Published online Aug 14, 2014. doi: 10.3748/wjg.v20.i30.10238
Revised: April 30, 2014
Accepted: May 25, 2014
Published online: August 14, 2014
Processing time: 176 Days and 1.6 Hours
Hepatocellular carcinoma (HCC) is one of the leading causes of death induced by cancer in the modern world and majority of the cases are related to chronic hepatitis B virus (HBV) infection. HBV-encoded X protein (HBx) is known to play a pivotal role in the pathogenesis of viral induced HCC. HBx is a multifunctional protein of 17 kDa which modulates several cellular processes by direct or indirect interaction with a repertoire of host factors resulting in HCC. HBX might interfere with several cellular processes such as oxidative stress, DNA repair, signal transduction, transcription, protein degradation, cell cycle progression and apoptosis. A number of reports have indicated that HBx is one of the most common viral ORFs that is often integrated into the host genome and its sequence variants play a crucial role in HCC. By mutational or deletion analysis it was shown that carboxy terminal of HBx has a likely role in protein-protein interactions, transcriptional transactivation, DNA repair, cell, signaling and pathogenesis of HCC. The accumulated evidence thus far suggests that it is difficult to understand the mechanistic nature of HBx associated HCC, and HBx mediated transcriptional transactivation and signaling pathways may be a major determinant. This article addresses the role of HBx in the development of HCC with particular emphasis on HBx mutants and their putative targets.
Core tip: The available evidence supports a well-defined for the hepatitis B virus-encoded X protein (HBx) protein in viral-induced hepatocellular carcinoma. The progression cell cycle, transactivation potential, compromised DNA repair, inhibition the tumor suppressor gene and senescence-related factors are few of the key pathways by which HBx is known to promote pathogenesis. The activation and inhibition of cellular calcium and tyrosine kinase signaling pathways are some of the other pathways modulated by HBx expression. Individually charged amino acids (K-130, V-131, D-120, 121) and or steches of amino acids (132-140) present in the carboxy-terminus of HBx protein seems to enhance the protein’s ability to deregulates cellular processes.
- Citation: Ali A, Abdel-Hafiz H, Suhail M, Al-Mars A, Zakaria MK, Fatima K, Ahmad S, Azhar E, Chaudhary A, Qadri I. Hepatitis B virus, HBx mutants and their role in hepatocellular carcinoma. World J Gastroenterol 2014; 20(30): 10238-10248
- URL: https://www.wjgnet.com/1007-9327/full/v20/i30/10238.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i30.10238
Throughout the world, hepatocellular carcinoma (HCC) is the sixth most commonly occurring cancer and is the third leading cause of death due to its complex nature[1]. Majority of the HCC cases occur as a result of Hepatitis B virus (HBV) infection[2]. HCC is more common in Asia-Pacific region and Africa. Due to high frequency of reported HBV cases, persistent HBV exposure triggers progression of HCC. More than 50% of the sufferers of HCC have history of HBV infection[2]. The estimated annual worldwide incidence is around 500000 and 1000000. In the last two decades, annual incidence in the United States has also increased by 80%[3]. HBV is the smallest ds DNA virus known to infect humans. It is the archetype of the hepadnavirus family and similar viruses infecting different hosts such as ducks (DHBV), ground squirrels (GSHV), and woodchucks (WHV). HBV genome encodes four proteins which include the envelope protein (S/Pre-S), the core protein (C/pre-C), the polymerase (P), and the X protein (HBx). On the basis of variation within the nucleotide sequence it is classified into eight genotypes named A-H[4]. Among them, B and C are mostly located in East Asia including China[5]. The HBx transcript is translated into a multifunctional protein which can regulate the activities of host cellular genes[6]. HBx triggers viral and cellular promoter indirectly by interaction with nuclear transcription factors[7-9]. Additionally, HBx is found to be involved in the activation of numerous signal transduction cascades that are linked to cell proliferation and survival[10]. HBx mutants, especially those with mutations in the COOH-terminal have been implicated in HCC[11]. Therefore, strategies involving targeting of HBx could help to achieve a significant level of therapeutic effect by inhibition of its functions which interferes with the cellular machinery and disrupts homeostasis.
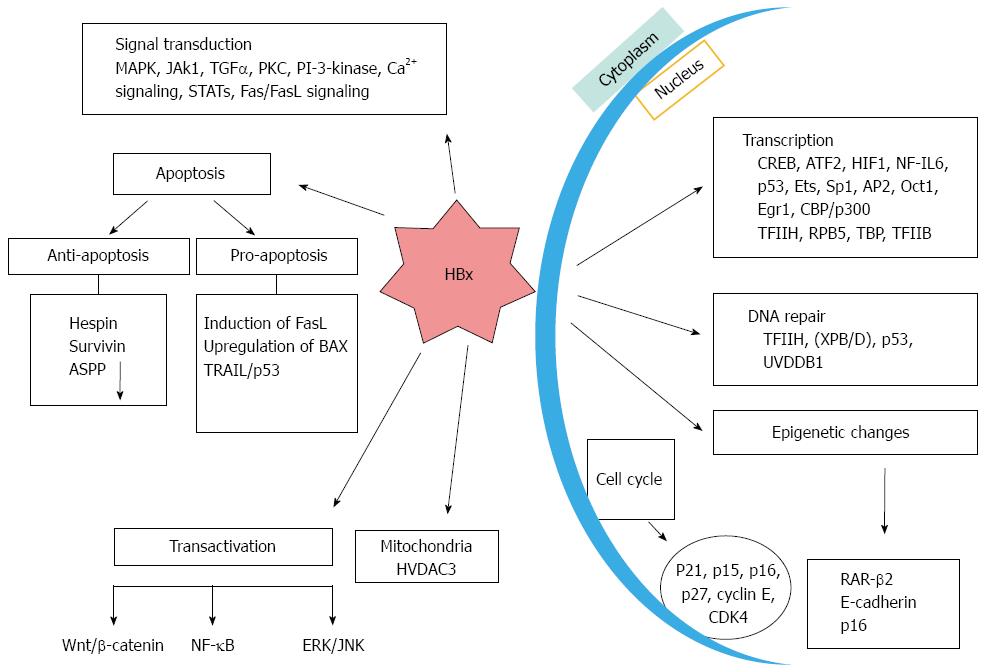
HBx is a 154-amino acid regulatory protein with a molecular mass of approximately 17 kDa. As amino acid sequence of this gene is not homologous to any known protein, it was assigned the name “HBx”[12]. It is one of the most conserved proteins among different HBV subtypes and is found in almost all viruses of Hepadnaviridae. It is mostly localized within the cytoplasm and upto some extent in the nucleus of hepatic cells. HBx protein also play an important regulatory role in viral replication and subsequent infection[13]. HBx protein consists of two functional domains[14]. The amino-terminal domain is mapped to the first 50 amino acids including the dimerization region which is required for dimerization activity. The C-terminal transactivation domain is located between amino acid 53 and 142 which interacts with Xenopus anti-photoreceptor-1 (XAP-1)/UV-damaged DNA binding protein (UVDDB) and p53[15,16]. HBx is multi-functional protein that can transactivate viral and cellular promoters and enhancers through protein-protein interactions with several well defined targets (Figure 1). Instead of binding directly with DNA, HBX activates transcription by getting involved with several nuclear transcription factors like RNA polymerase binding protein (RBP5), transcriptional factor IIB (TFIIB), transcriptional factor IIH (TFIIH), a subunit of RNA polymerase, cAMP response element-binding protein (CREB), CREB1-binding protein (CBP)/p300, activating transcription factor 2 (ATF-2), activating protein (AP)-2, AP-1, and nuclear factor kappa B (NF-κB)[17,18]. Moreover, HBx can modulate cytoplasmic signal transduction pathways including Ras-Raf-mitogen activated protein kinase (Ras-Raf-MAPK), Janus kinase/STAT (JNK/STAT), focal adhesion kinase (FAK), proline-rich tyrosine kinase 2 (Pyk2), protein kinase C (PKC) and Src-dependent phosphatiylinositol-3 kinase (PI3K/Akt)[17,18]. Activation through transactivation of cellular signaling molecules can lead to hepatic cells proliferation. In addition, it has the tendency to directly inactivate or indirectly down-regulate various tumor suppressors, such as p53, or senescence-related factors[17,18].
One of the key factors playing a role in the commencement and development of HCC is DNA damage and accumulation of errors[19]. Previously, we and others have shown that HBx protein inhibits DNA repair pathway. Acidic amino acids residues 120 and 121 of HBx were found to be critical for interaction with TFIIH and modulation of DNA repair process. It has been shown by in vitro and in vivo studies that HBx expression did not affect cell growth and tumor formation unless coupled with other factors such as exposure to hepatocarcinogen (diethynitrosamine) or UV irradiation[20]. HBx is not directly implicated in cancer progression but plays a major role as a promoting factor in HCC development. HBx protein is also known to interfere with nucleotide excision repair (NER) pathway through both p53-dependent and independent mechanisms. Studies have demonstrated that HBx protein inhibits cell cycle control checkpoints and facilitate accumulation of host mutations by interfering with NER[21-23]. HBx exerts these effects by binding to several proteins involved in DNA repair pathways and inhibiting their repair capacity. HBx de-regulated factors include human homologue of UV-DDB, XPB and XPD components of TFIIH. These are essential components for NER in both p53-proficient andp53-deficient hepatocytes[24,25]. HBx has been shown to repress XPB and XPD indirectly and inhibit the DNA binding properties of the transcription factor Sp1[22,26,27]. It has been earlier noted that HBx inactivates p53 and also impair DNA repair. The COOH-terminus of p53 serves as a scaffold for HBx binding which ultimately results in the localization of p53 from nucleolus to cytoplasm. This leads to uncontrolled cell cycle progression and DNA repair. HBx also represses transcriptional activity of p53 by disrupting cross talk among several cellular factors[28]. Furthermore, it also disrupts interaction of p53 with TFIIH resulting in compromised TFIIH induced helicase activity during assembly of the DNA repair complex[29,30]. HBx binding to the carboxy (COOH) terminal domain of p53 is known block its association with XPB and XPD.
HBx contributes to the progression of HCC by affecting apoptosis in different ways. HBx has been shown to either inhibit or have no effect on apoptosis[31-33]. Such contradictory manifestations are likely due to the differential pattern of HBx expression and depend upon the cell types involved. It has been suggested that increased quantity of HBx promote apoptosis, whereas low levels inhibits this phenomenon[34]. One of the examples of its anti-apoptotic feature involves complex formation and sequestration of p53 in the cytoplasm and blocking its entrance into the nucleus[35-38]. As a result, p53 fails to up-regulate its various downstream effector molecules like Bax, p21, or Fas, which are required in the apoptotic pathway thus leading to increased cell survival[35]. HBx also prevents p53-induced apoptosis through HBx-P3K-Akt-Bad pathway, by inhibiting caspase 3 function, associated with H-ras oncogene by induction of phosphatidyl inositol-3 kinase and AKt pathway[39]. Increase in the concentration of anti-apoptotic protein survivin is another example in this category which acts by p38/MAPK pathway[40]. HBx upregulation of survivin expression was found to be HURP-dependent. Another molecule which is highly expressed in hepatocellular carcinoma and mediates actions of HBx opposite to that of p53 is cyclooxygenase-2 (COX-2)[41]. Activation of COX-2/PGE2 pathway leads to elimination of p53-induced apoptosis imposed by HBX expression. In contrast, it is demonstrated by cell culture studied in HeLa, Hep3B, and HepG2 cells that HBx induces apoptosis directly by inducing cytoplasmic cytochrome c, caspase-3-like activity and nuclear fragmentation. HBx induced apoptosis is evident in HBx transgenic livers, primary hepatocytes and in mice having p53 null mutation. Down-regulation of C-myc expression by HBx increased the response of PLC/PRF/5 cells to TNF-α induced apoptosis[42,43]. TRAIL-induced apoptosis mediated by HBx has also been shown in the hepatocytes[44]. A recent report shows that only the complete HBx protein, and not the one with the truncated C-terminus, induces apoptosis, which further indicate that the COOH-terminal region is necessary for its pro-apoptotic functions[45].
The importance of epigenetic modification in hepatocellular carcinoma was revealed when researchers found out that the methylation of at least one tumor suppressor gene was present in majority of tumors studied. However, its prevalence in HBx induced HCCs is not clear[46]. Additional studies have shown that HBx can induce epigenetic changes and inactivate tumor suppressor genes and/or activate oncogenes thereby promoting the development of HCC[47]. Moreover, these hyper methylated genes show direct role in cell growth, cell cycle regulation, dedifferentiation, apoptosis and xenobiotic metabolism[48], altogether which contributes to HBV-related HCC progression. HBx may modify transcription of DNA methyltransferase (DNMT)1 and DNMT3 and may act as potentially critical epigenetic deregulating agent. These epigenetic changes induced by HBx in several tumor suppressor genes have been highlighted. For example, HBx represses the E-cadherin (a tumor suppressor gene) by inducing DNMT1 transcription[49,50] and also influences Wnt signaling pathway. Such repressive effect is brought about by Lysine-30 located in the transactivating domain of HBx. HBx induces hypermethylation of p16 which show positive correlation with HCC[51,52]. Activation of DNMT1 transcription by HBx results in hypermethylation of p16 and ultimately its repression[53]. This activation is mediated by pRb-E2f pathway. Genomic hypomethylation and regional hypermethylation of the tumor promoter gene IGFBP-3 are known contributors of HCC. Promoter region of IGFBP is hypermethylated by DNMT1, DNMT3A1 and DNMT3A2[53]. Activation of Ras signaling pathway is responsible for Induction of DNMT1[54]. Tumor suppressor p53 also display a key role in regulation of the DNMTs expression. It regulates apoptosis by interacting with apoptosis-stimulating protein of p53 (ASPP)1 and ASPP2 family of proteins[55,56]. ASPP2 has a much important role than ASPP1 in HBx protein induced HCC. Promoter hypermethylation of glutathione S-transferase P1, which helps in mounting a protective state against injury from oxidant carcinogens, is shown to be mediated by HBx. In summary the data is consistent with the hypothesis that epigenetic events induced by HBx play an important role in the pathogenesis of HCC, and also that of several other tumors. It is assumed that epigenetic down-regulation of retinoic acid receptor-beta 2 (RAR-β2) by HBx protein is an important step in HCC progression[57,58]. HBx decreases the expression of retinoic acid receptor-beta two (RAR-β2) in human HCC cells by hypermethylating its promoter through DNA methyltransferase 1 and 3a. HBx abolishes the crucial potent effect of retinoic acid to down-regulate G1-checkpoint regulators such as p16, p21, and p27. This ultimately leads to activation of E2F1 and the generation of HCC tumor[59].
Induction of Telomerase plays an important role in transformation of cancer cells as studies have shown that majority of hepatocellular carcinomas display a high level of telomerase activity[60]. Several studies were carried out to establish the role of HBx in telomerase but such effects has been controversial. HBx can increase the telomerase activity in cells transfected by virus, protein-positive HCC tissues or cultured HCC cells by increasing expression of TERT[61]. These studies demonstrate that HBx induced C-myc have a role in the process of telomerase activation and prolonging the life-span of normal cells. On the other hand Su et al[62] and his coworkers have shown that HBx inhibit telomerase activity and suggested that the controversial results could be attributed to various isoforms of HBx. They found that the transcriptional suppression of human telomerase achieved by HBx isoform is by increased binding of MAZ to its promoter[63,64]. However, more data is required to authenticate the function of telomerase in HBx induced HCC.
It is widely acknowledged that HBx protein acts as a potent transactivator and can affect various viral and cellular promoters and enhancers[65]. It regulates transcriptional activity basically via direct protein-protein interaction. The transactivation functions of HBx is exercised in cytoplasm by signaling pathways, and in the nucleus by DNA-binding proteins[66]. A wide variety of cellular and viral genes, class II and III promoters/enhancers, and proto-oncogenes such as c-jun, c-fos, and c-myc are upregulated by HBx. Besides, it activates number of transcriptional factors and genes like NFκB, AP-1, ATF/cAMP and CREB in the nucleus[67,68]. The repertoire of genes and proteins which are linked with proliferation of cells also include interleukin-8 (IL-8), tumor necrosis factor (TNF), transforming growth factor (TGF)-β1, and early growth response factor (EGRF)[69]. In cytoplasm HBx activates signal transduction cascades involving RAS/RAF/MAPK, JAK/STAT, and Src kinases[27,70]. Induction of RAS/RAF/MAPK by HBx leads to activation of AP-1 and NFκB transcription factors which causes the de-regulation of various checkpoints which control the cell cycle[71,72]. HBx may also affect angiogenic pathways which play an important role in HCC development. HBx can induce up-regulation of vascular endothelial growth factor (VEGF) besides upregulating metalloproteinases (MMPs, MMP2, MMP9 and MMP14) which can be instrumental in promoting invasion and metastasis[73]. Apart from this, there are several other ways by which HBx induces HCC like stimulation of a number of signaling pathways associated with enhancing motility and overall survival chance of tumor cells, up-regulation of inflammatory mediators and an increased possibility for men to develop liver cancer by influencing androgen receptor pathway. There are many reports which advocate that COOH-terminal of HBx is critical for its transactivational activity and that it regulates cell proliferation and viability[74]. Deletion of its COOH-terminal has been documented in patients with chronic HBV infection[36]. The carcinogenic potential of C-terminus truncated HBx has also been implicated in Wnt signaling pathways which may have influence upon the development of HCC[68].
MicroRNAs are noncoding RNAs which are involved in the regulation of gene expression. Additionally, they play crucial roles in numerous pathobiological processes including tumor formation. Therefore, their possible role in the causation of hepatocellular carcinoma is not unexpected. However, the question as to how HBx actually regulates miRNA expression during HCC has no easy answer. However, Wu et al[75] in 2014 have shown that HBx down regulates microRNA-15b via fucosyl transferase 2 induced Globo H (a cancer-associated carbohydrate antigen) expression ultimately influencing HCC proliferation. In a recently published article Zhang et al[76] 2013 have found that miR-205 is down regulated in 33 samples of HCC tissues in contrast to adjacent healthy areas of the liver. They suggested that miR-205 could be a potential tumor-suppressive gene in case of HCC. According to them, HBx inhibit tumor suppressor miR-205 and increases hepato carcinogenesis by hypermethylation of miR-205 promoter. Wei et al[77] have demonstrated HBx induces epigenetic repression of miR-132 by methylation of DNA and suggested that it could be a promising biochemical marker for HCC. They have also shown that miR-101 is down regulated by HBx which was in turn induced by HBX targeting the methyl transferase 3A (DNMT3A) gene. In a mouse model of liver cancer, it was observed that miR-148a is repressed by HBx and this leads to cancer growth and eventual metastasis. Expression of miR-148a in hepatoma cells reduces hematopoietic pre-B cell leukemia transcription factor-interacting protein. This caused the suppression of AKT and extracellular signal-regulated kinases (ERK) induced mammalian target of rapamycin inhibition involving AKT/ERK/Fork head box protein O4/ATF5 pathways[78]. Recently Qiu et al[79] have highlighted that HBx downregulates PDCD4 by upregulation of miR-21. HBx suppresses EGFR by miR-7 which confirms the role of HBX-miR-7-EGFR as a critical signaling pathway in controlling cell growth in HCC[80]. HBx perturbs the in vitro expression of miRNA in cancerous hepatocytes of the host liver, especially by down regulating the mi-16 family. Its suppression was c-Myc mediated and is a necessary requirement for the HBx-induced transformation of HepG2 cells in vitro[81]. Another microRNA, miR-152 is frequently down-regulated during HBx expression and it is also known to regulate DNMT1 in HBV-related HCC. Additionally, tumor-suppressive role of miR-152 in the epigenetic irregularity of HBV-related HCC has been observed[82]. Wild-type HBx and the high proliferation-inducing mutant HBx can influence the expression profile of miR-338-3p and miR-551b by its down regulation in L02 cells. Here, the cell growth inhibition occurs by direct modulation of cyclinD1, cyclinG1, and E2F[83,84]. In another study showing linkage of mi RNA with HBx, it was found that HBx could inhibit apoptosis of HepG2 cells through down-regulation of miR-192 which induces apoptosis of HepG2 cells[85].
In the HCC tissue samples, negative with hepatitis B surface antigen, there seem to be a preferential accumulation of X transcripts suggesting the possible role of HBX in neoplastic transformation[86]. Several mutants of HBx have been isolated from hepatic tissues and sera taken from sufferers of HCC[87,88]. The incidence of HBx mutations in such circumstances have been found to be reasonably high which underscores the point that the HBx mutants after all may be highly significant in the pathogenesis of hepatocellular carcinoma[89,90] (Table 1). Deletions of 3’ is one of the most frequently reported mutations of HBx, and such mutations are more frequently observed in hepatocarcinoma cells rather than untransformed liver cells which highlights the potential role of these mutations in tumor development. Poussin et al[91] have analyzed the HBx expression correlation in tumorous vs non-tumorous tissues of human origin. They observed that HBV X gene was truncated at its 3’ end in five out of nine tumors from the liver and only one out of eight liver tissues with no tumors. A high rate of mutation was revealed in X genes from three tumors upon Sequence analysis. This was in contrast to the lower rate of mutation observed in the X genes of adjoining non tumorous tissues. In an investigation carried out by Tu et al[45], sera and tissues were taken from tumor as well as healthy liver regions of patients with HBV related HCC. They conducted a detailed analysis of the structure and functioning of HBx sequences. With the exception of one, six out of seven tumor samples showed that the HBx sequences in these tissues and especially those harvested from HBV integrant have a deletion at COOH-terminal part. Many of these shortened sequences were divested of their transcriptional capacity. Additionally, their function of acting as a brake to cell division and transformation seemed to be compromised. In fact COOH shortened HBX improved the transforming character of ras and myc oncogenes[45]. Centromere protein A (CENP-A) is known to be frequently over-expressed in HCC. A positive correlation between HBx COOH terminus mutation and expression of CENP-A has been documented in HCC tissues[92]. Carboxy-terminal truncated HBx were frequently detected in HCC tissues by Ma et al[93]. They found that the truncated HBx is able to effectively transform immortalized liver cell line. Differential expression of key genes implicated in the control of cell cycle and apoptosis have been observed by them. Based on these studies, it can be concluded that the length of COOH terminus truncation is important for HBx’s transcription activity. The truncated HBx exhibiting deletion within 14 amino acids had no effect on its transactivation property[94]. Thus it could be concluded that the last 14 amino acids at the carboxy terminus of HBx does not contribute towards transactivation activity[94]. HBx is divided into six regions of which A, C, and E regions show a more conserved state. To decipher the regions of HBx which have a significant role in transactivation, Kumar et al[94] effected a range of 10 deletion mutants and four single point mutants which related to corresponding conserved cysteine residues. The mutant gene expression was also examined by the use of HBx specific monoclonal antibody. It was found by them that deletion of region A which happens to be extremely conserved did not in anyways affect transactivation. Also the non-substitution of the N terminal cysteine produced no difference to transactivation. Removal of the regions C and D however led to an important loss of function. Kumar et al[94] inferred and stated that residues 132-140 of region E and C137 seemed to be crucial for transactivation. They also showed that a fusion cassette containing residues 58-140 proved to be an efficient transactivator, suggesting an important role of these residues in transcriptional activity[94]. C-terminal deletional mutant of HBx (deleted at nucleotides 382-400) was shown to down-regulate a miRNA (miR338-3p) in HBx infected cells which prolonged cell proliferation and increased the risk of HCC development[83,84]. Wnt-5a is correlated with the HCC development. HBx mutants deprived of thirty amino acids from their C-terminus induced the expression of Wnt-5a gene by more than 10 folds in Huh7 cells. However, the expression of Wnt-5a was suppressed when the same cells were transfected with HBx mutant deleted with forty amino acids from the same C-terminus. Such changes in Wnt-5a expression levels were also observed in patients with HCC when compared with appropriate non-tumorous counterparts. This suggests that HBx mutants might play a role in the development and progression of HCC through the Wnt-5a pathway as HBx showed negative correlation with Wnt-5a[95]. Sirma et al[74] studied the influence of HBx and its natural mutants on cellular growth, proliferation and survival. They explained that the loss of antiproliferative and apoptotic role of HBx by the natural mutants may propel the hepatocytes to go into uncontrollable cell division and probably help in malignant transformation which could ultimately lead to HCC.
| No. | Putative mutation | Function/significance of the mutation | Ref. |
| 1 | Insert mutation at position 204 | Associated with the nuclear localization of HBx protein | [88] |
| 2 | Mutations at aa 127, 130, and 131 | HBx deletions may be implicated in HCC | [89] |
| 3 | Distal COOH-terminal region deletion | COOH-terminal truncated HBx plays critical role in the HCC carcinogenesis via the activation of cell proliferation | [45,93] |
| 4 | HBx-A31 mutation | Development of this mutant represent a strategy of escape immune surveillance and thus may contribute to the process of multiple-step HCC | [101] |
| 5 | Truncation mutation at 3′ end of HBx | Potential role in HBV related HCC | [91] |
| 6 | Point mutations at X gene codons 130 (AAG - ATG) and 131 (GTC - ATC) | Modification in the transactivation function of HBx | [102,103] |
| 7 | Deletion from 382 to 401 base pairs (HBxDelta127) | HBxÄ127 promotes hepatoma cell growth through activating SREBP-1c involving 5-LOX | [96,104] |
| 8 | HBx M130K and V131I (T-A) mutations in HBV genotype F | Associated with severe liver damage and HCC, can acts as prognostic marker for HCC | [49] |
| 9 | aa 52 to 65 and 88 to 154 | Regions are important for augmentation function of HBX in HBV replication | [105] |
| 10 | Five types of “hot-spot” mutations of genotype B or C | Affects antiproliferation and transactivation of genotype B or C of HBx | [106] |
| 11 | A1762T/G1764A | Associated with development of HCC in Thai patients | [107] |
| 12 | and G1899A mutationsNatural HBx mutant truncated 27 amino acids at the COOH terminal | Promotes cell proliferation | [108] |
| 13 | TP53 R249S mutation, genetic variations in HBx | Associated with diagnosis of HCC or liver cirrhosis | [109] |
| 14 | A truncated mutant (residues 58-140) of the HBx | Participate in transactivation function | [94] |
| 15 | Nucleotide change of codon 38 in the X gene of hepatitis B virus genotype C | Codon-38 change in genotype C is an independent risk factor for HCC development | [110] |
| 16 | Ser-101, Met-130 Mutation | Have differential effect on the expression of cyclin dependent kinase inhibitor | [111] |
| 17 | HBx 120 and 121 | Inhibits DNA repair | [112] |
Some point mutations have also been implicated in affecting the functions of HBx and HCC development. HBx with point mutations K130M/V131I has been shown to induce hypoxia-inducible factor 1α which has a role in the development of solid tumors under hypoxic environment conditions in HCC. A novel HBx associated mutation at amino acids L30F/S144A was recorded in 13 out of 44 HCC tissue samples[96]. Chen et al[88] highlighted more than 50 varying type of mutations involving the HBx gene. They found that HBx mutation in tissue and serum (12 and 9 sites respectively) showed significant association with HCC. This point to a possible role in HCC development. Insertion mutation at position 204: Insert 204AGGCCC was found to be coexisting with point mutation at 260 (G-->A) and 264 (G/C/T-->A). This opened a significant window in the understanding of HBX mutation in that, it also pointed that the nuclear localization of HBx protein in tumorous hepatocytes was closely associated with the afore mentioned characteristic alignment of HBx. The above mutation seems to have connection with the nuclear localization of HBx protein, suggesting a link with HCC development. With the help of innovative laser capture micro dissection technology, Iavarone et al[89] consistently identified mutations at aa 127, 130 and 131, though they are not able to find any clear point mutation profile between tumor and healthy tissue samples. On the other hand, deletion in HBx gene which was detected in several sufferers was more commonly associated with tumor derived sequences (6/18). This is in contrast to healthy tissue derived sequences in which the no is less (1 out of 20). The researchers concluded that there is a consistent presence of characteristic HBx encoding sequences in clonally proliferating cells. This strengthens the hypothesis that HBx deletions may be critically involved in the development of liver cancer. Another study which identified mutations linked with the progression of HCC is mutation of nucleotides T1653, T1689, and/or T1762/A1764. Double mutant T1762/A1764 along with the adjacent T1766/A1768 mutation was found to significantly elevate the risk of HCC in HBV-infected patients[90]. Recently, a natural mutant of HBx (HBxDelta127) was discovered with shortening of 27 amino acids at the COOH-terminal (deletion of 382 to 401 bp). This can potentially induce growth and proliferation of hepatoma cells. HBx Delta127 significantly enhanced the transcription of FAS in human hepatoma HepG2 and H7402 cells by activating SREBP-1c which in turn activates 5-LOX[97]. In an important discovery it was revealed that cancer associated HBx variants clearly blunted transactivation and proapoptotic functions. Although their ability to block p53-mediated apoptosis remained unaltered, suggesting that mutations in HBx may contribute to the development of HCC. This result is consistent with the hypothesis that certain mutations in HBx and p53 (at codon 249) may cooperate in contributing to liver carcinogenesis[98]. Diet incorporated fungal aflatoxin B1 has been instrumental in the causation of HCC in Africa and China region, yet how HBx mutants interplay with toxins is still not known. Exposure to aflatoxin B1 can result in induction of mutation in the p53 gene, a G to T transversion in codon 249 (p53ser249 mutation) inactivates the tumor suppressor gene. This mutation is found in majority of HCC patients in regions having high aflatoxin β1 exposure[99]. It was shown by Belloni et al[100] that HBV mutants that lack the ability to express HBx are replication deficient. Such replication incompetence is restored by exogenously expressing HBx transcomplements. Several evidences suggest that mutations in HBx contribute heavily towards the development of HCC. However, in what profound and exact manner these structural and point mutation affects functions and behavior of HBX need to be further explored.
HBx modulates a variety of cellular activities such as transcription, cell cycle progression, DNA damage repair, cell proliferation, and apoptosis. The localization of HBx is consistent with its dual specificity role. HBx pre-dominantly localizes in the cytoplasm and to some extent within the nucleus and mitochondria. This dynamic cellular distribution and compartmentalization could attribute to HBx multiple functions at different stages of viral infection and HCC progression. Although extensive study has been done to elucidate the implications of HBx in hepatocellular carcinoma, its precise role in carcinogenic manifestations is yet to be deciphered completely and is appropriate to be the main focus of the future studies. The difficulty in addressing its role arises due to the lack of convenient and clinically relevant models to study viral replication. However, shedding some light on its indirect effects, it can be concluded that HBx helps in the progression of HCC since it can modulate cellular micro-environment by acting on transcription factors, cell cycle regulators, apoptosis and DNA repair mechanisms. This conclusion can be further supported by the fact that the duck HBV, that lacks an X open reading frame, does not induce cancer in ducks. Thus, HBx represents one of the central players of oncogenesis in liver and could be an attractive therapeutic target for HCC suppression. HBx mutants especially those with mutation in the COOH-terminal end have been implicated in HCC, therefore therapeutic strategies targeting HBx could be effective at multiple stages of HCC development.
We thank King Fahd Medical Research Center (KFMRC) and Center of Genomic Medicine (CEGMR) for financial support. We thank Mr. Mohammad Sanaullah M.Y. Gazdar (Head KFMRC Library), Ms. Shylu Mathew of CEGMR, Dr. Zubair Alam of KFMRC and Dr. Zainab Younis and Ms. Nadia Rashid of IQ Institute of Infection and Immunity, Lahore Pakistan for helpful discussion and editing.
P- Reviewer: Gallego-Duran R, Luo GH, TanakaT S- Editor: Nan J L- Editor: A E- Editor: Wang CH
| 2. | Lupberger J, Hildt E. Hepatitis B virus-induced oncogenesis. World J Gastroenterol. 2007;13:74-81. [PubMed] |
| 3. | Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27:1485-1491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1196] [Cited by in RCA: 1326] [Article Influence: 82.9] [Reference Citation Analysis (0)] |
| 4. | Sun TT, Chu YY. Carcinogenesis and prevention strategy of liver cancer in areas of prevalence. J Cell Physiol Suppl. 1984;3:39-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 24] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 5. | Chen X, Dai B, Liu Z, Gao J, Ji Z, Guo J, Chen G, Deng Z, Shao Z. A novel B/C inter-genotype recombinant of hepatitis B virus identified in north-west China. J Gen Virol. 2014;95:153-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 6. | Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725-12734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 387] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 7. | Edamoto Y, Hara A, Biernat W, Terracciano L, Cathomas G, Riehle HM, Matsuda M, Fujii H, Scoazec JY, Ohgaki H. Alterations of RB1, p53 and Wnt pathways in hepatocellular carcinomas associated with hepatitis C, hepatitis B and alcoholic liver cirrhosis. Int J Cancer. 2003;106:334-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 158] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 8. | Tommasi S, Pinto R, Pilato B, Paradiso A. Molecular pathways and related target therapies in liver carcinoma. Curr Pharm Des. 2007;13:3279-3287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 9. | Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J Gastroenterol Hepatol. 2011;26 Suppl 1:144-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 214] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 10. | McCubrey JA, Steelman LS, Abrams SL, Lee JT, Chang F, Bertrand FE, Navolanic PM, Terrian DM, Franklin RA, D’Assoro AB. Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT pathways in malignant transformation and drug resistance. Adv Enzyme Regul. 2006;46:249-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 458] [Cited by in RCA: 504] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 11. | Ng SA, Lee C. Hepatitis B virus X gene and hepatocarcinogenesis. J Gastroenterol. 2011;46:974-990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 136] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 12. | Miller RH, Robinson WS. Common evolutionary origin of hepatitis B virus and retroviruses. Proc Natl Acad Sci USA. 1986;83:2531-2535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 135] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Murakami S. Hepatitis B virus X protein: a multifunctional viral regulator. J Gastroenterol. 2001;36:651-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 260] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 14. | Gong DY, Chen EQ, Huang FJ, Leng XH, Cheng X, Tang H. Role and functional domain of hepatitis B virus X protein in regulating HBV transcription and replication in vitro and in vivo. Viruses. 2013;5:1261-1271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 15. | Lin Y, Nomura T, Yamashita T, Dorjsuren D, Tang H, Murakami S. The transactivation and p53-interacting functions of hepatitis B virus X protein are mutually interfering but distinct. Cancer Res. 1997;57:5137-5142. [PubMed] |
| 16. | Becker SA, Lee TH, Butel JS, Slagle BL. Hepatitis B virus X protein interferes with cellular DNA repair. J Virol. 1998;72:266-272. [PubMed] |
| 17. | Feitelson MA, Lee J. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett. 2007;252:157-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 180] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 18. | Diao J, Garces R, Richardson CD. X protein of hepatitis B virus modulates cytokine and growth factor related signal transduction pathways during the course of viral infections and hepatocarcinogenesis. Cytokine Growth Factor Rev. 2001;12:189-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 125] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 19. | Qadri I, Fatima K, AbdeL-Hafiz H. Hepatitis B virus X protein impedes the DNA repair via its association with transcription factor, TFIIH. BMC Microbiol. 2011;11:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 20. | Slagle BL, Lee TH, Medina D, Finegold MJ, Butel JS. Increased sensitivity to the hepatocarcinogen diethylnitrosamine in transgenic mice carrying the hepatitis B virus X gene. Mol Carcinog. 1996;15:261-269. [PubMed] |
| 21. | Bellamy CO, Clarke AR, Wyllie AH, Harrison DJ. p53 Deficiency in liver reduces local control of survival and proliferation, but does not affect apoptosis after DNA damage. FASEB J. 1997;11:591-599. [PubMed] |
| 22. | Lee TH, Elledge SJ, Butel JS. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J Virol. 1995;69:1107-1114. [PubMed] |
| 23. | Capovilla A, Carmona S, Arbuthnot P. Hepatitis B virus X-protein binds damaged DNA and sensitizes liver cells to ultraviolet irradiation. Biochem Biophys Res Commun. 1997;232:255-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 24. | Qadri I, Conaway JW, Conaway RC, Schaack J, Siddiqui A. Hepatitis B virus transactivator protein, HBx, associates with the components of TFIIH and stimulates the DNA helicase activity of TFIIH. Proc Natl Acad Sci USA. 1996;93:10578-10583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 116] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 25. | Groisman IJ, Koshy R, Henkler F, Groopman JD, Alaoui-Jamali MA. Downregulation of DNA excision repair by the hepatitis B virus-x protein occurs in p53-proficient and p53-deficient cells. Carcinogenesis. 1999;20:479-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 62] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 26. | Benn J, Schneider RJ. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc Natl Acad Sci USA. 1995;92:11215-11219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 236] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 27. | Lee YI, Lee S, Lee Y, Bong YS, Hyun SW, Yoo YD, Kim SJ, Kim YW, Poo HR. The human hepatitis B virus transactivator X gene product regulates Sp1 mediated transcription of an insulin-like growth factor II promoter 4. Oncogene. 1998;16:2367-2380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 63] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Dewantoro O, Gani RA, Akbar N. Hepatocarcinogenesis in viral Hepatitis B infection: the role of HBx and p53. Acta Med Indones. 2006;38:154-159. [PubMed] |
| 29. | Madden CR, Finegold MJ, Slagle BL. Altered DNA mutation spectrum in aflatoxin b1-treated transgenic mice that express the hepatitis B virus x protein. J Virol. 2002;76:11770-11774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 30. | Smith ML, Chen IT, Zhan Q, O’Connor PM, Fornace AJ. Involvement of the p53 tumor suppressor in repair of u.v.-type DNA damage. Oncogene. 1995;10:1053-1059. [PubMed] |
| 31. | Terradillos O, Pollicino T, Lecoeur H, Tripodi M, Gougeon ML, Tiollais P, Buendia MA. p53-independent apoptotic effects of the hepatitis B virus HBx protein in vivo and in vitro. Oncogene. 1998;17:2115-2123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 136] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 32. | Kim H, Lee H, Yun Y. X-gene product of hepatitis B virus induces apoptosis in liver cells. J Biol Chem. 1998;273:381-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 164] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 33. | Miao J, Chen GG, Chun SY, Lai PP. Hepatitis B virus X protein induces apoptosis in hepatoma cells through inhibiting Bcl-xL expression. Cancer Lett. 2006;236:115-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 55] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 34. | Ye L, Dong N, Wang Q, Xu Z, Cai N, Wang H, Zhang X. Progressive changes in hepatoma cells stably transfected with hepatitis B virus X gene. Intervirology. 2008;51:50-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 35. | Wang XW, Gibson MK, Vermeulen W, Yeh H, Forrester K, Stürzbecher HW, Hoeijmakers JH, Harris CC. Abrogation of p53-induced apoptosis by the hepatitis B virus X gene. Cancer Res. 1995;55:6012-6016. [PubMed] |
| 36. | Elmore LW, Hancock AR, Chang SF, Wang XW, Chang S, Callahan CP, Geller DA, Will H, Harris CC. Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc Natl Acad Sci USA. 1997;94:14707-14712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 253] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 37. | Ueda H, Ullrich SJ, Gangemi JD, Kappel CA, Ngo L, Feitelson MA, Jay G. Functional inactivation but not structural mutation of p53 causes liver cancer. Nat Genet. 1995;9:41-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 255] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 38. | Wang XW, Forrester K, Yeh H, Feitelson MA, Gu JR, Harris CC. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci USA. 1994;91:2230-2234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 482] [Article Influence: 15.5] [Reference Citation Analysis (1)] |
| 39. | Lee YI, Kang-Park S, Do SI, Lee YI. The hepatitis B virus-X protein activates a phosphatidylinositol 3-kinase-dependent survival signaling cascade. J Biol Chem. 2001;276:16969-16977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 153] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 40. | Kuo TC, Chao CC. Hepatitis B virus X protein prevents apoptosis of hepatocellular carcinoma cells by upregulating SATB1 and HURP expression. Biochem Pharmacol. 2010;80:1093-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 41. | Cheng AS, Yu J, Lai PB, Chan HL, Sung JJ. COX-2 mediates hepatitis B virus X protein abrogation of p53-induced apoptosis. Biochem Biophys Res Commun. 2008;374:175-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 42. | Su F, Schneider RJ. Hepatitis B virus HBx protein sensitizes cells to apoptotic killing by tumor necrosis factor alpha. Proc Natl Acad Sci USA. 1997;94:8744-8749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 251] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 43. | Chan DW, Ng IO. Knock-down of hepatitis B virus X protein reduces the tumorigenicity of hepatocellular carcinoma cells. J Pathol. 2006;208:372-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 44. | Liu YG, Liu SX, Liang XH, Zhang Q, Gao LF, Han LH, Cao YL, Hou N, Du J, Sun WS. Blockade of TRAIL pathway ameliorates HBV-induced hepatocyte apoptosis in an acute hepatitis model. Biochem Biophys Res Commun. 2007;352:329-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 45. | Tu H, Bonura C, Giannini C, Mouly H, Soussan P, Kew M, Paterlini-Bréchot P, Bréchot C, Kremsdorf D. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 2001;61:7803-7810. [PubMed] |
| 46. | Yang B, Guo M, Herman JG, Clark DP. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am J Pathol. 2003;163:1101-1107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 288] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 47. | Tian Y, Yang W, Song J, Wu Y, Ni B. Hepatitis B virus X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis. Mol Cell Biol. 2013;33:2810-2816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 117] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 48. | Pogribny IP, Rusyn I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett. 2014;342:223-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 144] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 49. | Lee JO, Kwun HJ, Jung JK, Choi KH, Min DS, Jang KL. Hepatitis B virus X protein represses E-cadherin expression via activation of DNA methyltransferase 1. Oncogene. 2005;24:6617-6625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 152] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 50. | Liu J, Lian Z, Han S, Waye MM, Wang H, Wu MC, Wu K, Ding J, Arbuthnot P, Kew M. Downregulation of E-cadherin by hepatitis B virus X antigen in hepatocellullar carcinoma. Oncogene. 2006;25:1008-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 88] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 51. | Jung JK, Arora P, Pagano JS, Jang KL. Expression of DNA methyltransferase 1 is activated by hepatitis B virus X protein via a regulatory circuit involving the p16INK4a-cyclin D1-CDK 4/6-pRb-E2F1 pathway. Cancer Res. 2007;67:5771-5778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 151] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 52. | Zhu R, Li BZ, Li H, Ling YQ, Hu XQ, Zhai WR, Zhu HG. Association of p16INK4A hypermethylation with hepatitis B virus X protein expression in the early stage of HBV-associated hepatocarcinogenesis. Pathol Int. 2007;57:328-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 53. | Zhu YZ, Zhu R, Fan J, Pan Q, Li H, Chen Q, Zhu HG. Hepatitis B virus X protein induces hypermethylation of p16(INK4A) promoter via DNA methyltransferases in the early stage of HBV-associated hepatocarcinogenesis. J Viral Hepat. 2010;17:98-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 54. | Park IY, Sohn BH, Yu E, Suh DJ, Chung YH, Lee JH, Surzycki SJ, Lee YI. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132:1476-1494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 247] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 55. | Samuels-Lev Y, O’Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell. 2001;8:781-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 509] [Cited by in RCA: 505] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 56. | Zhao J, Wu G, Bu F, Lu B, Liang A, Cao L, Tong X, Lu X, Wu M, Guo Y. Epigenetic silence of ankyrin-repeat-containing, SH3-domain-containing, and proline-rich-region- containing protein 1 (ASPP1) and ASPP2 genes promotes tumor growth in hepatitis B virus-positive hepatocellular carcinoma. Hepatology. 2010;51:142-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 88] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 57. | Jung JK, Park SH, Jang KL. Hepatitis B virus X protein overcomes the growth-inhibitory potential of retinoic acid by downregulating retinoic acid receptor-beta2 expression via DNA methylation. J Gen Virol. 2010;91:493-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 58. | Niu D, Zhang J, Ren Y, Feng H, Chen WN. HBx genotype D represses GSTP1 expression and increases the oxidative level and apoptosis in HepG2 cells. Mol Oncol. 2009;3:67-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 48] [Article Influence: 2.8] [Reference Citation Analysis (1)] |
| 59. | Yokota T, Suda T, Igarashi M, Kuroiwa T, Waguri N, Kawai H, Mita Y, Aoyagi Y. Telomere length variation and maintenance in hepatocarcinogenesis. Cancer. 2003;98:110-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 24] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 60. | Zhang X, Dong N, Zhang H, You J, Wang H, Ye L. Effects of hepatitis B virus X protein on human telomerase reverse transcriptase expression and activity in hepatoma cells. J Lab Clin Med. 2005;145:98-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 48] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 61. | Liu YC, Chen CJ, Wu HS, Chan DC, Yu JC, Yang AH, Cheng YL, Lee SC, Harn HJ. Telomerase and c-myc expression in hepatocellular carcinomas. Eur J Surg Oncol. 2004;30:384-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 62. | Su JM, Lai XM, Lan KH, Li CP, Chao Y, Yen SH, Chang FY, Lee SD, Lee WP. X protein of hepatitis B virus functions as a transcriptional corepressor on the human telomerase promoter. Hepatology. 2007;46:402-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 63. | Her S, Bell RA, Bloom AK, Siddall BJ, Wong DL. Phenylethanolamine N-methyltransferase gene expression. Sp1 and MAZ potential for tissue-specific expression. J Biol Chem. 1999;274:8698-8707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 64. | Qu ZL, Zou SQ, Cui NQ, Wu XZ, Qin MF, Kong D, Zhou ZL. Upregulation of human telomerase reverse transcriptase mRNA expression by in vitro transfection of hepatitis B virus X gene into human hepatocarcinoma and cholangiocarcinoma cells. World J Gastroenterol. 2005;11:5627-5632. [PubMed] |
| 65. | Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. Hepatitis B virus-related hepatocellular carcinoma: paradigms for viral-related human carcinogenesis. Oncogene. 2006;25:3823-3833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 210] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 66. | Nakatake H, Chisaka O, Yamamoto S, Matsubara K, Koshy R. Effect of X protein on transactivation of hepatitis B virus promoters and on viral replication. Virology. 1993;195:305-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 55] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 67. | Chirillo P, Falco M, Puri PL, Artini M, Balsano C, Levrero M, Natoli G. Hepatitis B virus pX activates NF-kappa B-dependent transcription through a Raf-independent pathway. J Virol. 1996;70:641-646. [PubMed] |
| 68. | Kim HR, Lee SH, Jung G. The hepatitis B viral X protein activates NF-kappaB signaling pathway through the up-regulation of TBK1. FEBS Lett. 2010;584:525-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 69. | Andrisani OM, Barnabas S. The transcriptional function of the hepatitis B virus X protein and its role in hepatocarcinogenesis (Review). Int J Oncol. 1999;15:373-379. [PubMed] |
| 70. | Benn J, Schneider RJ. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc Natl Acad Sci USA. 1994;91:10350-10354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 333] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 71. | Benn J, Su F, Doria M, Schneider RJ. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J Virol. 1996;70:4978-4985. [PubMed] |
| 72. | Um HR, Lim WC, Chae SY, Park S, Park JH, Cho H. Raf-1 and protein kinase B regulate cell survival through the activation of NF-kappaB in hepatitis B virus X-expressing cells. Virus Res. 2007;125:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 73. | Liu LP, Liang HF, Chen XP, Zhang WG, Yang SL, Xu T, Ren L. The role of NF-kappaB in Hepatitis b virus X protein-mediated upregulation of VEGF and MMPs. Cancer Invest. 2010;28:443-451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 74. | Sirma H, Giannini C, Poussin K, Paterlini P, Kremsdorf D, Bréchot C. Hepatitis B virus X mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene. 1999;18:4848-4859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 161] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 75. | Wu CS, Yen CJ, Chou RH, Chen JN, Huang WC, Wu CY, Yu YL. Downregulation of microRNA-15b by hepatitis B virus X enhances hepatocellular carcinoma proliferation via fucosyltransferase 2-induced Globo H expression. Int J Cancer. 2014;134:1638-1647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 76. | Zhang T, Zhang J, Cui M, Liu F, You X, Du Y, Gao Y, Zhang S, Lu Z, Ye L. Hepatitis B virus X protein inhibits tumor suppressor miR-205 through inducing hypermethylation of miR-205 promoter to enhance carcinogenesis. Neoplasia. 2013;15:1282-1291. [PubMed] |
| 77. | Wei X, Tan C, Tang C, Ren G, Xiang T, Qiu Z, Liu R, Wu Z. Epigenetic repression of miR-132 expression by the hepatitis B virus x protein in hepatitis B virus-related hepatocellular carcinoma. Cell Signal. 2013;25:1037-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 94] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 78. | Xu X, Fan Z, Kang L, Han J, Jiang C, Zheng X, Zhu Z, Jiao H, Lin J, Jiang K. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest. 2013;123:630-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 141] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 79. | Qiu X, Dong S, Qiao F, Lu S, Song Y, Lao Y, Li Y, Zeng T, Hu J, Zhang L. HBx-mediated miR-21 upregulation represses tumor-suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene. 2013;32:3296-3305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 99] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 80. | Chen YJ, Chien PH, Chen WS, Chien YF, Hsu YY, Wang LY, Chen JY, Lin CW, Huang TC, Yu YL. Hepatitis B Virus-Encoded X Protein Downregulates EGFR Expression via Inducing MicroRNA-7 in Hepatocellular Carcinoma Cells. Evid Based Complement Alternat Med. 2013;2013:682380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 81. | Wu G, Yu F, Xiao Z, Xu K, Xu J, Tang W, Wang J, Song E. Hepatitis B virus X protein downregulates expression of the miR-16 family in malignant hepatocytes in vitro. Br J Cancer. 2011;105:146-153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 82. | Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 274] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 83. | Fu X, Tan D, Hou Z, Hu Z, Liu G. miR-338-3p Is Down-Regulated by Hepatitis B Virus X and Inhibits Cell Proliferation by Targeting the 3’-UTR Region of CyclinD1. Int J Mol Sci. 2012;13:8514-8539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 84. | Fu X, Tan D, Hou Z, Hu Z, Liu G, Ouyang Y, Liu F. The effect of miR-338-3p on HBx deletion-mutant (HBx-d382) mediated liver-cell proliferation through CyclinD1 regulation. PLoS One. 2012;7:e43204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 85. | Xie QH, He XX, Chang Y, Jiang X, Lin JS. [HBx gene down-regulates miR-192 expression and inhibits apoptosis of human hepatoma cell line HepG2]. Zhonghua Gan Zang Bing Za Zhi. 2011;19:857-860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 86. | Paterlini P, Poussin K, Kew M, Franco D, Brechot C. Selective accumulation of the X transcript of hepatitis B virus in patients negative for hepatitis B surface antigen with hepatocellular carcinoma. Hepatology. 1995;21:313-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 87. | Wang WL, London WT, Feitelson MA. Hepatitis B x antigen in hepatitis B virus carrier patients with liver cancer. Cancer Res. 1991;51:4971-4977. [PubMed] |
| 88. | Chen GG, Li MY, Ho RL, Chak EC, Lau WY, Lai PB. Identification of hepatitis B virus X gene mutation in Hong Kong patients with hepatocellular carcinoma. J Clin Virol. 2005;34:7-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 89. | Iavarone M, Trabut JB, Delpuech O, Carnot F, Colombo M, Kremsdorf D, Bréchot C, Thiers V. Characterisation of hepatitis B virus X protein mutants in tumour and non-tumour liver cells using laser capture microdissection. J Hepatol. 2003;39:253-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 63] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 90. | Guo X, Jin Y, Qian G, Tu H. Sequential accumulation of the mutations in core promoter of hepatitis B virus is associated with the development of hepatocellular carcinoma in Qidong, China. J Hepatol. 2008;49:718-725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 75] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 91. | Poussin K, Dienes H, Sirma H, Urban S, Beaugrand M, Franco D, Schirmacher P, Bréchot C, Paterlini Bréchot P. Expression of mutated hepatitis B virus X genes in human hepatocellular carcinomas. Int J Cancer. 1999;80:497-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 92. | Liu L, Li Y, Zhang S, Yu D, Zhu M. Hepatitis B virus X protein mutant upregulates CENP-A expression in hepatoma cells. Oncol Rep. 2012;27:168-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 93. | Ma NF, Lau SH, Hu L, Xie D, Wu J, Yang J, Wang Y, Wu MC, Fung J, Bai X. COOH-terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin Cancer Res. 2008;14:5061-5068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 138] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 94. | Kumar V, Jayasuryan N, Kumar R. A truncated mutant (residues 58-140) of the hepatitis B virus X protein retains transactivation function. Proc Natl Acad Sci USA. 1996;93:5647-5652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 117] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 95. | Liu X, Wang L, Zhang S, Lin J, Zhang S, Feitelson MA, Gao H, Zhu M. Mutations in the C-terminus of the X protein of hepatitis B virus regulate Wnt-5a expression in hepatoma Huh7 cells: cDNA microarray and proteomic analyses. Carcinogenesis. 2008;29:1207-1214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 46] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 96. | Wang Q, Zhang T, Ye L, Wang W, Zhang X. Analysis of hepatitis B virus X gene (HBx) mutants in tissues of patients suffered from hepatocellular carcinoma in China. Cancer Epidemiol. 2012;36:369-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 97. | Wang Q, Zhang WY, Ye LH, Zhang XD. A mutant of HBx (HBxDelta127) promotes hepatoma cell growth via sterol regulatory element binding protein 1c involving 5-lipoxygenase. Acta Pharmacol Sin. 2010;31:367-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 98. | Jiang W, Wang XW, Unger T, Forgues M, Kim JW, Hussain SP, Bowman E, Spillare EA, Lipsky MM, Meck JM. Cooperation of tumor-derived HBx mutants and p53-249(ser) mutant in regulating cell proliferation, anchorage-independent growth and aneuploidy in a telomerase-immortalized normal human hepatocyte-derived cell line. Int J Cancer. 2010;127:1011-1020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 99. | Hsu IC, Metcalf RA, Sun T, Welsh JA, Wang NJ, Harris CC. Mutational hotspot in the p53 gene in human hepatocellular carcinomas. Nature. 1991;350:427-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1066] [Cited by in RCA: 969] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 100. | Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, Raimondo G, Levrero M. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci USA. 2009;106:19975-19979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 400] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 101. | Yeh CT, Shen CH, Tai DI, Chu CM, Liaw YF. Identification and characterization of a prevalent hepatitis B virus X protein mutant in Taiwanese patients with hepatocellular carcinoma. Oncogene. 2000;19:5213-5220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 61] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 102. | Hsia CC, Yuwen H, Tabor E. Hot-spot mutations in hepatitis B virus X gene in hepatocellular carcinoma. Lancet. 1996;348:625-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 54] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 103. | Hsia CC, Nakashima Y, Tabor E. Deletion mutants of the hepatitis B virus X gene in human hepatocellular carcinoma. Biochem Biophys Res Commun. 1997;241:726-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 104. | Wang Q, Zhang W, Liu Q, Zhang X, Lv N, Ye L, Zhang X. A mutant of hepatitis B virus X protein (HBxDelta127) promotes cell growth through a positive feedback loop involving 5-lipoxygenase and fatty acid synthase. Neoplasia. 2010;12:103-115. [PubMed] |
| 105. | Tang H, Delgermaa L, Huang F, Oishi N, Liu L, He F, Zhao L, Murakami S. The transcriptional transactivation function of HBx protein is important for its augmentation role in hepatitis B virus replication. J Virol. 2005;79:5548-5556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 170] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 106. | Lin X, Xu X, Huang QL, Liu YQ, Zheng DL, Chen WN, Lin JY. Biological impacts of “hot-spot” mutations of hepatitis B virus X proteins are genotype B and C differentiated. World J Gastroenterol. 2005;11:4703-4708. [PubMed] |
| 107. | Tangkijvanich P, Sa-Nguanmoo P, Mahachai V, Theamboonlers A, Poovorawan Y. A case-control study on sequence variations in the enhancer II/core promoter/precore and X genes of hepatitis B virus in patients with hepatocellular carcinoma. Hepatol Int. 2010;4:577-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 108. | Zhang H, Shan CL, Li N, Zhang X, Zhang XZ, Xu FQ, Zhang S, Qiu LY, Ye LH, Zhang XD. Identification of a natural mutant of HBV X protein truncated 27 amino acids at the COOH terminal and its effect on liver cell proliferation. Acta Pharmacol Sin. 2008;29:473-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 43] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 109. | Gouas DA, Villar S, Ortiz-Cuaran S, Legros P, Ferro G, Kirk GD, Lesi OA, Mendy M, Bah E, Friesen MD. TP53 R249S mutation, genetic variations in HBX and risk of hepatocellular carcinoma in The Gambia. Carcinogenesis. 2012;33:1219-1224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 110. | Muroyama R, Kato N, Yoshida H, Otsuka M, Moriyama M, Wang Y, Shao RX, Dharel N, Tanaka Y, Ohta M. Nucleotide change of codon 38 in the X gene of hepatitis B virus genotype C is associated with an increased risk of hepatocellular carcinoma. J Hepatol. 2006;45:805-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 111. | Kwun HJ, Jang KL. Natural variants of hepatitis B virus X protein have differential effects on the expression of cyclin-dependent kinase inhibitor p21 gene. Nucleic Acids Res. 2004;32:2202-2213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 112. | Qadri I, Maguire HF, Siddiqui A. Hepatitis B virus transactivator protein X interacts with the TATA-binding protein. Proc Natl Acad Sci USA. 1995;92:1003-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 157] [Article Influence: 5.2] [Reference Citation Analysis (0)] |