care have enabled more extended anatomic and non-anatomic resections. LDLT could shorten the waiting time and lower the dropout rate, particularly in patients with hepatocellular carcinoma[22]. LDLT involves two patients, the recipient and the living donor. Despite the gravity of the procedure for LDLT donors, few studies have investigated the effect of LDLT on the subsequent quality of life of the donors[23]. Hepatic steatosis increases the risk of primary dysfunction after liver transplantation[10], and liver resection includes amplified postoperative morbidity and mortality[24,25]. Lipid accumulation interferes with hepatic energy homeostasis and induces hepatocellular damage, affecting hepatocellular recovery after liver resection[26]. Evolving knowledge regarding hepatic steatosis combined with its increasing prevalence emphasizes the critical need to understand the implications of liver resection in the development of steatosis. The basic pathogenesis of hepatic steatosis, particularly ethanol-induced hepatic steatosis and the mechanisms behind it after liver resection, are being elucidated. In this study, the 28-d ethanol treatment induced severe ethanol-induced hepatic steatosis and liver function impairment in rats; these effects were compared to those in the rats without liver resection, and the results suggested an enhanced susceptibility to ethanol-induced hepatic steatosis and liver dysfunction after liver resection in rats.
Chronic alcohol consumption is a major risk factor for the development of liver disease, and ethanol-induced hepatic steatosis occurs in 90% of those who consume alcohol in excess of 60 g/d[27]. The metabolism of ethanol by hepatocytes initiates a pathogenic process involving the production of protein-aldehyde adducts, lipid peroxidation, immunologic activity, and cytokine release[28]. Ethanol-induced hepatic steatosis has been regarded as entirely benign; however, as occurs in the spectrum of non-alcoholic steatohepatitis disorders, certain pathological features such as giant mitochondria, perivenular fibrosis, and microvesicular fat might be associated with progressive liver injury[14,28]. The mechanisms by which ethanol induces fatty liver are extremely complex and not completely understood. Ethanol is a direct hepatotoxin, and ADH1-related ethanol metabolism by hepatocytes induces the reduction of NAD+ to NADH, which could inhibit the NAD+-requiring tricarboxylic acid cycle and β-oxidation of fatty acid[11]. The ADH family consists of numerous enzymes that are able to catalyze the reversible oxidation of a wide variety of xenobiotics and endogenous ethanol to their corresponding aldehydes[29]. ADH1, the key alcohol-metabolizing enzyme in the conversion of ethanol to its metabolite, acetaldehyde, particularly during the elimination phase, is expressed predominantly in the liver[17,30,31]; its gene expression is strongly induced by ethanol treatment[21]. Our results showed that the 28-d ethanol treatment induced hepatic steatosis accompanied by the desensitization of Adh1 gene expression because of the post-hepatectomy ethanol treatment and a higher residual blood ethanol concentration in the PH-ethanol rats. No residual blood ethanol was detected after a 5-h fast, and the up-regulation of the Adh1 gene and protein expressions was demonstrated at the end of the 28-d pair-feeding period in the Sham-ethanol rats. Collectively, these results might suggest that ethanol hepatotoxicity directly induced fatty livers in the PH-ethanol rats, whereas alterations of ethanol metabolism related to ADH1 induced fatty liver in the rats without liver resection.
To determine the effects of PH on ethanol-induced hepatic steatosis after liver resection, the gene expressions of Fas, Srebp-1, Ppar-α, Pai-1, Tnf-α and Adh1 were assessed before and 1 wk after liver resection in the rats that had been subjected to the preliminary examination in this study. The results showed that the post-resection recovery of the Adh1 gene expression was more sluggish than for the other relevant genes, liver weight (Figure 1A), and liver function (Figure 1B). The incomplete recovery of ADH1 from liver resection might be responsible for the down-regulation of the Adh1 gene expression after hepatic surgery that induced the ethanol elimination delays and the higher residual blood ethanol concentrations, resulting in the increase in direct hepatotoxicity of the liquid ethanol diet. In a mouse model of PH, a protein expression study showed that ADH1 slowly decreased after PH and was significantly down-regulated in the late stages of regeneration (i.e., 72 h after PH)[32]. In this study, the Adh1 gene (Figure 6B) and protein (Figure 6C) expressions were fully recovered at the end of the 28-d pair-feeding period in the control-liquid-diet fed rats that had undergone liver resection. Further study is required to clarify when the Adh1 gene and/or protein expression recovers fully from liver resection.
Ethanol-induced fatty liver is accompanied by a substantial increase of the mature SREBP-1 protein levels as well as the activation of the SREBP-1 target hepatic lipogenic genes, such as Fas[14,33]. Ethanol-induced PPAR-α inhibition could lead to acetyl-CoA carboxylase induction and malonyl-CoA decarboxylase suppression, followed by the elevation of the malonyl-CoA level and the inhibition of carnitine acyltransferase 1 activity, resulting in the reduction of fatty acid oxidation[34,35]. Consistent with these previous reports, we observed that 28-d ethanol administration induced hepatic steatosis associated with the up-regulation of Srebp-1 (Figure 5A) and the down-regulation of Ppar-α (Figure 5B) in the Sham-ethanol rats and not in the PH-ethanol rats (Figure 5A and B). Liver resection likely plays a major role in these discrepant results; however, further study is required. Those discrepant results led us to assess a separate mechanism by which alcohol might cause fatty liver under conditions of PH-related injury and/or inflammation, namely, via the increased release of mediators that alter lipid metabolism such as TNF-α and PAI-1. The results showed that chronic ethanol consumption induced significant up-regulation of the Pai-1 mRNA expression (Figure 5C) and TNF-α overproduction (Figure 5D), especially in the PH group rats. TNF-α, a pro-inflammatory cytokine, potentially plays a major role in causing steatosis after alcohol exposure by increasing the hepatic lipid synthesis, decreasing the lipoprotein lipase activity, and/or inhibiting the fatty acid oxidation in rat hepatocytes[36,37]. One mechanism by which TNF-α could mediate this effect is by inducing PAI-1 expression[38]. PAI-1 is an acute-phase protein that is typically expressed only in adipocytes and endothelial cells; it could be highly expressed by most cells in response to stress[39], and plasma PAI-1 levels have been shown to be strongly related to liver steatosis[19]. Our results indicated that AFLD is involved in the progression of alcoholic liver disease via the up-regulation of hepatic Pai-1 mRNA expression and TNF-α overproduction in rats with liver resections. Evaluation of the mechanisms underlying PAI-1 gene up-regulation and TNF-α overproduction as possible therapeutic targets, particularly for subjects having undergone hepatic surgery, is important.
Our data suggest that the incomplete recovery of hepatic Adh1 gene expression after liver resection and the desensitization of Adh1 expression because of post-hepatectomy ethanol treatment likely played roles in the delayed ethanol elimination, leading to the enhanced susceptibility to ethanol-induced hepatic steatosis and liver function impairment in the rats that underwent liver resection. Pre-surgical counseling of patients and living liver donors regarding the enhanced risk of liver dysfunction after liver resection, particularly in conjunction with alcohol consumption, and the use of evidenced-based medical care after liver resection should be prioritized as medical and surgical advances in facilitating effective hepatic surgery and liver transplantation.
The ADH1 antibody was kindly provided by Dr. Takeshi Haseba, Department of Legal Medicine, Nippon Medical School, 1-1-5 Sendagi Bunkyo-ku, Tokyo 113-8602, Japan.
Hepatic surgery is the curative treatment for patients with primary or secondary malignant liver tumors, and the procedure is performed in living donor liver transplantations to overcome the shortage of cadaver organ donations, particularly in Asia. The prevalence rates of alcohol abuse and obesity are reaching epidemic proportions globally, and the effect of fatty liver, which frequently accompanies these conditions, on postoperative outcomes is not well understood.
The consumption of alcohol is extensive, and alcohol is a well-known hepatotoxin. Alcohol abstention is difficult for some individuals. One mechanism by which ethanol induces fatty liver is that hepatic class-1 alcohol dehydrogenase (ADH1)-related ethanol metabolism. In this study, the authors demonstrate that the desensitization to post-hepatectomy ethanol treatment and slow recovery from liver surgery in ADH1 gene expression could enhance the susceptibility to ethanol-induced fatty liver after liver resection in rats.
Recent reports have highlighted the effects of fatty liver on the postoperative complications, mortality, and morbidity in patients with liver surgery. This is the first study to report that the recovery of ADH1 gene fall behind other genes in rats had been performed liver resection. Furthermore, desensitization to post-hepatectomy ethanol treatment in ADH1 gene could enhance the susceptibility to ethanol-induced fatty liver after liver surgery in rats.
By understanding the basic pathogenic mechanism of ethanol-induced fatty liver after liver resection in rats, this study may represent a future strategy for the individuals considering hepatic surgery and/or liver resection as living donors in living donor liver transplantation.
ADH1-related ethanol metabolism induces the reduction of NAD+ to NADH, which could inhibit the NAD+-requiring tricarboxylic acid cycle and the β-oxidation of fatty acids. Such a mechanism is thought to be the important in ethanol-induced fatty liver.
The authors examined the fatty liver, liver function, lipid metabolism-related regulators’ genes, and the major enzyme gene (ADH1) for ethanol metabolism in rats treated ethanol after liver resection. It revealed that the desensitization to post-hepatectomy ethanol treatment and slow recovery from liver surgery in ADH1 gene expression could enhance the susceptibility to ethanol-induced fatty liver after liver resection in rats. The results are interesting and may represent the cell biology and molecular mechanisms of ethanol-induced fatty liver in subjects that had been conducted into liver resection.
P- Reviewer: Chen LZ S- Editor: Qi Y L- Editor: A E- Editor: Zhang DN
| 1. | Aljiffry M, Abdulelah A, Walsh M, Peltekian K, Alwayn I, Molinari M. Evidence-based approach to cholangiocarcinoma: a systematic review of the current literature. J Am Coll Surg. 2009;208:134-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 179] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 2. | Li Y, Liu L, Wang B, Wang J, Chen D. Metformin in non-alcoholic fatty liver disease: A systematic review and meta-analysis. Biomed Rep. 2013;1:57-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 217] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 3. | Petrowsky H, Busuttil RW. Evolving surgical approaches in liver transplantation. Semin Liver Dis. 2009;29:121-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 4. | González-Chamorro A, Loinaz Segurola C, Moreno González E, Jiménez Romero C, González-Pinto Arrillaga I, Gomez Sanz R, Garcia Garcia I, Manzanera Diaz M, Alonso Casado O. Graft mass and volume calculation in living related donors for liver transplantation. Hepatogastroenterology. 1998;45:510-513. [PubMed] |
| 5. | Kaido T, Uemoto S. Does living donation have advantages over deceased donation in liver transplantation? J Gastroenterol Hepatol. 2010;25:1598-1603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 6. | Middleton PF, Duffield M, Lynch SV, Padbury RT, House T, Stanton P, Verran D, Maddern G. Living donor liver transplantation--adult donor outcomes: a systematic review. Liver Transpl. 2006;12:24-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 195] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 7. | Ringe B, Strong RW. The dilemma of living liver donor death: to report or not to report? Transplantation. 2008;85:790-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 60] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 8. | Veteläinen R, van Vliet A, Gouma DJ, van Gulik TM. Steatosis as a risk factor in liver surgery. Ann Surg. 2007;245:20-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 226] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 9. | Angulo P. Nonalcoholic fatty liver disease. N Engl J Med. 2002;346:1221-1231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3655] [Cited by in RCA: 3718] [Article Influence: 161.7] [Reference Citation Analysis (2)] |
| 10. | Said A. Non-alcoholic fatty liver disease and liver transplantation: outcomes and advances. World J Gastroenterol. 2013;19:9146-9155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 26] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 11. | Grunnet N, Kondrup J. The effect of ethanol on the beta-oxidation of fatty acids. Alcohol Clin Exp Res. 1986;10:64S-68S. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 53] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 12. | Molotkov A, Duester G. Genetic evidence that retinaldehyde dehydrogenase Raldh1 (Aldh1a1) functions downstream of alcohol dehydrogenase Adh1 in metabolism of retinol to retinoic acid. J Biol Chem. 2003;278:36085-36090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 119] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 13. | Higgins GM, Anderson RM. Experimental pathology of the liver. I. Restoration of the liver of the white rat following partial surgical removal. Arch Pathol. 1931;12:186-202. |
| 14. | Liu J, Takase I, Hakucho A, Okamura N, Fujimiya T. Carvedilol attenuates the progression of alcohol fatty liver disease in rats. Alcohol Clin Exp Res. 2012;36:1587-1599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 15. | Seth D, D’Souza El-Guindy NB, Apte M, Mari M, Dooley S, Neuman M, Haber PS, Kundu GC, Darwanto A, de Villiers WJ. Alcohol, signaling, and ECM turnover. Alcohol Clin Exp Res. 2010;34:4-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 16. | Zeng T, Xie KQ. Ethanol and liver: recent advances in the mechanisms of ethanol-induced hepatosteatosis. Arch Toxicol. 2009;83:1075-1081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 17. | Kedishvili NY, Gough WH, Davis WI, Parsons S, Li TK, Bosron WF. Effect of cellular retinol-binding protein on retinol oxidation by human class IV retinol/alcohol dehydrogenase and inhibition by ethanol. Biochem Biophys Res Commun. 1998;249:191-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 40] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 18. | Watanabe K, Togo S, Takahashi T, Matsuyama R, Yamamoto H, Shimizu T, Makino H, Matsuo K, Morioka D, Kubota T. PAI-1 plays an important role in liver failure after excessive hepatectomy in the rat. J Surg Res. 2007;143:13-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 19. | Alessi MC, Bastelica D, Mavri A, Morange P, Berthet B, Grino M, Juhan-Vague I. Plasma PAI-1 levels are more strongly related to liver steatosis than to adipose tissue accumulation. Arterioscler Thromb Vasc Biol. 2003;23:1262-1268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 137] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 20. | Bosron WF, Magnes LJ, Li TK. Kinetic and electrophoretic properties of native and recombined isoenzymes of human liver alcohol dehydrogenase. Biochemistry. 1983;22:1852-1857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 106] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 21. | Park PH, Lim RW, Shukla SD. Gene-selective histone H3 acetylation in the absence of increase in global histone acetylation in liver of rats chronically fed alcohol. Alcohol Alcohol. 2012;47:233-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 22. | Zimmerman MA, Baker T, Goodrich NP, Freise C, Hong JC, Kumer S, Abt P, Cotterell AH, Samstein B, Everhart JE. Development, management, and resolution of biliary complications after living and deceased donor liver transplantation: a report from the adult-to-adult living donor liver transplantation cohort study consortium. Liver Transpl. 2013;19:259-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 23. | Parikh ND, Ladner D, Abecassis M, Butt Z. Quality of life for donors after living donor liver transplantation: a review of the literature. Liver Transpl. 2010;16:1352-1358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 82] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 24. | Behrns KE, Tsiotos GG, DeSouza NF, Krishna MK, Ludwig J, Nagorney DM. Hepatic steatosis as a potential risk factor for major hepatic resection. J Gastrointest Surg. 1998;2:292-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 306] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 25. | Belghiti J, Hiramatsu K, Benoist S, Massault P, Sauvanet A, Farges O. Seven hundred forty-seven hepatectomies in the 1990s: an update to evaluate the actual risk of liver resection. J Am Coll Surg. 2000;191:38-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 800] [Cited by in RCA: 798] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 26. | Huang J, Rudnick DA. Elucidating the metabolic regulation of liver regeneration. Am J Pathol. 2014;184:309-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 110] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 27. | McCullough AJ, O’Shea RS, Dasarathy S. Diagnosis and management of alcoholic liver disease. J Dig Dis. 2011;12:257-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 28. | Mailliard ME, Sorrell MF. Alcoholic liver disease. Harrison’s Principles of internal medicine. New York: McGraw-Hill Medical Publishing Division 2005; 1855-2566. |
| 29. | Pastino GM, Flynn EJ, Sultatos LG. Genetic polymorphisms in ethanol metabolism: issues and goals for physiologically based pharmacokinetic modeling. Drug Chem Toxicol. 2000;23:179-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 30. | Alamillo JM, Cárdenas J, Pineda M. Purification and molecular properties of urate oxidase from Chlamydomonas reinhardtii. Biochim Biophys Acta. 1991;1076:203-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 31. | Birley AJ, James MR, Dickson PA, Montgomery GW, Heath AC, Martin NG, Whitfield JB. ADH single nucleotide polymorphism associations with alcohol metabolism in vivo. Hum Mol Genet. 2009;18:1533-1542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 62] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 32. | Hsieh HC, Chen YT, Li JM, Chou TY, Chang MF, Huang SC, Tseng TL, Liu CC, Chen SF. Protein profilings in mouse liver regeneration after partial hepatectomy using iTRAQ technology. J Proteome Res. 2009;8:1004-1013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 33. | You M, Crabb DW. Recent advances in alcoholic liver disease II. Minireview: molecular mechanisms of alcoholic fatty liver. Am J Physiol Gastrointest Liver Physiol. 2004;287:G1-G6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 171] [Article Influence: 8.1] [Reference Citation Analysis (1)] |
| 34. | Ringseis R, Muschick A, Eder K. Dietary oxidized fat prevents ethanol-induced triacylglycerol accumulation and increases expression of PPARalpha target genes in rat liver. J Nutr. 2007;137:77-83. [PubMed] |
| 35. | Wan YJ, Morimoto M, Thurman RG, Bojes HK, French SW. Expression of the peroxisome proliferator-activated receptor gene is decreased in experimental alcoholic liver disease. Life Sci. 1995;56:307-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 71] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 36. | Feingold KR, Grunfeld C. Tumor necrosis factor-alpha stimulates hepatic lipogenesis in the rat in vivo. J Clin Invest. 1987;80:184-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 304] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 37. | Nachiappan V, Curtiss D, Corkey BE, Kilpatrick L. Cytokines inhibit fatty acid oxidation in isolated rat hepatocytes: synergy among TNF, IL-6, and IL-1. Shock. 1994;1:123-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 54] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 38. | Fearns C, Loskutoff DJ. Induction of plasminogen acti-vator inhibitor 1 gene expression in murine liver by lipopolysaccharide. Cellular localization and role of endogenous tumor necrosis factor-alpha. Am J Pathol. 1997;150:579-590. [PubMed] |