Published online Oct 14, 2014. doi: 10.3748/wjg.v20.i38.13658
Revised: April 29, 2014
Accepted: May 26, 2014
Published online: October 14, 2014
Processing time: 226 Days and 20.1 Hours
Gastric cancer (GC) is the fourth most common cancer worldwide and ranks second in global cancer mortality statistics. Perioperative chemotherapy plays an important role in the management and treatment of advanced stage disease. However, response to chemotherapy varies widely, with some patients presenting no or only minor response to treatment. Hence, chemotherapy resistance is a major clinical problem that impacts on outcome. Unfortunately, to date there are no reliable biomarkers available that predict response to chemotherapy before the start of the treatment, or that allow modification of chemotherapy resistance. MicroRNAs (miRNAs) could provide an answer to this problem. miRNAs are involved in the initiation and progression of a variety of cancer types, and there is evidence that miRNAs impact on resistance towards chemotherapeutic drugs as well. This current review aims to provide an overview about the potential clinical applicability of miRNAs as biomarkers for chemoresistance in GC. The authors focus in this context on the potential of miRNAs to predict sensitivity towards different chemotherapeutics, and on the potential of miRNAs to modulate sensitivity and resistance towards chemotherapy in GC.
Core tip: MicroRNAs (miRNAs) are a relatively new class of gene expression regulators, being involved in cancer initiation and progression. There is evidence that miRNAs impact on resistance towards various chemotherapeutics and are accessible and detectable in different tissue types, including blood samples, with great stability. Taken together, miRNAs seem to have great potential as new biomarkers for diagnostic and therapeutic approaches. This current review aims to provide an overview about the potential clinical applicability of miRNAs as biomarkers for chemoresistance in gastric cancer (GC), focusing on prediction and modulation of sensitivity and resistance towards chemotherapeutic drugs in GC.
- Citation: Matuszcak C, Haier J, Hummel R, Lindner K. MicroRNAs: Promising chemoresistance biomarkers in gastric cancer with diagnostic and therapeutic potential. World J Gastroenterol 2014; 20(38): 13658-13666
- URL: https://www.wjgnet.com/1007-9327/full/v20/i38/13658.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i38.13658
Gastric cancer (GC) is the fourth most common cancer worldwide and ranks second in global cancer mortality statistics[1]. The prognosis of GC is poor with an overall 5-year survival rate of less than 35%[2,3], mostly caused by locally advanced disease at the date of initial diagnosis, reduced response to neoadjuvant or adjuvant therapy or tumor recurrence after surgical resection. Surgery is the preferred primary treatment option for the early stage of GC. Unfortunately, over 60% of the patients present advanced stage disease at the time point of diagnosis. In these cases, perioperative chemotherapy is widely applied based on the data of the MAGIC[4] and the ACCORD trials[5], including for example platinum- and fluoropyrimidine-based regimen or three-drug protocols with additional application of taxanes or anthracyclines[4,6,7]. These therapies are recommended nowadays even for patients with uT2 tumors[8,9]. However, response of the individual patient to this perioperative treatment varies widely, and some tumors are resistant to chemotherapeutic treatment completely. Hence, one main obstacle in the success of perioperative chemotherapy is the development of multidrug resistance (MDR) in GC[10]. Based on the current literature, there are four major mechanisms that contribute to drug resistance in cancer cells: (1) decreased uptake of water soluble drugs[10]; (2) changes in intracellular pathways that affect the potential of cytotoxic drugs to kill cells, including alterations in the cell cycle, DNA repair, apoptosis pathways, metabolism/elimination of drugs, or others[10-12]; (3) increased energy-dependent efflux of hydrophobic drugs mediated via overexpression of a family of energy-dependent transporters (known as ATP-binding cassette transporters) such as P-glycoprotein 1 (P-gp, ABCB1) or breast cancer resistance protein (ABCG2) amongst others[10]; and (4) intracellular detoxifiers such as antioxidants (e.g., glutathione)[13,14]. Unfortunately, from a clinical point of view, there are so far no reliable biomarkers available that allow a prediction of response to chemotherapy in the individual patient before the start of treatment[15]. This leads to the problem that a number of patients undergo futile treatment with potential severe side-effects but without any benefit. If response to chemotherapy can be predicted before treatment, therapy could be better tailored via identification of patients that in fact profit from chemotherapy.
In this context, microRNAs (miRNAs) could provide a new approach for better clinical decision making. miRNAs are a relatively novel class of regulatory molecules that control translation and stability of messenger RNA (mRNAs) on a post-transcriptional level via interaction with the 3’-untranslated region (UTRs) of target mRNAs, that finally leads to destabilization and/or inhibition of their translation[16]. So far, over 2500 human miRNAs have been identified according to the latest release (June 2013) of miRBase database[17], and each single miRNA can potentially target up to hundreds of mRNAs[18]. miRNAs are involved in the regulation of almost all physiological processes such as cell development, differentiation, proliferation and apoptosis[19,20]. But more importantly, miRNAs have been found to impact on pathogenesis of a variety of human cancers. In this context, miRNAs can be roughly categorized into miRNAs that promote tumor development and growth (so-called oncomiRs), and miRNAs that inhibit tumor progression (so-called tumor-suppressor-miRs)[21,22]. Regarding GC, several (in vitro and in vivo) studies demonstrated according to the findings in other cancer types that tumors or GC cell lines present aberrant miRNA expression pattern compared to controls (normal gastric cell lines or samples from healthy patients)[23-31]. In conclusion, this data highly suggests that miRNAs may have an enormous potential as diagnostic[32], prognostic[33-35], or even therapeutic biomarkers[36,37].
This current review article now aims to summarize the evidence available so far about the impact of miRNAs on MDR in GC. We aim to specifically highlight from a clinical point of view, that miRNAs might help to predict tumor response to conventional chemotherapy, thereby providing a new approach as diagnostic biomarkers in GC. Moreover, we intend to show that miRNAs might further provide an enormous potential as therapeutic tools in the battle against GC by modulating and/or reversing MDR.
The National Institutes of Health Biomarkers Definition Working Group defined a biomarker as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention”[38].
This definition of “a characteristic that is objectively measured and evaluated” implicates that molecular biomarkers which might be useful for prediction of the success of chemotherapy in cancer patients should (1) be easily detectable and assessable in clinical patient samples; and (2) be stably expressed and refractory to degradation in these samples in order to allow proper analysis on clinical samples. Only if these characteristics are present, miRNAs can further be assessed regarding their potential as biomarkers for response prediction.
And in fact, regarding the aspect of easily accessible clinical samples, there is a large body of evidence that shows that miRNAs are indeed detectable in a number of different sample types. For example, miRNAs can be found and analyzed in fresh frozen samples such as tumor biopsies or resection specimens, or in paraffin-embedded tissues[39]. Even more interesting from a clinical point of view, miRNAs can be detected as so-called “circulating miRNAs” in a broad variety of human body fluids including urine[23], saliva[39], amniotic fluid[39] and pleura fluid[40] in healthy volunteers and cancer patients[25,40-43]. These circulating miRNAs are considered believed to originate either from immunocytes[44], as byproducts of dead/dying cells[44-46], or to be secreted actively from tumor cells via microvesicles such as exosomes[44-47].
Regarding the stability of miRNA expression in clinical samples, Lu et al[48] reported that miRNAs are quite stable in different tissue samples (paraffin-embedded sections, rapid frozen samples) and present unique expression levels in different tissue types (salvia, blood, urine). Moreover, circulating miRNAs have been shown to be highly stable in the peripheral circulation caused for example by binding to RNA-binding proteins[18,49-51] or by their excretion in microvesicles[51-54]. In addition, Fang et al[41] reported that miRNAs present tissue-specific expression patterns with a reproducible and consistent expression. Finally, studies demonstrated that circulating miRNAs are fairly resistant to different pH solutions or repeated freeze-thawing cycles, and they are stable for 24 h at room temperature[55,56].
Taken together, miRNA expression patterns can successfully be assessed and analyzed in a variety of different samples, including biopsy or resection specimen, or blood samples, what supports the hypothesis that miRNAs could be used as clinically relevant diagnostic or therapeutic molecular biomarkers.
Another aspect of using miRNAs as potential biomarkers for chemoresistance in GC includes their possible impact on MDR in general. Only if there are data available which support that miRNAs modulate drug resistance in general, these molecules might be useful for evaluation of response prediction in GC patients.
And in fact, miRNAs have been proposed to play an important role in the development of MDR in cancer progression[55-58]. Despite the fact that this field of research is still in its infancy, it has been demonstrated that miRNAs are highly related to cancer progression (including growth apoptosis, invasion and metastasis) and are responsible for cancer-related inflammation, anticancer drug resistance and regulation of cancer stem cells[59]. Most interestingly, a number of miRNAs (such as members of the let-7 family, miR-16, miR-21 or miR-451 amongst others) have been confirmed to impact on more than one anticancer drug and/or to play a role in chemotherapy resistance in more than just one tumor type[57,60-62]. In addition, several anti-cancer-drugs target the same cancer-related genes that are targeted and regulated by miRNAs, implicating a link between chemotherapy induced cancer cell death and a modulation of chemotherapy resistance via miRNAs[63-65]. However, it has to be noted in this respect that the regulation of known cancer-related genes by miRNAs is much more frequently reported simply because these genes are well studied, and not all miRNAs show similar sensitivity phenotypes in different cancer types. Finally, a number of transcripts from drug resistance-related genes may be targeted by more than one miRNA[66].
Taken together, the evidence available so far on miRNA analysis in different sample types, their stability in various tissues, and their involvement in chemotherapy resistance in general highly supports the hypothesis that miRNAs might be perfect candidates for biomarkers for chemoresistance in GC in clinical settings.
As miRNA research is so far still mostly limited to in vitro experiments, its clinical applicability at this stage is somewhat limited, and there are a number of hurdles that need to be overcome before these molecules can be used in standard clinical settings.
With regards to sample acquisition and miRNA extraction for examples from blood samples, it has been demonstrated that several aspects and factors impact on results of miRNA analyses. These include the appropriate selection of samples (e.g., plasma or serum), collection tubes (EDTA, citrate, heparin), extraction methods (phenol/chloroform, silica: distinct differences in required fluid volume, yield, procedural contaminants etc.), quality and quantity control, fasting or blood draw timing, all potentially. In addition, there are a number of different miRNA profiling methods including qRT-PCR, microarrays, sequence specific hybridization in solution followed by miRNA molecule counting based on reporter probes, and direct sequencing available, all of which with more or less relevant advantages and disadvantages regarding sensitivity and specificity, absolute quantification/accuracy and flexibility and throughput[67]. In general, it can be postulated, that there is a lack of standardized of experimental techniques at this stage, and this impacts clearly on the possibility to compare data of different studies.
With regards to miRNAs as therapeutic tools, in vivo data on animal experiments are limited, and there are to our best knowledge so far only two clinical studies on a potential use of miRNAs as therapeutic tools in human beings available or on its way. Both studies do not refer to GC patients. In a phase 2a clinical trial, Janssen et al[68] evaluated the safety and efficacy of Miravirsen® (a locked nucleic acid-modified DNA phosphorothioate antisense oligonucleotide that sequesters mature miR-122 in a highly stable heteroduplex, thereby inhibiting its function) in 36 patients with chronic HCV genotype 1 infection. The authors could demonstrate prolonged dose-dependent reductions in HCV RNA levels without evidence of viral resistance and without dose-limiting adverse events and no escape mutations in the miR-122 binding sites of the HCV genome[68]. It is worth to mention that in two phase I safety studies in humans with Miravirsen® showed that Miravirsen® was well tolerated with no dose-limiting toxicities[69,70].
The second clinical trial (phase I trial), which is currently recruiting healthy patients as a control group, uses MRX34 (a liposome-formulated mimic of the tumor suppressor miR-34) in patients with inoperable primary liver cancer or metastatic cancer with liver involvement[71]. In this trail, the safety, pharmacokinetics and pharmacodynamics MRX34 is being evaluated in healthy patients[72].
Taken together, there are many hurdles that need to be overcome before miRNAs can be safely applied as clinical biomarkers or therapeutic tools. Standardization of technical approaches, confirmation of in vitro results in animal experiments and finally safety assessment and treatment trials in humans have to show whether these molecules find their way into the daily clinic. However, first results are promising indeed.
There are numbers of potential applications for miRNAs as clinically relevant biomarkers. In the context of this review, we focus on two distinct aspects: miRNAs as diagnostic biomarkers, and miRNAs as therapeutic biomarkers.
MiRNAs as diagnostic biomarkers have to meet various requirements, some of which have been discussed in detail in the section above. However, the main requirement of a diagnostic biomarker in cancer is to distinguish between different “subgroups” of patients. For example, patients with cancer should reliably be distinguishable from patients with precancerous conditions or from non-cancer controls[73-76]. In this context, a “simple comparison” of miRNA expression pattern between different “subgroups” of patients is needed in order to allocate patients into the respective “subgroups”, and at this stage, there is no need for further information about the exact impact of the respective miRNAs on tumor growth, metastasis, etc.
In contrast to this “simple analytic” approach, the use of miRNAs as therapeutic biomarkers mandates different key features: these miRNAs might not be differentially expressed between different subgroups of patients (such as responders and non-responders to chemotherapy). However, manipulation of the levels of these miRNAs should lead to changes in the cellular response of tumors to anticancer treatment, increasing thereby for example drug resistance. This means that for therapeutic biomarkers, there is a need for further knowledge about the exact mechanisms by which the respective miRNAs finally affect cancer cell behavior. The objective in the establishment of a therapeutic biomarker is to provide a more individualized therapy approach for every tumor type in the future[73-75].
The potential to predict response to chemotherapy treatment is a promising clinical application of miRNAs. Successful prediction of treatment response would allow a more tailored, individual therapy planning, as patients who do not respond to this treatment modality would not have to undergo futile treatment with potential severe side-effects. However, response prediction of chemoresistance implies that miRNAs exhibit significant and reproducible differences in expression between patients that respond well to chemotherapy compared to patients that show no or minimal response to this treatment option. With other words, miRNA expression pattern should differ between chemotherapy resistant GC tumors and sensitive tumors.
And in fact, some first authors reported results from in vitro experiments that showed differing expression pattern of several miRNAs in chemotherapy resistant GC cell lines when compared to sensitive controls (Table 1), with some miRNAs being upregulated in resistant cancer cells, and others being downregulated. For example, Wang et al[77] demonstrated an upregulation of miR-19a/b in a vincristine resistant GC cell line (SCG7901/VCR) and a doxorubicin resistant GC cell line (SGC7901/ADR). In addition, Yang et al[78] reported that miR-21 was upregulated in cisplatin resistant GC cells (SCG7901/DDP) vs controls. Furthermore, miR-106a was shown to be upregulated in a cisplatin resistant GC cell line (SCG7901/DDP)[79]. Finally, miR-195 and miR-378 were found to be upregulated in 5-azazytidine resistant GC cell lines (MGC803, SGC7901 and AGS)[80].
| miRNAs | Regulation | Drug resistance cell lines | Target | Ref. |
| miR-15b | Down ↓ | Vincristine | PTEN | [62] |
| miR-16 | Down ↓ | Vincristine | PTEN | [62] |
| miR-19a/b | Up ↑ | Doxorubicin, vincristine | PTEN | [77] |
| miR-21 | Up ↑ | Cisplatin | PTEN | [78] |
| miR-106a | Up ↑ | Cisplatin | PTEN | [79] |
| miR-181b | Down ↓ | Cisplatin, vincristine | BCL-2 | [82] |
| miR-497 | Down ↓ | Cisplatin, vincristine | BCL-2 | [83] |
| miR-200b/c | Down ↓ | Cisplatin, vincristine | BCL-2, XIAP | [83] |
| miR-508-5p | Down ↓ | Vincristine, doxorubicin | ABCB1, ZNRD1 | [81] |
Xia et al[62] on the other hand showed that miR-15b and miR-16 both were downregulated in a vincristine resistant GC cell line (SGC7901), and Shang et al[81] showed that miR-508-5p was downregulated in two GC cell lines resistant towards doxorubicin (SGC7901/ADR) and vincristine (SGC7901/VCR). Furthermore, Zhu et al[82] demonstrated in vincristine resistant GC cell lines miR-181b, miR-200b/c and miR-497 to be downregulated compared to controls[83].
Finally, Wu et al[84] analyzed miRNA expression patterns in hydroxycamptothecin (HCPT)-resistant and HCPT-sensitive GC cell lines. miR-224 and miR-338-3p were only expressed in HCPT-resistant cells, and miR-141, miR-200a, miR-200b, miR-372, and miR-373 were only expressed in HCPT-sensitive cells[84].
Even more interesting from a clinical point of view, there is some first evidence that alterations in miRNA expression pattern might be able to discriminate between responders and non-responders in clinical patient samples (in vivo). Kim et al[85] for example presented data based on biopsy samples from 90 GC patients which were collected prior to chemotherapy. The authors identified a signature of several miRNAs (namely miR-363, miR-518f, miR-519e, miR-520a, and miR-520d) that was correlated to resistance to cisplatin and 5-fluorouracil therapy[85]. With regards to blood based circulating miRNAs, the current literature draws a promising picture: Zhu et al[86] showed in a recent review including 22 studies with a sample size ranging from 37 to 164, that a total of 35 circulating miRNAs were differentially expressed between GC patients and healthy controls. Most interestingly, 6 of these miRNAs (miR-21[78], -27a[87], -106a[79], -195[80], -200c[83] and -378[80]) were shown to be deregulated in chemotherapy resistant GC cell lines as demonstrated in the in vitro experiments in the paragraph above, and to be potentially involved in MDR.
The application of miRNAs as modifiers of chemotherapy sensitivity is an even more interesting approach for a potential clinical use of these molecules, especially as additive treatment with conventional chemotherapeutics. This implicates that artificial manipulation of miRNA levels lead to increased sensitivity or reduced resistance towards various chemotherapeutic drugs. And again, there is growing evidence that modulation of miRNA expression in fact affects resistance of various tumors to chemotherapeutic treatment. However, as this field of research is still very young, results on GC are limited and refer so far only to in vitro experiments. In vivo studies on human patients with GC are not available yet.
Similarly to the data presented in the section about miRNAs with diagnostic potential, some miRNAs impact on sensitivity towards chemotherapy if their levels were artificially upregulated, others if their levels were downregulated. For example, upregulation of miR-21 or miR-106a was demonstrated to increase cisplatin resistance of GC cells[78], and Deng et al[80] showed that upregulation of miR-195 or miR-378 led to an enhanced 5-azacytidine resistance in normal gastric cells. Upregulation of miR-449 or miR-508-5p was demonstrated to positively impact on sensitivity towards cisplatin (miR-449) respectively vincristine or doxorubicin (miR-508-5p)[81,88]. Interestingly, in accordance with these findings about the modulation of sensitivity towards chemotherapeutic drugs via miRNAs, Bandres et al[89] reported that upregulation of miR-451 led to an increased sensitivity of cancer cells towards radiotherapy by down-regulating the macrophage migration inhibitory factor (MIF).
Only one research group reported on the effect of miRNA downregulation on chemotherapy resistance: Zhao et al[87] found that increased doxorubicin sensitivity in GC cells is connected with downregulation of miR-27a.
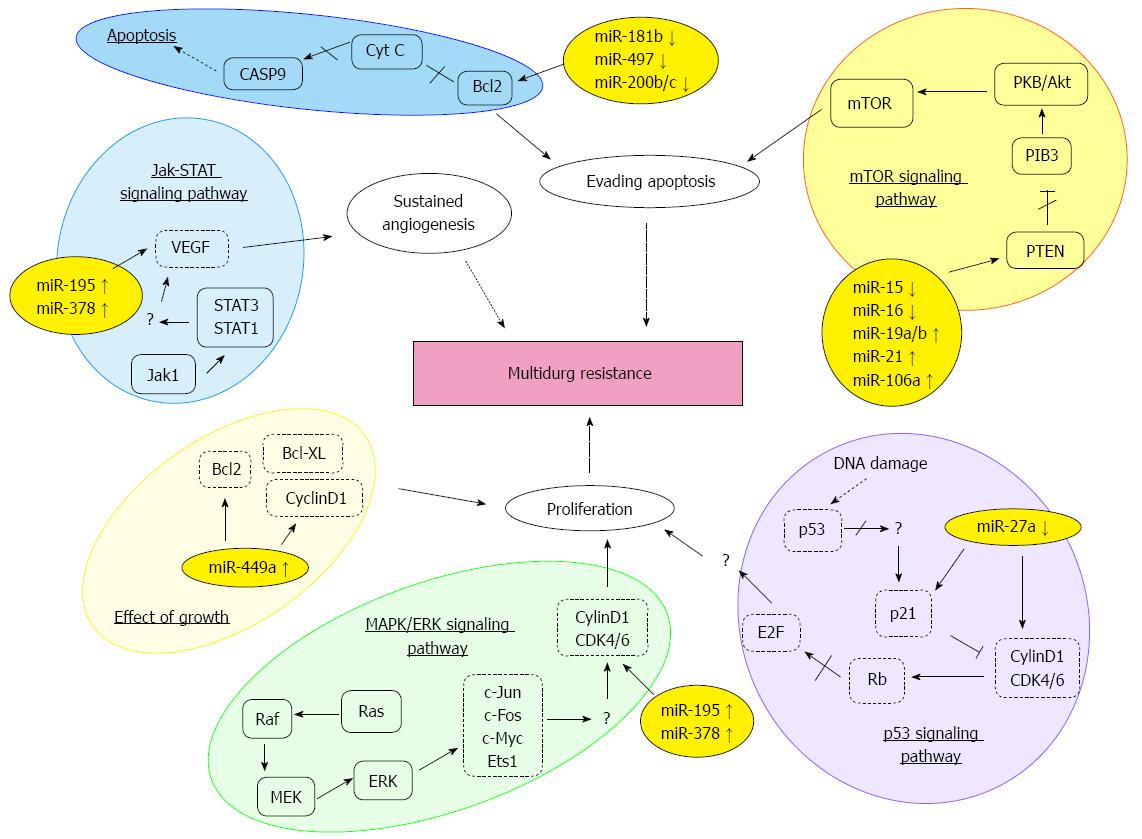
The authors of the aforementioned studies performed additional experiments in order to elucidate the underlying mechanisms by which the respective miRNAs finally impacted on sensitivity and resistance towards the different chemotherapeutic drugs. Table 2 and Figure 1 present an overview about the results of these investigations, and highlight the downstream targets and pathways that mediate the effects of manipulated miRNA expression in GC cells and their response to anticancer treatment.
| miRNAs | Regulation | Drug resistance | Target in GC | Drug treatment | Ref. |
| miR-21 | Up ↑ | Up ↑ | PTEN | Cisplatin | [78] |
| miR-27a | Down ↓ | Down ↓ | Cyclin-D1, p21 | Vincristine, 5-fluouracil, cisplatin, doxorubicin | [87] |
| miR-106a | Up ↑ | Up ↑ | PTEN | Cisplatin | [79] |
| miR-195 | Up ↑ | Up ↑ | CDK6, VEGF | 5-azacytidine | [80] |
| miR-378 | Up ↑ | Up ↑ | CDK6, VEGF | 5-azacytidine | [80] |
| miR-449a | Up ↑ | Down ↓ | BCL-2, CCDN1 | Cisplatin | [88] |
| miR-508-5p | Up ↑ | Down ↓ | ABCB1, ZNRD1 | Vincristine, doxorubicin | [81] |
MiRNAs are an astonishing new class of regulators of global gene expression, and they affect tumor initiation and progression in a large number of malignancies. Moreover, these molecules were shown to impact on sensitivity towards various chemotherapeutic agents in a variety of cancers. Based on the fact that miRNAs play an important role in the development and regulation of MDR, and in addition are accessible and detectable in different tissue types including blood samples with great stability, these molecules seems to have great potential as new biomarkers for diagnostic and therapeutic approaches. In the context of this current review, we presented an overview about the data available so far on this new aspect of miRNAs as biomarkers for chemoresistance in GC. Despite the fact that data on this topic is still somewhat limited, the first reports on miRNAs as potential clinical biomarkers that predict sensitivity towards chemotherapeutic drug are very promising. We found a number of studies that demonstrated various miRNAs to be indeed associated with chemoresistance by presenting different expression patterns between resistant and sensitive tumors, both in vitro and in vivo. Furthermore, several research groups could provide first evidence that manipulation of the expression of certain miRNAs lead to changes in the sensitivity and resistance of GC towards chemotherapeutics in vitro. Taken together, this data draw a very promising picture of miRNAs as potential new biomarkers for chemoresistance in GC with diagnostic and therapeutic potential. This would help to tailor cancer therapy more individually, and to specifically select patients for chemotherapy that would profit from this treatment option. Further research on this highly interesting and clinically most relevant aspect of a potential use of miRNAs is highly warranted.
P- Reviewer: Hann HW, Vynios D, Wiemer EAC S- Editor: Ding Y L- Editor: A E- Editor: Ma S
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25538] [Article Influence: 1824.1] [Reference Citation Analysis (7)] |
| 3. | Robb WB, Mariette C. Predicting the response to chemotherapy in gastric adenocarcinoma: who benefits from neoadjuvant chemotherapy? Recent Results Cancer Res. 2012;196:241-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 4. | Cunningham D, Allum WH, Stenning SP, Thompson JN, Van de Velde CJ, Nicolson M, Scarffe JH, Lofts FJ, Falk SJ, Iveson TJ. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N Engl J Med. 2006;355:11-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4899] [Cited by in RCA: 4600] [Article Influence: 242.1] [Reference Citation Analysis (0)] |
| 5. | Boige V, Pignon J, Saint-Aubert B, Lasser P, Conroy T, Bouch O, Segol P, Bedenne L, Rougier P, Ychou M. Final results of a randomized trial comparing preoperative 5-fluorouracil (F)/cisplatin (P) to surgery alone in adenocarcinoma of stomach and lower esophagus (ASLE): FNLCC ACCORD07-FFCD 9703 trial. J Clin Oncol. 2007;25 Suppl:4510. |
| 6. | Ychou M, Boige V, Pignon JP, Conroy T, Bouché O, Lebreton G, Ducourtieux M, Bedenne L, Fabre JM, Saint-Aubert B. Perioperative chemotherapy compared with surgery alone for resectable gastroesophageal adenocarcinoma: an FNCLCC and FFCD multicenter phase III trial. J Clin Oncol. 2011;29:1715-1721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1216] [Cited by in RCA: 1501] [Article Influence: 107.2] [Reference Citation Analysis (0)] |
| 7. | Kasper S, Schuler M. Targeted therapies in gastroesophageal cancer. Eur J Cancer. 2014;50:1247-1258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 8. | Oba K, Paoletti X, Alberts S, Bang YJ, Benedetti J, Bleiberg H, Catalano P, Lordick F, Michiels S, Morita S. Disease-free survival as a surrogate for overall survival in adjuvant trials of gastric cancer: a meta-analysis. J Natl Cancer Inst. 2013;105:1600-1607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 9. | Ronellenfitsch U, Schwarzbach M, Hofheinz R, Kienle P, Kieser M, Slanger TE, Jensen K. Perioperative chemo(radio)therapy versus primary surgery for resectable adenocarcinoma of the stomach, gastroesophageal junction, and lower esophagus. Cochrane Database Syst Rev. 2013;5:CD008107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 83] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 10. | Szakács G, Jakab K, Antal F, Sarkadi B. Diagnostics of multidrug resistance in cancer. Pathol Oncol Res. 1998;4:251-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 33] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 11. | Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1096] [Cited by in RCA: 1204] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 12. | Johnstone RW, Ruefli AA, Tainton KM, Smyth MJ. A role for P-glycoprotein in regulating cell death. Leuk Lymphoma. 2000;38:1-11. [PubMed] |
| 13. | Harfe BD. MicroRNAs in vertebrate development. Curr Opin Genet Dev. 2005;15:410-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 274] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 14. | Hwang W, Hwang Y, Lee S, Lee D. Rule-based multi-scale simulation for drug effect pathway analysis. BMC Med Inform Decis Mak. 2013;13 Suppl 1:S4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 15. | Pietrantonio F, De Braud F, Da Prat V, Perrone F, Pierotti MA, Gariboldi M, Fanetti G, Biondani P, Pellegrinelli A, Bossi I. A review on biomarkers for prediction of treatment outcome in gastric cancer. Anticancer Res. 2013;33:1257-1266. [PubMed] |
| 16. | David S, Meltzer SJ. MicroRNA involvement in esophageal carcinogenesis. Curr Opin Pharmacol. 2011;11:612-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 17. | Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152-D157. [PubMed] |
| 18. | Li L, Yang F, Wang X, Hu J, Yang L, Tang C, Wu Y, Miao K, Liu R, Shou T. Effect of 15-hydroxyprostaglandin dehydrogenase gene on the proliferation of gastric cancer cell murine forestomach carcinoma. Exp Ther Med. 2014;7:290-294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 19. | Barbarotto E, Schmittgen TD, Calin GA. MicroRNAs and cancer: profile, profile, profile. Int J Cancer. 2008;122:969-977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 136] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 20. | Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8847] [Cited by in RCA: 9282] [Article Influence: 464.1] [Reference Citation Analysis (0)] |
| 21. | Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5384] [Cited by in RCA: 5605] [Article Influence: 295.0] [Reference Citation Analysis (0)] |
| 22. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5705] [Cited by in RCA: 6029] [Article Influence: 317.3] [Reference Citation Analysis (0)] |
| 23. | Allegra A, Alonci A, Campo S, Penna G, Petrungaro A, Gerace D, Musolino C. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review). Int J Oncol. 2012;41:1897-1912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 269] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 24. | Guo LH, Li H, Wang F, Yu J, He JS. The Tumor Suppressor Roles of miR-433 and miR-127 in Gastric Cancer. Int J Mol Sci. 2013;14:14171-14184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 25. | Hou Z, Xie L, Yu L, Qian X, Liu B. MicroRNA-146a is down-regulated in gastric cancer and regulates cell proliferation and apoptosis. Med Oncol. 2012;29:886-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 26. | Song YX, Yue ZY, Wang ZN, Xu YY, Luo Y, Xu HM, Zhang X, Jiang L, Xing CZ, Zhang Y. MicroRNA-148b is frequently down-regulated in gastric cancer and acts as a tumor suppressor by inhibiting cell proliferation. Mol Cancer. 2011;10:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 195] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 27. | Peng W, Chen ZY, Wang L, Wang Z, Li J. MicroRNA-199a-3p is downregulated in gastric carcinomas and modulates cell proliferation. Genet Mol Res. 2013;12:3038-3047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 28. | Xiong X, Ren HZ, Li MH, Mei JH, Wen JF, Zheng CL. Down-regulated miRNA-214 induces a cell cycle G1 arrest in gastric cancer cells by up-regulating the PTEN protein. Pathol Oncol Res. 2011;17:931-937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 29. | Chen G, Shen ZL, Wang L, Lv CY, Huang XE, Zhou RP. Hsa-miR-181a-5p expression and effects on cell proliferation in gastric cancer. Asian Pac J Cancer Prev. 2013;14:3871-3875. [PubMed] |
| 30. | Xu XD, He XJ, Tao HQ, Zhang W, Wang YY, Ye ZY, Zhao ZS. Abnormal expression of miR-301a in gastric cancer associated with progression and poor prognosis. J Surg Oncol. 2013;108:197-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 31. | Zhang X, Li X, Tan Z, Liu X, Yang C, Ding X, Hu X, Zhou J, Xiang S, Zhou C. MicroRNA-373 is upregulated and targets TNFAIP1 in human gastric cancer, contributing to tumorigenesis. Oncol Lett. 2013;6:1427-1434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 32. | Wang P, Zhuang L, Zhang J, Fan J, Luo J, Chen H, Wang K, Liu L, Chen Z, Meng Z. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol Oncol. 2013;7:334-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 162] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 33. | Chistiakov DA, Chekhonin VP. Contribution of microRNAs to radio- and chemoresistance of brain tumors and their therapeutic potential. Eur J Pharmacol. 2012;684:8-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 34. | de la Chapelle A, Jazdzewski K. MicroRNAs in thyroid cancer. J Clin Endocrinol Metab. 2011;96:3326-3336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 103] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 35. | Marcucci G, Radmacher MD, Mrózek K, Bloomfield CD. MicroRNA expression in acute myeloid leukemia. Curr Hematol Malig Rep. 2009;4:83-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 36. | Millan MJ. MicroRNA in the regulation and expression of serotonergic transmission in the brain and other tissues. Curr Opin Pharmacol. 2011;11:11-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 37. | Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 722] [Cited by in RCA: 789] [Article Influence: 60.7] [Reference Citation Analysis (0)] |
| 38. | Biomarkers Definition Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89-95. |
| 39. | Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143-159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1316] [Cited by in RCA: 1260] [Article Influence: 96.9] [Reference Citation Analysis (0)] |
| 40. | Xiao YF, Yong X, Fan YH, Lü MH, Yang SM, Hu CJ. microRNA detection in feces, sputum, pleural effusion and urine: novel tools for cancer screening (Review). Oncol Rep. 2013;30:535-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 41. | Fang Y, Fang D, Hu J. MicroRNA and its roles in esophageal cancer. Med Sci Monit. 2012;18:RA22-RA30. [PubMed] |
| 42. | Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733-1741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1810] [Cited by in RCA: 2083] [Article Influence: 138.9] [Reference Citation Analysis (0)] |
| 43. | Bouyssou JM, Manier S, Huynh D, Issa S, Roccaro AM, Ghobrial IM. Regulation of microRNAs in cancer metastasis. Biochim Biophys Acta. 2014;1845:255-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 44. | Ma D, Tao X, Gao F, Fan C, Wu D. miR-224 functions as an onco-miRNA in hepatocellular carcinoma cells by activating AKT signaling. Oncol Lett. 2012;4:483-488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 45. | Rayner KJ, Hennessy EJ. Extracellular communication via microRNA: lipid particles have a new message. J Lipid Res. 2013;54:1174-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 46. | Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3218] [Cited by in RCA: 3552] [Article Influence: 208.9] [Reference Citation Analysis (0)] |
| 47. | Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087-2092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1048] [Cited by in RCA: 1029] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 48. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7369] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 49. | Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223-7233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1356] [Cited by in RCA: 1520] [Article Influence: 108.6] [Reference Citation Analysis (0)] |
| 50. | Turchinovich A, Burwinkel B. Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA Biol. 2012;9:1066-1075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 189] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 51. | Kim T, Reitmair A. Non-Coding RNAs: Functional Aspects and Diagnostic Utility in Oncology. Int J Mol Sci. 2013;14:4934-4968. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 52. | Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8246] [Cited by in RCA: 9782] [Article Influence: 543.4] [Reference Citation Analysis (0)] |
| 53. | Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G, Wang Z. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 923] [Cited by in RCA: 1094] [Article Influence: 78.1] [Reference Citation Analysis (0)] |
| 54. | Quackenbush JF, Cassidy PB, Pfeffer LM, Boucher KM, Hawkes JE, Pfeffer SR, Kopelovich L, Leachman SA. Isolation of circulating microRNAs from microvesicles found in human plasma. Methods Mol Biol. 2014;1102:641-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 55. | Mo MH, Chen L, Fu Y, Wang W, Fu SW. Cell-free Circulating miRNA Biomarkers in Cancer. J Cancer. 2012;3:432-448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 118] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 56. | Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1009] [Cited by in RCA: 1128] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 57. | Zheng T, Wang J, Chen X, Liu L. Role of microRNA in anticancer drug resistance. Int J Cancer. 2010;126:2-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 183] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 58. | Cho S, Lu M, He X, Ee PL, Bhat U, Schneider E, Miele L, Beck WT. Notch1 regulates the expression of the multidrug resistance gene ABCC1/MRP1 in cultured cancer cells. Proc Natl Acad Sci USA. 2011;108:20778-20783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 86] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 59. | Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13:e249-e258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 624] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 60. | Hummel R, Watson DI, Smith C, Kist J, Michael MZ, Haier J, Hussey DJ. Mir-148a improves response to chemotherapy in sensitive and resistant oesophageal adenocarcinoma and squamous cell carcinoma cells. J Gastrointest Surg. 2011;15:429-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 61. | Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, Pogribny IP. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7:2152-2159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 479] [Cited by in RCA: 500] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 62. | Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 531] [Cited by in RCA: 561] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 63. | Marin JJ, Briz O, Monte MJ, Blazquez AG, Macias RI. Genetic variants in genes involved in mechanisms of chemoresistance to anticancer drugs. Curr Cancer Drug Targets. 2012;12:402-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 64. | Marin JJ, Romero MR, Martinez-Becerra P, Herraez E, Briz O. Overview of the molecular bases of resistance to chemotherapy in liver and gastrointestinal tumours. Curr Mol Med. 2009;9:1108-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 65. | Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol. 2005;205:275-292. [PubMed] |
| 66. | Akao Y, Nakagawa Y, Naoe T. MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 2007;26:311-320. [PubMed] |
| 67. | Moldovan L, Batte KE, Trgovcich J, Wisler J, Marsh CB, Piper M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J Cell Mol Med. 2014;18:371-390. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 289] [Cited by in RCA: 322] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 68. | Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685-1694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1665] [Cited by in RCA: 1700] [Article Influence: 141.7] [Reference Citation Analysis (0)] |
| 69. | Lindow M, Kauppinen S. Discovering the first microRNA-targeted drug. J Cell Biol. 2012;199:407-412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 209] [Cited by in RCA: 220] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 70. | Hildebrandt-Eriksen ES, Bagger YZ, Knudsen TB. A unique therapy for HCV inhibits microRNA-122 in humans and results in HCV RNA suppression in chronically infected chimpanzees: results from primate and first-in-human studies. Hepatology. 2009;50 Suppl:Abstract 228. |
| 71. | Zhao Y, Liang YR. [In situ hybridization study of HBV DNA in chronic active hepatitis]. Zhonghua Bing Li Xue Zazhi. 1990;19:42-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 380] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 72. | A Multicenter Phase I Study of MRX34, MicroRNA miR-RX34 Liposome Injectable Suspension. Accessed on 1 November 2013. Available from: http://clinicaltrials.gov/ct2/show/NCT01829971?term=NCT01829971&rank=1/. |
| 73. | Lindner K, Haier J, Hummel R. MicroRNAs and Their Clinical Impact on Resistance to Anticancer Treatment. MicroRNAs: Key Regulators of Oncogenesis 2014; Springer, 2014: 369-386. |
| 74. | Collins S. Oncology Biomarker Diagnostics: Where We Are, Where We Need To Be: Identifying cancer biomarkers that are clinically useful is difficult. So is obtaining funding to get biomarker diagnostics to market. Biotechnol Healthc. 2010;7:22-25. [PubMed] |
| 75. | Bhatt AN, Mathur R, Farooque A, Verma A, Dwarakanath BS. Cancer biomarkers - current perspectives. Indian J Med Res. 2010;132:129-149. [PubMed] |
| 76. | Mäbert K, Cojoc M, Peitzsch C, Kurth I, Souchelnytskyi S, Dubrovska A. Cancer biomarker discovery: current status and future perspectives. Int J Radiat Biol. 2014;90:659-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 81] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 77. | Wang F, Li T, Zhang B, Li H, Wu Q, Yang L, Nie Y, Wu K, Shi Y, Fan D. MicroRNA-19a/b regulates multidrug resistance in human gastric cancer cells by targeting PTEN. Biochem Biophys Res Commun. 2013;434:688-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 140] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 78. | Yang SM, Huang C, Li XF, Yu MZ, He Y, Li J. miR-21 confers cisplatin resistance in gastric cancer cells by regulating PTEN. Toxicology. 2013;306:162-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 173] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 79. | Fang Y, Shen H, Li H, Cao Y, Qin R, Long L, Zhu X, Xie C, Xu W. miR-106a confers cisplatin resistance by regulating PTEN/Akt pathway in gastric cancer cells. Acta Biochim Biophys Sin (Shanghai). 2013;45:963-972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 80. | Deng H, Guo Y, Song H, Xiao B, Sun W, Liu Z, Yu X, Xia T, Cui L, Guo J. MicroRNA-195 and microRNA-378 mediate tumor growth suppression by epigenetical regulation in gastric cancer. Gene. 2013;518:351-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 81. | Shang Y, Zhang Z, Liu Z, Feng B, Ren G, Li K, Zhou L, Sun Y, Li M, Zhou J. miR-508-5p regulates multidrug resistance of gastric cancer by targeting ABCB1 and ZNRD1. Oncogene. 2014;33:3267-3276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 151] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 82. | Zhu W, Shan X, Wang T, Shu Y, Liu P. miR-181b modulates multidrug resistance by targeting BCL2 in human cancer cell lines. Int J Cancer. 2010;127:2520-2529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 238] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 83. | Zhu W, Zhu D, Lu S, Wang T, Wang J, Jiang B, Shu Y, Liu P. miR-497 modulates multidrug resistance of human cancer cell lines by targeting BCL2. Med Oncol. 2012;29:384-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 145] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 84. | Wu XM, Shao XQ, Meng XX, Zhang XN, Zhu L, Liu SX, Lin J, Xiao HS. Genome-wide analysis of microRNA and mRNA expression signatures in hydroxycamptothecin-resistant gastric cancer cells. Acta Pharmacol Sin. 2011;32:259-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 85. | Kim CH, Kim HK, Rettig RL, Kim J, Lee ET, Aprelikova O, Choi IJ, Munroe DJ, Green JE. miRNA signature associated with outcome of gastric cancer patients following chemotherapy. BMC Med Genomics. 2011;4:79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 117] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 86. | Zhu X, Lv M, Wang H, Guan W. Identification of circulating microRNAs as novel potential biomarkers for gastric cancer detection: a systematic review and meta-analysis. Dig Dis Sci. 2014;59:911-919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 87. | Zhao X, Yang L, Hu J. Down-regulation of miR-27a might inhibit proliferation and drug resistance of gastric cancer cells. J Exp Clin Cancer Res. 2011;30:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 107] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 88. | Hu J, Fang Y, Cao Y, Qin R, Chen Q. miR-449a Regulates proliferation and chemosensitivity to cisplatin by targeting cyclin D1 and BCL2 in SGC7901 cells. Dig Dis Sci. 2014;59:336-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 89. | Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res. 2009;15:2281-2290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 284] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 90. | DIANA miRPath v2. 0. Available from: http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=mirpath/index. |