Published online Dec 28, 2014. doi: 10.3748/wjg.v20.i48.18240
Revised: May 24, 2014
Accepted: July 11, 2014
Published online: December 28, 2014
Processing time: 260 Days and 1.3 Hours
AIM: To assess the role of circulating tumor cells (CTCs) and cancer stem cells (CSCs) in hepatitis C virus (HCV)-associated liver disease.
METHODS: Blood and/or tissue samples were obtained from HCV (genotype 4)-associated hepatocellular carcinoma patients (HCC; n = 120), chronic hepatitis C patients (CH; n = 30) and 33 normal control subjects (n = 33). Serum levels of alpha-fetoprotein (AFP), alkaline phosphatase, and alanine and aspartate aminotransferases were measured. Cytokeratin 19 (CK19) monoclonal antibody was used to enumerate CTCs, and CD133 and CD90 were used to enumerate CSCs by flow cytometry. The expression levels of the CSCs markers (CD133 and CD90) as well as telomerase, melanoma antigen encoding gene 1 (MAGE1) and MAGE3 were assessed by RT-PCR and quantitative real-time polymerase chain reactions. The number of CTCs and/or the expression levels of CK19, CD133, telomerase, MAGE1 and MAGE3 were correlated to the standard clinicopathologic prognostic factors and disease progression.
RESULTS: Levels of AFP, alkaline phosphatase and aspartate aminotransferase were significantly different among the HCC, CH and control groups (P < 0.001), whereas alanine aminotransferase differed significantly between patient (HCC and CH) and control groups (P < 0.001). At the specified cutoff values determine by flow cytometry, CK19 (49.8), CD90 (400) and CD133 (73) were significantly higher in the blood of HCC patients compared to those in the CH and control groups (P < 0.001). On the other hand, CD133 at a 69.5 cutoff was significantly higher in the CH compared to the control group (P≤ 0.001). Telomerase, MAGE1 and MAGE3 RNA were expressed in 55.71%, 60.00% and 62.86% of the HCC patients, respectively, but were not detected in patients in the CH or control groups, which were statistically significant (Ps < 0.001). The expression levels of telomerase, CD90, MAGE3, CD133 and CK19 were all significantly associated with high tumor grade and advanced stage in HCC patients (all Ps < 0.05).
CONCLUSION: CTC counts and AFP, CK19, telomerase, and MAGE1/MAGE3 expression predict disease progression in patients with HCV, whereas telomerase, MAGE3, CD90, CD133 and CK19 are prognostic markers in HCC.
Core tip: Recent studies have shown that cancer stem and circulating tumor cells contribute to tumor development and progression and can predict patient outcome. Although there are various methods for enumeration of circulating tumor cells, this study demonstrates that flow cytometry is a sensitive, rapid and easy technique that can be used to follow chronic hepatitis C virus patients for early detection of hepatocellular carcinoma. Additionally, telomerase, melanoma antigen encoding gene 3 and cancer stem cell markers (CD90, CD133, CK19) are prognostic indicators in hepatocellular carcinoma patients.
- Citation: Bahnassy AA, Zekri ARN, El-Bastawisy A, Fawzy A, Shetta M, Hussein N, Omran D, Ahmed AAS, El-Labbody SS. Circulating tumor and cancer stem cells in hepatitis C virus-associated liver disease. World J Gastroenterol 2014; 20(48): 18240-18248
- URL: https://www.wjgnet.com/1007-9327/full/v20/i48/18240.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i48.18240
Hepatocellular carcinoma (HCC) is the fifth most common cancer in men and the seventh in women. The main etiological factors are hepatitis B virus (HBV) and hepatitis C virus (HCV) infection, heavy alcohol consumption and aflatoxin B1[1,2]. With a population of approximately 85 million, the prevalence of HCV in Egypt is estimated to be over 18%, leaving HCC as a major health problem[3]. Until recently, the choice of therapy and prognosis largely depended on the severity of liver function, radiologic findings, and alpha-fetoprotein (AFP) levels. However these parameters are insufficient for predicting patient outcome, and therefore, individually-based biologic markers are needed[4].
Circulating tumor cells (CTCs) can be detected in blood or lymphatic vessels of cancer patients. The presence of CTCs has been associated with a high incidence of metastasis and increased resistance to therapy in some solid tumors. Thus, detection of CTCs has considerable clinical significance for monitoring treatment response and predicting recurrence[5], progression-free and overall survivals. CTCs therefore represent an interesting source of biological information to understand dissemination, drug resistance and treatment-induced cell death[6]. However, only a few studies have addressed the role of CTCs in HCC. This could be attributed to the paucity of CTCs in patient blood, which makes them difficult to detect, as well as the debate concerning detection methods and the relative lack of specific HCC markers[7].
Telomerase expression in liver tissues or peripheral blood has been used as a diagnostic and prognostic marker for HCC[8]. Telomerase is part of a protein/RNA complex involved in the extension of telomeres during the cell cycle, which stabilizes genomic integrity as well as cancer cell immortality. Some studies have shown semi-quantitative radioactive enzyme-linked immunosorbent assays are not sufficiently sensitive for quantifying telomerase activity in the blood of HCC patients[9]. Similarly, melanoma antigen encoding gene 1 (MAGE1) and MAGE3, which belong to a family of at least 12 members, were detected in a variety of tumors, and correlated with HCC prognosis. Moreover, MAGE1 and MAGE3 have been used to detect CTCs in HCC patients[10].
More recent studies have shown that cancer stem cells (CSCs) contribute to tumor growth, cancer relapse and poor response to treatment. These small, undifferentiated, progenitor cells are capable of self-renewal, production of heterogeneous progenies and resistance to chemotherapy. Thus, they are considered to be responsible for the acquisition of aggressive phenotypes. CSCs were detected in many tumor types using specific markers, such as c-kit, CD133, CD90, CD44, CD326 and OV6, and it has been proposed that they are the origin of CTCs[11,12]. However, few studies have correlated the clinical features of HCC patients with the expression of CSC markers. Therefore, we sought to assess the contribution of CSCs and CTCs in patients with HCV-associated chronic hepatitis (CH) and HCC via determination of the stem cell-related and liver-specific markers, including CD133, CD90, MAGE1/MAGE3, telomerase and cytokeratin 19 (CK19). We also evaluated the efficiency of flow cytometry as a method for the enumeration of CTCs in comparison to the commonly used techniques.
We prospectively collected peripheral blood samples from HCC patients (n = 70) who attended the National Cancer Institute and Kasr Al-Aini School of Medicine, Cairo University clinics between June and December 2010. HCC was histologically diagnosed whenever surgical specimens were available. Otherwise, diagnosis of HCC was based on computed tomography and elevated AFP levels. HCC patients were classified according to the sixth edition of the International Union against Cancer tumor-node-metastasis staging system[13] and the Milan criteria[14]. We also included post-HCV-CH patients (n = 30) who were diagnosed by clinical examination, abdominal ultrasound, laboratory investigations and liver biopsy. Age- and sex-matched healthy volunteers (n = 33) served as a control group. They all had normal values of serum alanine aminotransferase (ALT) and were sero-negative for hepatitis B surface markers (HBsAg, HBeAg and HBcAb) and HBV antibodies. Fresh tissue samples from an additional 50 pathologically confirmed HCC patients (26 men and 24 women) were also included as a confirmatory set to validate the data.
All cases were newly diagnosed cases that had not received prior chemotherapy. Patients were subjected to full clinical examinations, radiologic investigations (including abdominal ultrasonography and triphasic computed tomography) and laboratory investigations. All studied cases (HCC and CH) were HCV-positive and HBV-negative as confirmed by polymerase chain reaction (PCR) and serologic tests (Table 1). Written informed consent was obtained from all participants prior to enrollment in the study, which conformed to the ethical guidelines of the 2004 Declaration of Helsinki. The study protocol was approved by the Institutional Review Boards of the National Cancer Institute and Kasr Al-Aini School of Medicine.
| Gene | Primers sequences | Annealing temp.(°C) | Base pairs |
| MAGE1 | First round: | 60 | 421 |
| 5’-CGG CCG AAG GAA CCT GAC CCA G-3’ | |||
| 5’-GCT GGA ACC CTC ACT GGG TTG CC-3’ | |||
| Second round (nested): | 60 | 299 | |
| 5’-ACA GAG GAG CAC CAA GGA GAA G-3’ | |||
| 5’-AGT TGA TGG TAG TGG GAA AGG C-3’ | |||
| MAGE3 | First round: | 60 | 725 |
| 5’-TGG AGG ACC AGA GGC CCC C-3’ | |||
| 5’-GGA CGA TTA TCA GGA GGC CTG C-3’ | |||
| Second round (nested): | 60 | 371 | |
| 5’-CGG AGG AGC ACT GAA GGA GAA G-3’ | |||
| 5’-CCT CCT CTT CTT CGT TGC TGG-3’ | |||
| CK-19 | 5’-CCC GCG ACT ACA GCC ACTA-3’ | 60 | 745 |
| 5’-GCT CAT GCG CAG AGC CT-3’ | |||
| CD133 | P1: 5’-AAT TCT AAT ACG ACT CAC TAT AGG GAA GAA CAG GGA TGA TGT TGG GTC TCA-3’ | ||
| P2: 5’-TTT CAA GGA CTT GCG AAC TCT CTT GA-3’ | |||
| MB: 5’-CGA TCC AAG GAC AAG GCG TTC ACA GGA TCG-3’ | |||
| Telomerase | 5’-TGA CAC CTC ACC TCA CCC AC-3’ | 60 | 96 |
| 5’-CAC TGT CTT CCG CAA GTT CAC-3’ | |||
| β-actin | 5’-ACA CTG TGC CCA ACG AGG-3’ | 56 | 540 |
| 5’-AGG GGC CGG TCA TAC T-3’' |
Peripheral blood samples (two samples, 7.5 mL each) were collected from patient and control subjects in CellSave blood collection tubes (Immunicon Inc., Huntingdon Valley, PA, United States) containing EDTA and a cellular preservative. Samples from vein punctures were collected after discarding the first 0.5 mL to avoid skin-plug contamination. From each subject, one tube was used for assessment of CTCs and the other was used for RNA and DNA extraction.
Total viral DNA/RNA isolation was performed using QIAamp MinElute Virus Spin Kit (Qiagen, Venlo, Limburg, Germany). HBV core proteins were analyzed by PCR as previously described[15]. HCV detection and quantification were done using a StepOne Real-Time PCR system (Applied Biosystems of Thermo Fisher Scientific Inc., Waltham, MA, United States).
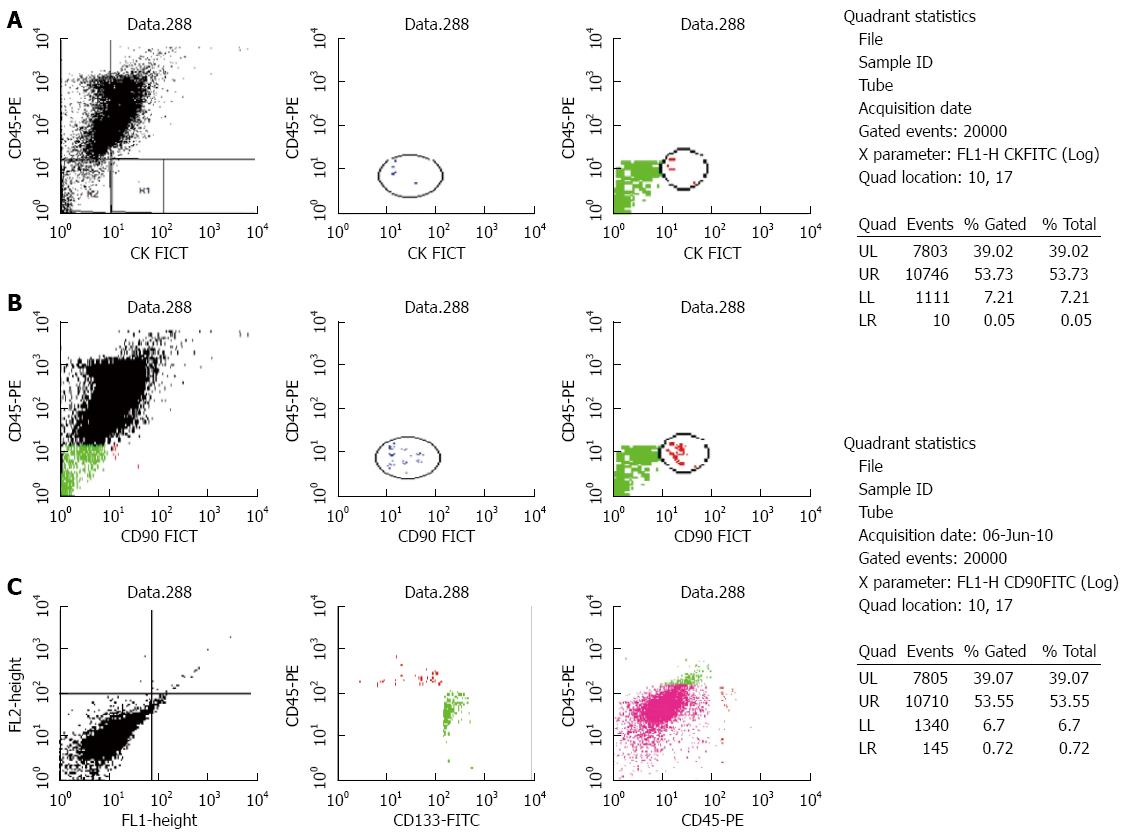
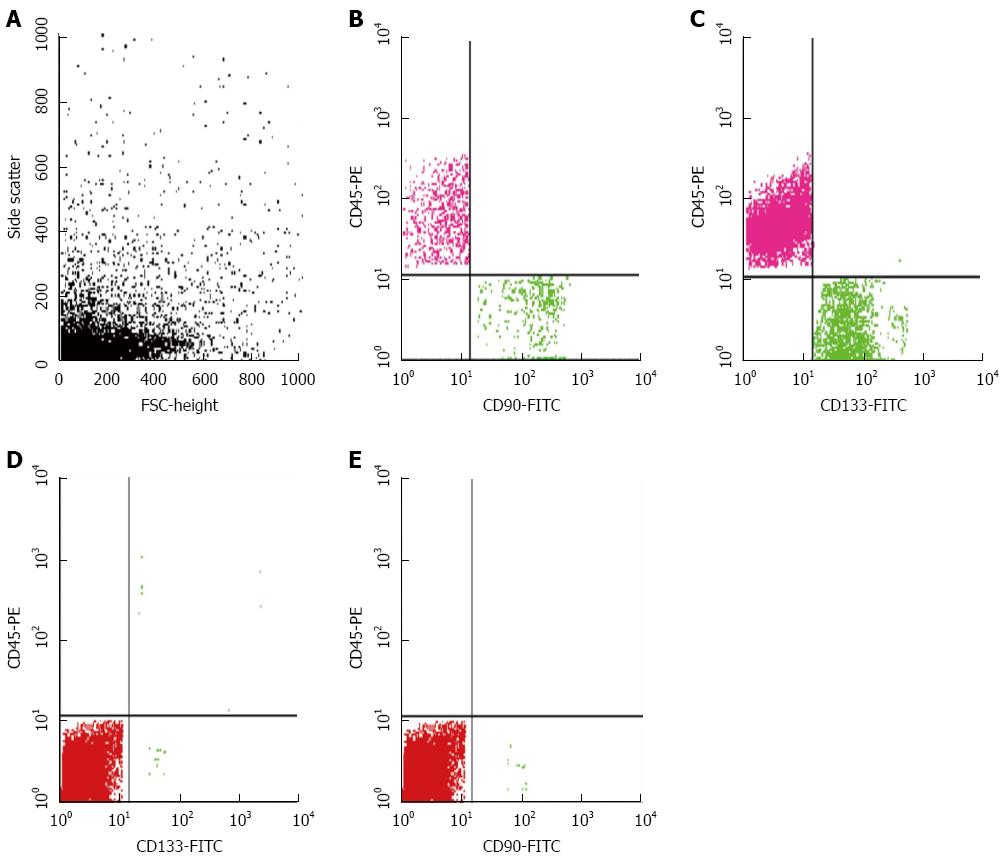
Peripheral blood mononuclear cells were obtained by gradient density centrifugation using Ficoll-Hypaque 1077 (Sigma-Aldrich, St. Louis, MO, United States). The separated cells were stained using the following fluorescent-labeled antibodies (three sets in three separate tubes): CK19-FITC/CD45-PE, CD45-PE/CK19-FITC/CD133-PerCP and CD45-PE/CD90-FITC (MACS; Milteny Biotec, Bergisch Gladbach, Germany) according to manufacturer’s protocols. CK19 is a well-known epithelial marker, whereas CD90 and CD133 are CSCs markers. Cells (≥ 30000/sample) were acquired after flow cytometry and counted using the Cell Quest software. The number of CD45-/CK19+ cells was considered as the number of CTCs, whereas the number of CD45-/CK19+/CD133+ and CD45-/CD90+ cells determined as the number of CSCs. Three successive readings were recorded for each sample and the mean was calculated and expressed as the number of CTCs or CSCs/7.5 mL of blood. A sample of normal lymphocytes was included in each run as a negative control. A cut-off of 4 ± 1 CTCs/7.5 mL was chosen to define the test as positive[16].
RNA was extracted from the separated peripheral blood mononuclear cells and from the tissues using the RNeasy kit (Qiagen). The extracted RNA (3-4 μg) was reverse transcribed using the GeneAmp Gold RNA PCR Reagent Kit (Applied Biosystems). Nested PCR was performed to detect MAGE1 and MAGE3 expression with HepG2 and huh7 cell lines as a positive control. Quantitative real-time PCR (qRT-PCR) was used to evaluate MAGE1/3, CK19 and CD133 expression (Table 1) using SYBR Green[17,18]. The expression of markers was normalized to β-actin and expressed as relative expression units for CD133 and CK or as absolute quantification for MAGE1 and MAGE3. Correlative cycle threshold values were recorded and a standard curve was drawn. A gene expression difference was considered to be valid if the trend of change of a gene measured by qRT-PCR agreed with that determined by RT-PCR. Real-time PCR assays were carried out in duplicate for each sample and mean values were used for the calculation of the mRNA levels[19].
Statistical analyses were performed using SPSS version 15 software (SPSS Inc., Chicago, IL, United States). mean ± SD were computed for the quantitative data. Data means were compared using a non-parametric t-test (Mann-Whitney test) or ANOVA (Kruskal-Wallis test). χ2 analysis was used to compare qualitative data. Variables were cross-tabulated in all possible combinations against each other. The receiver operating characteristic curve was constructed by calculating true and false positive fractions of the marker at several cutoff points. The best cutoff value, which differentiates between normal and diseased cases or between benign and malignant cases, was calculated as the value that maximizes the sum of sensitivity and specificity at which the highest predictive values were reached[20].
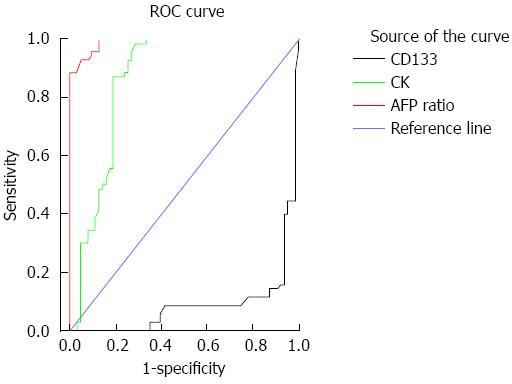
The serum levels of AFP, alkaline phosphatase and aspartate aminotransferase were significantly higher in HCC compared to CH and control groups (Ps < 0.05) (Table 2). Alanine aminotransferase levels were significantly higher in HCC and CH patients compared to the controls, whereas serum creatinine was significantly higher only in HCC patients (Ps < 0.05). The sensitivity and specificity of the AFP ratio are 95.7 and 90.5%; respectively, with an area under the curve of 0.99 (standard error = 0.005, 95%CI: 0.98-1.00, cut off point = 19.2) (Figure 1).
| HCC (n = 70) | CH (n = 30) | Control (n = 33) | ||
| Age, mean (range) | 57.32 (20-74)a | 47.70 (43-60)a | 51.09 (31-75) | |
| Gender (male/female) | 67/3 | 21/9 | 33/0 | |
| AFP | Mean (range) | 9547.54 (15-117032)ac | 11.02 (1-30)a | 7.45 (4-10) |
| Median | 1481.5ac | 8.9a | 7.9 | |
| Total bilirubin | Mean (range) | 2.66 (0.8-8.6)a | 4.73 (0.6-114.0) | 0.60 (0.3-1.0) |
| Median | 2.0a | 1 | 0.6 | |
| ALT | Mean (range) | 71.39 (17-189)a | 54.23 (10-200)a | 24.91 (16-37) |
| Median | 57.5a | 34.5a | 25 | |
| AST | Mean (range) | 122.94 (35-530)ac | 54.47 (12-140)a | 28.45 (17-39) |
| Median | 88.5ac | 39.5a | 28 | |
| Creatinine | Mean (range) | 1.09 (0.4-2.0)ac | 0.84 (0.5-1.1) | 0.81 (0.4-1.4) |
| Median | 1.0ac | 0.9 | 0.8 | |
| Alkaline phosphatase | Mean (range) | 240.50 (76-580)ac | 138.90 (80-220)a | 44.39 (15-71) |
| Median | 218.5ac | 132.5a | 44 | |
The median numbers of CTCs and CSCs were significantly different in HCC patients compared to the CH and control groups (P < 0.001). CK19+ cells were significantly higher in HCC and CH patients compared to the controls (P≤ 0.001), however there was no significant difference between HCC and CH groups. The number of CD90+ cells was significantly higher in HCC and CH patients compared to the controls (P≤ 0.01) with a significant difference between the HCC and CH groups (P≤ 0.01). CD133 was significantly higher in CH patients compared to HCC and control groups (P≤ 0.001) and it was also higher in HCC patients compared to controls (P≤ 0.001) (Figures 2 and 3; Table 3).
We also assessed the differences among the groups using various CK19 and CD133 cutoffs. The positivity rate (number of positive cases ≥ cutoff) for CK19 (73%) was significantly higher in HCC compared to the other two groups (Ps < 0.001), whereas CD133 (69.5%) was significantly lower in HCC than in CH (P < 0.001), but not controls. CD90 (49.8%) was significantly higher in HCC compared to the other two groups (Ps < 0.001). Therefore at the specific cutoffs, CK19 and CD90 in the blood of HCV-infected patients should be considered good markers for detection of HCC in CH patients with high sensitivity (87.1% and 82.5%, respectively) and specificity (81.0% and 89.6%, respectively), whereas CD133 had only 40.0% sensitivity and 6.3% specificity (Figure 1).
Telomerase expression was detected in 39/70 (55.71%) of the HCC patients, but in none of the CH patients or controls (P < 0.001) (Table 4). Similarly, MAGE1 and MAGE3 were expressed in 60% (42/70) and 62.9% (44/70) of HCC cases, respectively, but were not detectable in CH or control samples (P < 0.001).
| Gene | HCC (n = 70) | CH (n = 30) | Control (n = 33) | P value | χ2 | |
| Telomerase | +++ | 26 (37.14) | 0 (0.00) | 0 (0.00) | < 0.001 | 49.663 |
| ++ | 13 (18.57) | 0 (0.00) | 0 (0.00) | |||
| - | 31 (44.29) | 30 (100) | 33 (100) | |||
| MAGE1 | +++ | 9 (12.86) | 0 (0.00) | 0 (0.00) | < 0.001 | 55.246 |
| ++ | 33 (47.14) | 0 (0.00) | 0 (0.00) | |||
| - | 28 (40.00) | 30 (100) | 33 (100) | |||
| MAGE3 | +++ | 24 (34.29) | 0 (0.00) | 0 (0.00) | < 0.001 | 55.246 |
| ++ | 20 (28.57) | 0 (0.00) | 0 (0.00) | |||
| - | 26 (37.14) | 30 (100) | 33 (100) | |||
In the HCC group, advanced disease stage and high tumor grade were significantly associated with the expression levels of telomerase, MAGE3, CD133, CK19 and CD90, (Ps < 0.05), but not MAGE1 (Table 5).
| Gene | Stage | Grade | |||||
| Early (n = 18) | Late (n = 52) | P value | Low (n = 43) | High (n = 27) | P value | ||
| Telomerase | +++ | 2 (11.11) | 24 (46.15) | 0.019 | 8 (18.60) | 18 (66.67) | < 0.001 |
| ++ | 6 (33.33) | 7 (13.46) | 11 (25.58) | 2 (7.41) | |||
| - | 10 (55.56) | 21 (40.37) | 24 (55.81) | 7 (25.93) | |||
| MAGE1 | +++ | 0 (0.00) | 9 (17.31) | 0.099 | 3 (6.98) | 6 (22.22) | 0.123 |
| ++ | 8 (44.44) | 25 (48.08) | 20 (46.51) | 13 (48.15) | |||
| - | 10 (55.56) | 18 (34.62) | 20 (46.51) | 8 (29.63) | |||
| MAGE3 | +++ | 2 (11.11) | 22 (42.31) | < 0.001 | 6 (13.95) | 18 (66.67) | < 0.001 |
| ++ | 13 (72.22) | 7 (13.46) | 19 (44.19) | 1 (3.70) | |||
| - | 3 (16.67) | 23 (44.23) | 18 (41.86) | 8 (29.63) | |||
| CD133 | + | 0 (0.00) | 28 (53.85) | < 0.001 | 9 (20.93) | 19 (70.37) | < 0.001 |
| - | 18 (100.00) | 24 (46.15) | 34 (79.07) | 8 (29.63) | |||
| CK19 | + | 10 (55.56) | 52 (100.00) | < 0.001 | 35 (81.40) | 27 (100.00) | < 0.017 |
| - | 8 (44.44) | 0 (0.00) | 8 (18.60) | 0 (0.00) | |||
| CD90 | + | 6 (33.30) | 31 (59.60) | 0.015 | 9 (20.93) | 19 (70.37) | < 0.001 |
| - | 12 (66.70) | 11 (21.15) | 34 (79.07) | 8 (29.63) | |||
HCC is one of the most aggressive tumors that usually presents in a late stage with limited therapeutic options[21]. Some recent studies mentioned that the aggressive behavior of HCC could be partially attributed to the presence of malignant hepatocytes that gained entry into circulation, either before or during surgery. Consequently, they recommended the use of neo-adjuvant chemotherapy. Therefore, identification of accurate techniques for detection of these small populations of cells in patients’ blood together with the search for sensitive biological biomarkers are highly recommended for better patient management[22].
In the present study, we validated the utility of flow cytometry for cell immuno-phenotyping as a rapid and highly sensitive technique for the follow-up of HCV-infected patients at different disease stages. This was achieved through detecting the interaction of CK19 antibody with its antigen, which is present in the cytoplasm of hepatocytes after negative selection of the non-epithelial (CD45) cells. The number of CSCs was then determined using the liver CSC markers, CD90 and CD133. The possible prognostic and predictive values of CTC and CSC markers in monitoring HCV-infected patients was assessed by comparing their expression with standard prognostic factors, and their utility for early detection of HCC was also evaluated.
Our data show that flow cytometry supplemented with qRT-PCR was able to identify a significantly higher number of CTCs (CD45-/CK19+) and CSCs (CD45-/CK19+ or CD45-/CK19+/CD90+) in the blood of HCC patients compared to CH and control groups with 96% concordance between the two techniques. This confirms the utility of flow cytometry in enumerating CTCs, and thus it can be used to monitor CH patients for early detection of HCC, as it is sensitive and easy, relatively less expensive, and more rapid compared to the currently used techniques such as PCR or Cell Search.
The data also show that the number of CD45-/CK19+ and CD45-/CD90+ cells were significantly higher in HCC patients than in the other two groups, whereas CD133+ cells was significantly higher in CH patients. One possible explanation for this finding is that CD133+ cells also represent a subset of (normal) stem cells that are released from the bone marrow into circulation during the early inflammatory stage of HCV-associated liver disease to repair hepatic damage, compensate for the cell loss and prevent or eliminate fibrosis. However, with failure to clear viral infection and/or repair the damage, the CD133+ cells, having the plasticity and the ability of unlimited proliferation, will set the stage for the development of HCC on top of the chronically inflamed, and possibly cirrhotic, liver.
The first evidence for the existence of a CSC population in the liver was reported by Haraguchi et al[23]. Subsequent studies identified liver CSC markers such as CD133, CD90, epithelial cell adhesion molecule, and CD44[12]. CD133 was detected in HCC cell lines and human tissues, suggesting a stem cell origin for HCC. In addition, CD133 expression in HCC samples was associated with poor prognosis[22]. This is in accordance with our results, as we were able to detect CD133+ cells by two methods in the blood and tissues of HCC and, to a lesser extent, CH patients. However, CD133 expression may contribute to a poor prognosis in HCC patients, as it was significantly associated with large tumor size and advanced disease stage, which indicates that it is a good prognostic marker for HCC.
Similarly, CD90 was highly expressed in the HCC cases compared to CH or control groups. It was significantly associated with an aggressive tumor phenotype, suggesting a prognostic and predictive value. This is in accordance with some previous studies on HCC cell lines and human samples, where CD90 was highly expressed in malignant hepatocytes and the presence of CD90+/CD44+ cells contributed to an aggressive phenotype with more frequent metastatic lesions in the lung[24].
In an attempt to identify sensitive diagnostic marker(s) that can help to differentiate between CH and HCC in HCV-infected patients and thus permit early detection of HCC, we calculated the best cutoffs for CK19, CD90 and CD133 and we found that, in addition to high serum AFP levels, CK19+ (≥ 73) and/or CD90+ (≥ 49.8) cells in patients’ blood, as measured by flow cytometry, can differentiate between the two groups. This provides evidence that both CK19 and CD90 could be used as biomarkers to predict HCC in CH patients. Moreover, CK19 and CD90 above the determined cutoffs, were also associated with an aggressive tumor phenotype and advanced stage, demonstrating their prognostic capability.
Some previous studies have also shown that CK19 can predict HCC with high sensitivity (87%) and specificity (100%), and can thus be used as a prognostic factor which is associated with increased metastatic potential and early recurrence[25]. In addition, HCC animal models have shown that 10/10000 CTCs can initiate new metastatic deposits and even after curative resection, thus the tumor recurrence rates remain high[26]. An interesting finding in some of these studies was the expression of AFP mRNA in the isolated CTCs.
Currently, AFP is the generally accepted serologic marker for liver diseases. In our study, AFP was significantly higher in HCC patients compared to CH and control groups. An elevated AFP (> 400 ng/mL) level was associated with advanced disease stage in HCC patients. Some previous studies showed that AFP has a low specificity for diagnosing HCC to the extent that the American Association for the Study of Liver Diseases-Practice Guidelines Committee has recently recommended ultrasound examination alone (without AFP) to be used for HCC surveillance[27]. However, the interpretation of ultrasound is operator dependent and can be difficult in persons who are obese or have underlying cirrhosis. Therefore, other reliable biomarkers are required to complement ultrasound and AFP for proper diagnosis and early detection of HCC[28].
Members of the MAGE family were expressed in tumors of different histologic types but not in normal tissues, providing attractive targets for cancer immunotherapy[29]. MAGE1 and MAGE3 transcripts were detected in cultured HCC cells but not in normal hepatocytes, and thus they were considered tumor-specific markers[30]. Further studies showed that MAGE-1 and MAGE-3 transcripts were present in 46%-80% and 42%-68%, respectively, of HCC tissue samples, but not in the surrounding non-cancerous tissues, CH, cirrhotic or normal tissues[29,31]. This confirms the sensitivity of MAGE1/MAGE3 as specific markers for the diagnosis and early detection of HCC in CH patients. Our results provide additional evidence to the literature as MAGE1 and MAGE3 were detected in a majority of HCC patients, but not of those with CH or controls.
Our data also show that telomerase was detected in 42.3% of samples from HCC patients, compared to only 10% of CH patients and none of the controls. This could be attributed to the presence of free plasma DNA, or CTCs derived from the original tumor that have the same genetic aberrations, or to an aggressive clone that escaped into the circulation[32]. A previous study by Miura et al[33] demonstrated the presence of telomerase mRNA in CTCs and sera of HCC patients, and another study illustrated that shortened telomeres induce chromosomal instability in hepatocytes, which is an early important event in hepatocarcinogenesis that could be detected in the preneoplastic lesions of the dysplastic nodules[32].
We conclude that CTCs and CSCs play important roles in the development and progression of HCV-associated HCC. Enumeration of CTCs by flow cytometry using CK and CD90 has high sensitivity and specificity and is likely clinically useful in improving prognostic accuracy and monitoring therapeutic outcomes of HCV-infected patients. In addition, aberrant expression of HCC-specific and CSC markers (CD90, MAGE3, telomerase, CD133 and CK19) contributes to poor prognosis in HCC patients and should be assessed to provide better management of those patients. However, further studies are still needed to confirm the utility of these biomarkers in personalized medicine and targeted therapy as well as to clarify the possibility of using CD90 as a marker for early detection of HCC in HCV-infected patients, as it increased significantly with disease progression from CH to HCC.
Hepatitis C virus (HCV)-genotype 4-associated chronic hepatitis and hepatocellular carcinoma (HCC) are prevalent in Egypt. Recent studies demonstrate that cancer stem cells (CSCs) contribute to the development and progression of tumors, whereas circulating tumor cells (CTCs) predict patient outcome. Although various methods for enumeration of CTCs have been described, the use of flow cytometry for detection in HCC has not been examined.
Flow cytometry provides a rapid, sensitive and easy technique for enumeration of CTCs and CSCs using specific markers such as CK, CD133, CD90, melanoma antigen encoding gene 1 (MAGE1) and MAGE3, especially when supported by quantitative polymerase chain reaction. This method can assist in the follow-up of chronic-HCV patients for the early detection of HCC. In addition, telomerase, MAGE3 and CSC markers (CD90, CD133, CK19) could be used as prognostic and predictive markers for HCC.
Hepatocarcinogenesis can be better understood by examining the roles of CSCs and CTCs in HCV-associated chronic hepatitis and HCC. Therefore this study may represent an advancement in such knowledge by identifying accurate biomarkers, as well as rapid and sensitive techniques for detection of these markers. These findings may promote better patient management.
This study indicated that the expressions of telomerase, CD90, CD133, MAGE3, and CK19 were significantly associated with high-grade and advanced stage in HCC patients. It concluded that telomerase, CD90, CD133, MAGE3, and CK19 are prognostic markers in HCC patients.
P- Reviewer: Sutti S, Wang GY, Watashi K S- Editor: Ding Y L- Editor: AmEditor E- Editor: Wang CH
| 1. | Hussain K, El-Serag HB. Epidemiology, screening, diagnosis and treatment of hepatocellular carcinoma. Minerva Gastroenterol Dietol. 2009;55:123-138. [PubMed] |
| 2. | Idilman R, De Maria N, Colantoni A, Van Thiel DH. Pathogenesis of hepatitis B and C-induced hepatocellular carcinoma. J Viral Hepat. 1998;5:285-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 83] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 3. | AbdulQawi K, Youssef A, Metwally MA, Ragih I, AbdulHamid M, Shaheen A. Prospective study of prevalence and risk factors for hepatitis C in pregnant Egyptian women and its transmission to their infants. Croat Med J. 2010;51:219-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 4. | McMahon BJ. The natural history of chronic hepatitis B virus infection. Hepatology. 2009;49:S45-S55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 519] [Cited by in RCA: 557] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 5. | Mocellin S, Hoon D, Ambrosi A, Nitti D, Rossi CR. The prognostic value of circulating tumor cells in patients with melanoma: a systematic review and meta-analysis. Clin Cancer Res. 2006;12:4605-4613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 6. | Strijbos MH, Gratama JW, Kraan J, Lamers CH, den Bakker MA, Sleijfer S. Circulating endothelial cells in oncology: pitfalls and promises. Br J Cancer. 2008;98:1731-1735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 74] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 7. | Chiappini F. Circulating tumor cells measurements in hepatocellular carcinoma. Int J Hepatol. 2012;2012:684802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 8. | Wu W, Yao DF, Qiu LW, Wu XH, Yao M, Su XQ, Zou L. Abnormal expression of hepatomas and circulating telomerase and its clinical values. Hepatobiliary Pancreat Dis Int. 2005;4:544-549. [PubMed] |
| 9. | Phatak P, Burger AM. Telomerase and its potential for therapeutic intervention. Br J Pharmacol. 2007;152:1003-1011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 63] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 10. | Caballero OL, Zhao Q, Rimoldi D, Stevenson BJ, Svobodová S, Devalle S, Röhrig UF, Pagotto A, Michielin O, Speiser D. Frequent MAGE mutations in human melanoma. PLoS One. 2010;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 11. | Rountree CB, Ding W, He L, Stiles B. Expansion of CD133-expressing liver cancer stem cells in liver-specific phosphatase and tensin homolog deleted on chromosome 10-deleted mice. Stem Cells. 2009;27:290-299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 84] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | Yoon SK. The biology of cancer stem cells and its clinical implication in hepatocellular carcinoma. Gut Liver. 2012;6:29-40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 13. | Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL. AJCC Cancer Staging Manual. 7th ed. New York, NY: Springer 2010; 299-323. |
| 14. | Kamath PS, Wiesner RH, Malinchoc M, Kremers W, Therneau TM, Kosberg CL, D’Amico G, Dickson ER, Kim WR. A model to predict survival in patients with end-stage liver disease. Hepatology. 2001;33:464-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3462] [Cited by in RCA: 3679] [Article Influence: 153.3] [Reference Citation Analysis (0)] |
| 15. | Kazemi-Shirazi L, Petermann D, Müller C. Hepatitis B virus DNA in sera and liver tissue of HBsAg negative patients with chronic hepatitis C. J Hepatol. 2000;33:785-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 67] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 16. | Komeda T, Fukuda Y, Sando T, Kita R, Furukawa M, Nishida N, Amenomori M, Nakao K. Sensitive detection of circulating hepatocellular carcinoma cells in peripheral venous blood. Cancer. 1995;75:2214-2219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 17. | Leelawat K, Narong S, Udomchaiprasertkul W, Wannaprasert J, Treepongkaruna SA, Subwongcharoen S, Ratanashu-ek T. Prognostic relevance of circulating CK19 mRNA in advanced malignant biliary tract diseases. World J Gastroenterol. 2012;18:175-181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 18. | Mehra N, Penning M, Maas J, Beerepoot LV, van Daal N, van Gils CH, Giles RH, Voest EE. Progenitor marker CD133 mRNA is elevated in peripheral blood of cancer patients with bone metastases. Clin Cancer Res. 2006;12:4859-4866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 66] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 19. | Hosgood HD, Baris D, Zhang Y, Zhu Y, Zheng T, Yeager M, Welch R, Zahm S, Chanock S, Rothman N. Caspase polymorphisms and genetic susceptibility to multiple myeloma. Hematol Oncol. 2008;26:148-151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 36] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 20. | Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39:561-577. [PubMed] |
| 21. | el-Zayadi AR, Badran HM, Barakat EM, Attia Mel-D, Shawky S, Mohamed MK, Selim O, Saeid A. Hepatocellular carcinoma in Egypt: a single center study over a decade. World J Gastroenterol. 2005;11:5193-5198. [PubMed] |
| 22. | Yeh CT, Kuo CJ, Lai MW, Chen TC, Lin CY, Yeh TS, Lee WC. CD133-positive hepatocellular carcinoma in an area endemic for hepatitis B virus infection. BMC Cancer. 2009;9:324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 23. | Haraguchi N, Inoue H, Tanaka F, Mimori K, Utsunomiya T, Sasaki A, Mori M. Cancer stem cells in human gastrointestinal cancers. Hum Cell. 2006;19:24-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 89] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 24. | Zhu F, Zheng CJ, Han LY, Xie B, Jia J, Liu X, Tammi MT, Yang SY, Wei YQ, Chen YZ. Trends in the exploration of anticancer targets and strategies in enhancing the efficacy of drug targeting. Curr Mol Pharmacol. 2008;1:213-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 25. | Attallah AM, El-Far M, Abdel Malak CA, Zahran F, Farid K, Omran MM, Zagloul H, El-Deen MS. Evaluation of cytokeratin-1 in the diagnosis of hepatocellular carcinoma. Clin Chim Acta. 2011;412:2310-2315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 26. | Hermanek P, Hutter RV, Sobin LH, Wittekind C. International Union Against Cancer. Classification of isolated tumor cells and micrometastasis. Cancer. 1999;86:2668-2673. [PubMed] |
| 27. | Masuda T, Miyoshi E. Cancer biomarkers for hepatocellular carcinomas: from traditional markers to recent topics. Clin Chem Lab Med. 2011;49:959-966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 28. | Lok AS, Everhart JE, Wright EC, Di Bisceglie AM, Kim HY, Sterling RK, Everson GT, Lindsay KL, Lee WM, Bonkovsky HL. Maintenance peginterferon therapy and other factors associated with hepatocellular carcinoma in patients with advanced hepatitis C. Gastroenterology. 2011;140:840-849; quiz e12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 158] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 29. | Zerbini A, Pilli M, Soliani P, Ziegler S, Pelosi G, Orlandini A, Cavallo C, Uggeri J, Scandroglio R, Crafa P. Ex vivo characterization of tumor-derived melanoma antigen encoding gene-specific CD8+cells in patients with hepatocellular carcinoma. J Hepatol. 2004;40:102-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 59] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 30. | Salwa HT, Sara HAA, Noha AEl, Manal AH, Amany MM, Mourad MH. Multiple molecular markers MAGE-1, MAGE-3 and AFP mRNAs expression nested PCR assay for sensitive and specific detection of circulating hepatoma cells: Enhanced detection of hepatocellular carcinoma. Egypt J Med Human Genet. 2013;14:21-28. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 31. | Mou DC, Cai SL, Peng JR, Wang Y, Chen HS, Pang XW, Leng XS, Chen WF. Evaluation of MAGE-1 and MAGE-3 as tumour-specific markers to detect blood dissemination of hepatocellular carcinoma cells. Br J Cancer. 2002;86:110-116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 32. | Plentz RR, Caselitz M, Bleck JS, Gebel M, Flemming P, Kubicka S, Manns MP, Rudolph KL. Hepatocellular telomere shortening correlates with chromosomal instability and the development of human hepatoma. Hepatology. 2004;40:80-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 97] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 33. | Miura T, Suzuki N, Nakamura J, Yamada S, Miura T, Yanagi M, Usuda H, Emura I, Takahashi T. Hepatocellular carcinoma, with portal thrombus after viral eradication, disappeared by 5-fluorouracil and interferon. World J Hepatol. 2010;2:416-418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |