Published online Jul 14, 2014. doi: 10.3748/wjg.v20.i26.8653
Revised: March 17, 2014
Accepted: April 27, 2014
Published online: July 14, 2014
Processing time: 168 Days and 15.4 Hours
AIM: To perform plasma free amino acid (PFAA) profiling of esophageal squamous cell carcinoma (ESCC) patients at different pathological stages and healthy subjects.
METHODS: Plasma samples from ESCC patients (n = 51) and healthy control adults (n = 60) were analyzed by high-performance liquid chromatography (HPLC). The ESCC patients included moderate/poorly-differentiation (n = 24), lymph node metastasis (n = 17) and clinical stage > Ib2 (n = 36). Partial least squares discriminant analysis was performed to demonstrate that the PFAA metabolic patterns enabled discrimination between ESCC patients and controls, and the Student t test was applied to assess significant differences in PFAA concentrations between the two groups.
RESULTS: There were significant differences in the PFAA profiles between controls and ESCC patients. Compared with healthy controls, the levels of Asp, Glu, Gly, His, Thr, Tau, Ala, Met, Ile, Leu, and Phe were decreased in ESCC patients, but Cys was increased. There exists a strong correlation between PFAA profiles and clinicopathological characteristics in ESCC patients. The levels of many PFAAs (i.e., Glu, Asp, Ser, Gly, Tau, Ala, Tyr, Val, Ile, and Leu) were related to pathological grading, lymph node metastasis, and ESCC clinical stage. Very good discrimination between ESCC patients and control subjects was achieved by multivariate modeling of plasma profiles.
CONCLUSION: HPLC-based plasma profiling analysis was shown to be an effective approach to differentiate between ESCC patients and controls. PFAA profiles may have potential value for screening or diagnosing ESCC.
Core tip: Recently, metabolomics-based techniques have been developed to identify cancer-related metabolic signatures for early cancer detection. However, studies on esophageal squamous cell carcinoma (ESCC) remain limited. This study used high-performance liquid chromatography to quantitatively study plasma free amino acid (PFAA) changes in ESCC patient, and analyze the correlation between PFAA profiles and clinicopathological characteristics of ESCC. The results showed that most of the amino acids were differentially expressed in ESCC patients and control subjects, and were related to the ESCC clinicopathological characteristics. The study suggests that PFAA profiling is of potential value for screening or diagnosing ESCC.
- Citation: Ma H, Hasim A, Mamtimin B, Kong B, Zhang HP, Sheyhidin I. Plasma free amino acid profiling of esophageal cancer using high-performance liquid chromatography spectroscopy. World J Gastroenterol 2014; 20(26): 8653-8659
- URL: https://www.wjgnet.com/1007-9327/full/v20/i26/8653.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i26.8653
Esophageal cancer (EC) is the 8th most common cancer worldwide, and ranked 6th as the leading cause of cancer-related deaths[1]. Approximately 386000 individuals worldwide die from EC every year, with 160000 cases in China[2]. Esophageal squamous cell carcinoma (ESCC) is the most common histologic EC type in China. Xinjiang is one of the high-risk areas in China, with an adjusted mortality rate of Kazakh EC as high as 155.9/100000 in Toli county of northern Xinjiang[3], which is higher than different ethnicities of the same area and the average rate in other nations. EC has become the leading cause of cancer deaths in the Kazakh ethnic group. Because of the lack of specific signs or symptoms at an early stage, the majority of patients present at an advanced stage. Despite recent advances in multimodal treatment, overall 5-year survival rates for advanced EC are only 5%-15% due to lymph node metastasis and local recurrence[4,5]. Currently, conventional endoscopic detection combining biopsy is the main method for EC diagnosis; however, it is associated with significant limitations because dysplastic and early carcinomatous lesions are not macroscopically visible. Therefore, a screening method that facilitates early detection of EC is of crucial clinical importance.
Amino acids are biologically important organic compounds, which play central roles both as building blocks of proteins and as intermediates in metabolism. The free amino acids are distributed throughout the body to participate in metabolism, and are termed the amino acid pool. Malignancy is characterized by the fast speed of cell multiplication and enhanced metabolism, showing that malignant cells require a large number of amino acids from the amino acid pool to synthesize protein and nucleic acid[6]. Consequently, changes in the plasma free amino acid (PFAA) profile might reflect the cancer-induced protein metabolism in tumors, skeletal muscle, and liver in cancer patients. However, previous studies reported that the PFAA profile is not the same in different cancers[7]. Miyagi et al[8] investigated the differences in PFAA profiles from five types of cancer patients, including lung cancer, gastric cancer, colorectal cancer, breast cancer, and prostate cancer. The results showed that some of the amino acids (such as tyrosine, glycine, leucine, phenylalanine) had a close link with the specific cancers, indicating that PFAA profiles correlate with the organ-site origin among the five different cancers. Regarding ESCC, few studies have focused on the quantitation of PFAA profiles.
Recently, metabolomics has developed rapidly as a branch of systems biology. Metabolomics studies the global assessment and validation of endogenous small-molecule biochemicals (metabolites) within a biological system, including cells, tissues, or organisms[9,10]. These metabolites may directly or indirectly interact with molecular targets and thereby influence the risk and complications associated with various diseases, including cancer[11]. Metabolomics-based methods have been widely used in disease diagnosis, biomarker screening, gene modifications, drug efficacy and toxicity[12-14]. The main analytical techniques for metabolomic studies are based on nuclear magnetic resonance (NMR), gas chromatography/liquid chromatography-mass spectrometry and high-performance liquid chromatography (HPLC). HPLC as an outstanding metabolomic tool together with multivariate statistics is widely used in metabolite identification and quantification. Compared with other analytical techniques, HPLC has already shown excellent discrimination and high sensitivity, with minimal effort and at a reasonable cost[15].
Previously, we described the application of 1HNMR-based metabolomics in differentiating the metabolic profiles of plasma in ESCC patients and healthy controls. We discovered that a series of metabolites in glucose metabolism, fatty acid metabolism and the tricarboxylic acid cycle showed an altered expression level in ESCC patient’s plasma[16]. In this study, we used HPLC to quantitatively detect PFAA profiles of ESCC patients and healthy people, and systematically analyzed the discriminating metabolites in the different pathological stages of ESCC. Our study may be helpful for improving non-invasive ESCC screening or diagnosis and for providing novel insights about EC metabolism.
This study was approved by the Ethics Committee of the Medical University of Xinjiang. Written informed consent was obtained from all participants before study participation. Plasma samples were collected preoperatively from Kazakh patients from Xinjiang with ESCC (n = 51) at the Department of Thoracic Surgery of the First Affiliated Hospital in Medical University of Xinjiang, between June 2010 and March 2012. Patients were between 37 and 80 years old (mean age: 57 years), and had not received chemoradiation therapy. Tumors were pathologically confirmed and classified according to World Health Organization classification standards. The patients included 27 cases of well-differentiated and 24 cases of moderately/poorly-differentiated ESCC. Of these patients, 17 cases were positive for lymph node metastasis, 15 cases had stage ≤ Ib2 disease and 36 cases had stage > Ib2 disease. Controls (n = 60) were healthy age- and sex-matched male and female volunteers. Exclusion criteria included prior or concurrent neoplasm, or cardiovascular, hepatic, renal, or inflammatory disease. Blood samples were collected from patients and controls between 7-8 am, were centrifuged at 4000 rpm for 10 min, and plasma samples were isolated and stored at -80 °C until use.
Plasma samples were thawed in a 4 °C water bath, and vortexed with 400 μL of methyl cyanide for 30 s to precipitate proteins. This solution was again centrifuged at 12000 rpm for 15 min, and the separated supernatant was transferred to 1.5 mL microtubes and vortexed for 30 s. Ten microliter aliquots of supernatant were sampled by amino acid-calibrated HPLC (Alliance® e2695 HPLC unit; Waters Corp., Milford, MA, United States). Ten microliters of plasma supernatant were mixed in 60 μL of boric acid solution (pH 8.8) and vortexed for 30 s, followed by warming in a 55 °C water bath for 15 min. Five microliters of each sample were injected into the HPLC system for analysis. Each standard was individually run on the gradient noted in the LC parameters program (reaction temperature 35 °C, gradient elution analysis). The extracted phenols were detected by an octadecylsilyl column with a gradient elution and programmable fluorescence wavelength detector, column size 250 mm × 4.6 mm, flow rate 1.0 mL per detection, excitation wavelength 235 nm, emission wavelength 395 nm, injection volume 5 μL). Two consecutive runs with the same retention times were taken, and the means were plotted for calibration graphs. From the chromatograms thus obtained, the areas of the peaks were recorded with the standards and used for calibration.
The following 19 amino acids were analyzed: aspartate (Asp), glutamate (Glu), asparagine (Asn), serine (Ser), glycine (Gly), histidine (His), arginine (Arg), threonine (Thr), taurine (Tau), alanine (Ala), proline (Pro), cysteine (Cys), tyrosine (Tyr), valine (Val), methionine (Met), lysine (Lys), isoleucine (Ile), leucine (Leu), and phenylalanine (Phe). The absolute concentration of each amino acid was expressed in μmol/L.
The metabolic data were analyzed statistically using SPSS v.17.0 software (SPSS Inc., Chicago, IL, United States), and the mean amino acid concentrations standard deviations (SDs) were calculated to determine PFAA profiles for both ESCC patients and controls. The Student t-test was used to assess significant differences in the PFAA concentrations between cancer patients and controls. P-values < 0.05 were considered statistically significant. After the chromatographic peak area was normalized, partial least squares-discriminant analysis (PLS-DA) was performed to construct plasma amino acid metabolic profiles of ESCC patients and controls.
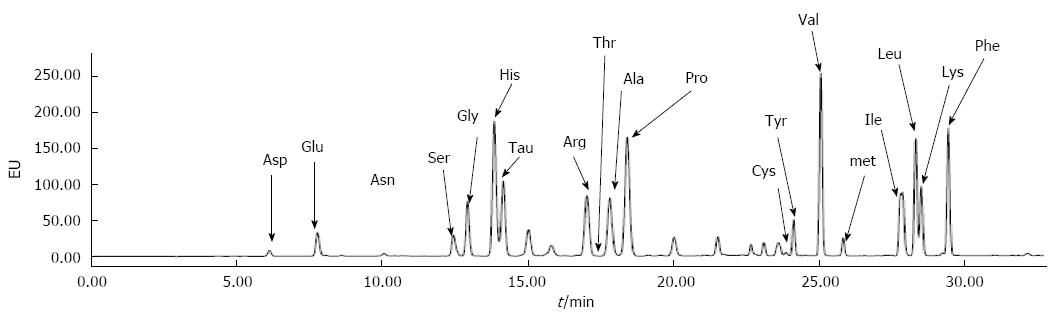
The representative chromatograms of human plasma amino acids are shown in Figure 1. The method demonstrated good chromatographic separation of all the amino acids within a short analysis time. Retention times for Asp, Glu, Asn, Ser, Gly, His, Tau, Arg, Thr, Ala, Pro, Cys, Tyr, Val, Met, Ile, Leu, Lys, and Phe were approximately 6.1, 7.7, 10.1, 12.4, 12.8,13.8, 14.1, 17.0, 17.3, 17.8, 18.2, 23.9, 24.1, 25.0, 25.8, 27.9, 28.3, 28.5 and 29.4 min, respectively. There was no interference from other amino acids or other endogenous plasma compounds.
Univariate analysis was used to compare the PFAA profiles of controls vs ESCC patients. The levels of each amino acid are shown in Table 1. There were significant differences in the PFAA profiles between the controls and the ESCC patients. Compared with healthy controls, the plasma concentrations of all PFAAs were decreased in ESCC patients, except for Cys. The plasma concentrations of Asp, Glu, Gly, His, Thr, Tau, Ala, Met, Ile, Leu, and Phe were significantly decreased (P < 0.05) in ESCC patients compared with controls.
| Amino acid (μmol/L) | Control | ESCC | P value |
| Aspartate | 0.04 ± 0.03 | 0.06 ± 0.07 | < 0.001 |
| Glutamate | 0.37 ± 0.30 | 0.07 ± 0.06 | < 0.001 |
| Asparagine | 0.06 ± 0.04 | 0.05 ± 0.04 | 0.36 |
| Serine | 0.27 ± 0.51 | 0.12 ± 0.12 | 0.51 |
| Glycine | 0.14 ± 0.09 | 0.07 ± 0.03 | < 0.001 |
| Histidine | 0.28 ± 0.17 | 0.16 ± 0.11 | < 0.001 |
| Arginine | 0.09 ± 0.05 | 0.09 ± 0.15 | 0.77 |
| Threonine | 0.21 ± 0.21 | 0.08 ± 0.08 | < 0.001 |
| Taurine | 0.26 ± 0.18 | 0.13 ± 0.12 | < 0.001 |
| Alanine | 0.98 ± 1.06 | 0.19 ± 0.18 | < 0.001 |
| Proline | 0.08 ± 0.06 | 0.06 ± 0.07 | 0.19 |
| Cystine | 0.01 ± 0.02 | 0.03 ± 0.04 | 0.01 |
| Tyrosine | 0.07 ± 0.06 | 0.06 ± 0.03 | 0.50 |
| Valine | 0.24 ± 0.16 | 0.23 ± 0.29 | 0.80 |
| Methionine | 0.12 ± 0.19 | 0.02 ± 0.01 | < 0.001 |
| Lysine | 0.09 ± 0.05 | 0.08 ± 0.07 | 0.45 |
| Isoleucine | 0.29 ± 0.33 | 0.12 ± 0.08 | < 0.001 |
| Leucine | 0.21 ± 0.16 | 0.12 ± 0.08 | < 0.001 |
| Phenylalanine | 0.12 ± 0.07 | 0.07 ± 0.03 | < 0.001 |
We further evaluated the association of PFAA profiles with tumor invasiveness, metastasis, and differentiation. All ESCC cases were classified by different pathologic criteria, including lymph node metastasis, differentiation grade, and clinical stage. Statistical analysis showed that the concentrations of most amino acids were significantly associated with lymph node metastasis, differentiation grade, and clinical stage (P < 0.05 in all cases). When comparing positive lymph node metastases vs negative node metastases, except for Arg, Thr, and Cys, the concentrations of the remaining tested amino acids were decreased (Table 2). The PFAA levels were significantly lower in patients with late-stage disease (> Ib2) vs early-stage disease (≤ Ib2), except for Asp, Arg, Pro, Cys, and Lys (Table 2). The PFAAs were also lower in all cancer patients with middle/poorly differentiated cancers vs well differentiated cancers, except for His, Pro, Met, and Phe (Table 2).
| Pathological differentiation | Lymph node metastasis | Clinical stage | |||||||
| Amino acid | Mid/poor | Well | P value | Positive | Negative | P value | ≤Ib2 | > Ib2 | P value |
| Aspartate | 0.06 ± 0.07 | 0.09 ± 0.04 | 0.04 | 0.00 ± 0.00 | 0.01 ± 0.01 | 0.01 | 0.01 ± 0.01 | 0.00 ± 0.00 | 0.14 |
| Glutamate | 0.03 ± 0.02 | 0.07 ± 0.05 | < 0.001 | 0.03 ± 0.02 | 0.09 ± 0.06 | < 0.001 | 0.11 ± 0.04 | 0.06 ± 0.06 | < 0.001 |
| Asparagine | 0.07 ± 0.04 | 0.18 ± 0.16 | < 0.001 | 0.03 ± 0.02 | 0.06 ± 0.04 | 0.01 | 0.09 ± 0.06 | 0.04 ± 0.02 | < 0.001 |
| Serine | 0.11 ± 0.08 | 0.22 ± 0.11 | < 0.001 | 0.05 ± 0.03 | 0.16 ± 0.14 | < 0.001 | 0.23 ± 0.19 | 0.08 ± 0.04 | 0.01 |
| Glycine | 0.06 ± 0.04 | 0.09 ± 0.02 | < 0.001 | 0.10 ± 0.08 | 0.19 ± 0.10 | < 0.001 | 0.22 ± 0.12 | 0.14 ± 0.09 | 0.01 |
| Histidine | 0.06 ± 0.03 | 0.12 ± 0.22 | 0.22 | 0.04 ± 0.03 | 0.08 ± 0.03 | < 0.001 | 0.09 ± 0.03 | 0.06 ± 0.03 | < 0.001 |
| Arginine | 0.05 ± 0.05 | 0.12 ± 0.09 | < 0.001 | 0.06 ± 0.04 | 0.10 ± 0.19 | 0.35 | 0.14 ± 0.28 | 0.07 ± 0.04 | 0.34 |
| Threonine | 0.10 ± 0.10 | 0.17 ± 0.13 | 0.02 | 0.05 ± 0.06 | 0.10 ± 0.09 | 0.06 | 0.06 ± 0.07 | 0.06 ± 0.07 | < 0.001 |
| Taurine | 0.12 ± 0.12 | 0.28 ± 0.20 | < 0.001 | 0.06 ± 0.06 | 0.17 ± 0.13 | < 0.001 | 0.20 ± 0.13 | 0.11 ± 0.10 | 0.01 |
| Alanine | 0.04 ± 0.03 | 0.09 ± 0.09 | 0.02 | 0.07 ± 0.07 | 0.26 ± 0.18 | < 0.001 | 0.30 ± 0.22 | 0.15 ± 0.14 | 0.02 |
| Proline | 0.02 ± 0.04 | 0.03 ± 0.05 | 0.552 | 0.04 ± 0.02 | 0.07 ± 0.08 | 0.02 | 0.09 ± 0.10 | 0.05 ± 0.05 | 0.18 |
| Cystine | 0.04 ± 0.02 | 0.09 ± 0.03 | < 0.001 | 0.02 ± 0.05 | 0.03 ± 0.05 | 0.74 | 0.03 ± 0.04 | 0.02 ± 0.05 | 0.59 |
| Tyrosine | 0.12 ± 0.06 | 0.36 ± 0.38 | < 0.001 | 0.04 ± 0.02 | 0.08 ± 0.03 | < 0.001 | 0.10 ± 0.03 | 0.05 ± 0.02 | < 0.001 |
| Valine | 0.02 ± 0.01 | 0.03 ± 0.01 | < 0.001 | 0.09 ± 0.06 | 0.30 ± 0.33 | 0.01 | 0.45 ± 0.47 | 0.14 ± 0.07 | 0.02 |
| Methionine | 0.07 ± 0.10 | 0.09 ± 0.03 | 0.36 | 0.02 ± 0.01 | 0.03 ± 0.01 | < 0.001 | 0.04 ± 0.01 | 0.02 ± 0.01 | < 0.001 |
| Lysine | 0.07 ± 0.03 | 0.19 ± 0.06 | < 0.001 | 0.05 ± 0.04 | 0.09 ± 0.08 | 0.03 | 0.09 ± 0.03 | 0.07 ± 0.08 | 0.33 |
| Isoleucine | 0.06 ± 0.04 | 0.18 ± 0.07 | < 0.001 | 0.05 ± 0.03 | 0.16 ± 0.07 | < 0.001 | 0.21 ± 0.07 | 0.09 ± 0.05 | < 0.001 |
| Leucine | 0.05 ± 0.02 | 0.09 ± 0.02 | < 0.001 | 0.05 ± 0.04 | 0.15 ± 0.08 | < 0.001 | 0.20 ± 0.08 | 0.08 ± 0.05 | < 0.001 |
| Phenylalanine | 0.00 ± 0.00 | 0.01 ± 0.01 | 0.12 | 0.04 ± 0.02 | 0.08 ± 0.03 | < 0.001 | 0.10 ± 0.02 | 0.05 ± 0.02 | < 0.001 |
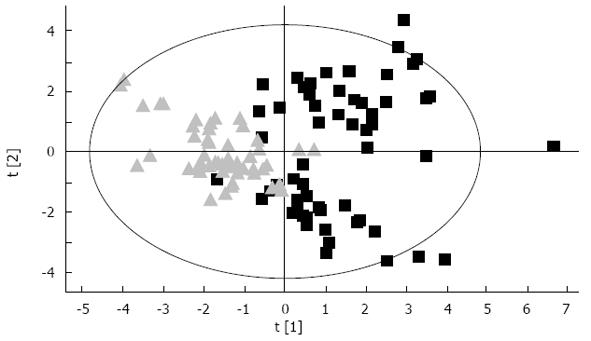
SIMCA-P 11 (Umetrics Inc., Umea, Sweden) software is used for PLS-DA of the data from HPLC. The scores plot shows each set of two groups scattering into different regions, representing a good separation of the cancer from non-cancer cases with corresponding plasma amino acid pattern, and suggested that the ESCC have a specific profile distinguishable from healthy controls (Figure 2).
EC is one of the most frequent malignancies in China, and is a serious threat to human health. Effectively improving EC prognoses is directly dependent upon early diagnosis and treatment. Tumor biological markers such as carcinoembryonic antigen and epithelial membrane antigen are not useful for early EC diagnosis, because of low sensitivity and specificity. HPLC is one of the most powerful chromatographic techniques, and can often easily achieve separations and analyses that would be difficult or impossible using other forms of chromatography.
In the present study, we performed PFAA profiling using HPLC spectroscopy, and identified useful metabolic markers for ESCC screening or diagnosis. The results showed that most of the amino acids were differentially expressed in ESCC patients and control subjects, and were related to the ESCC clinicopathological characteristics. The PLS-DA model identified patients with ESCC and healthy controls, suggesting that PFAA profiling is useful for screening or diagnosis of ESCC.
In previous studies, the changes in PFAA profiles in cancer patients were sometimes inconsistent[17]. Some discrepancies exist between our current study and other prior reports. For example, Shingyoji et al[18] reported that HPLC-determined plasma Gly levels are increased in lung cancer patients. However, plasma Gly was decreased in ESCC patients in our study. This difference may be due not only to sample size but also to cancers from different organs having different PFAA profiles. There are also many similarities between our results and previous studies. Our study showed a significant reduction in gluconeogenic amino acids (such as Ala, His, and Gly) in ESCC patients, which is similar to that seen in esophageal adenocarcinoma plasma metabolomics as well as in other cancers[14,19]. This reveals a typical signature in cancer patients, and it has previously been proven that tumors rely on glycolysis as a main source of energy, even in the presence of oxygen[20].
Recent research has shown that several non-essential amino acids (i.e., Glu, Ser, and Gly) play critical roles in cancer metabolism[21]. In the present study, the levels of Gly, Ala, Glu, and Asp were significantly decreased in ESCC patients vs healthy controls. Gly is the best discriminating amino acid because it is an important intermediate in folate metabolism, which is especially altered in colon cancer[22]. Due to its recently demonstrated bowel-protective effects and its easy administration, Gly is a highly interesting candidate for therapeutic approaches[23]. It has been reported that Gly consumption and expression of the mitochondrial Gly biosynthetic pathway proteins are strongly correlated with rapid cancer cell proliferation[24]. In our study, plasma Gly concentrations were decreased in ESCC patients, and were associated with poorly-differentiation, non-lymph node metastasis, and late stage disease (> Ib2). These results further confirm that disturbances in Gly metabolism can promote tumor growth and proliferation.
Ala was described as a precursor of gluconeogenesis because it is the key protein-derived glucose precursor used by the liver[25]. Tessem et al[26] used 1H high-resolution magic angle spinning spectroscopy approach to investigate the potential role of Ala as metabolic biomarker in prostate biopsy tissues, and found that Ala levels were significantly higher in prostate cancer vs benign prostate tissues. In our ESCC patients, especially those with late-stage disease, Ala levels were decreased significantly compared with healthy controls. This observation may reflect that malignant tumors are associated with increased glycolytic flux. This is consistent with the need for increased protein synthesis in tumors, which leads to increased Ala consumption by the tumor tissues, causing low plasma Ala levels in ESCC. Additionally, the transamination to Ala from pyruvate is catalyzed by the enzyme alanine transaminase (ALT); therefore, further studies are needed to establish the activity of ALT and find the underlying biochemical pathways for Ala use in ESCC. Previous work suggests that Glu oxidation may be a major source of respiratory energy for tumor cells[27]. Our present results showed that Glu levels was decreased in plasma of ESCC patients, which is in agreement with other studies that showed Glu transamination primarily to pyruvate to form Ala[28]. This increased Ala concentration may enhance gluconeogenesis by proliferating cancer cells.
The branched-chain amino acids, Leu, Ile, and Val, are essential amino acids that serve both as important energy substrates and as precursors for the synthesis of other amino acids and proteins[29]. In the present study, the Leu and Ile levels in the plasma of patients with ESCC were significantly lower than those in normal controls. The cause may be that a large number of amino acids were consumed as a substrate for synthesis of proteins and nucleic acids and other substances. As the source of one carbon unit, histidine supplies the formanimo to tetrahydrofolate, which participates in the synthesis of purines and pyrimidines. Because tumor cells have an active nucleic acid metabolism, histidine is excessively absorbed into tumor tissue, and consumption is markedly increased, leading to lower levels in the body.
Furthermore, we noticed that the levels of a large group of PFAAs (Glu, Asp, Ser, Gly, Tau, Ala, Tyr, Val, Ile, and Leu) were decreased in poorly-differentiated, lymph node metastasizing, late stage (> Ib2) ESCC vs well-differentiated, non-lymph node metastasizing, and early stage (≤ Ib2) disease. The results showed that PFAA profiles were strongly correlated with ESCC tumor progression, and may probably reflect an increased demand for amino acids to supply the growing tumor. Thus, PFAA profiles may be valuable molecular markers for tumor progression, recurrence, and prognosis.
In conclusion, PFAA profiles in ESCC patients are significantly different from those in healthy persons, using HPLC spectroscopy. Additionally, there exists a strong correlation between PFAA profiles and clinicopathological characteristics in ESCC patients. We believe that the non-invasive HPLC-based metabolic method has a potential role in improving early diagnosis and screening of high-ESCC risk populations, and in helping judge disease prognosis. In prospective studies, we will further verify our results using a larger sample size, and aim to identify the underlying mechanisms involved in the coupling of amino acid metabolism to altered reprogramming of gene expression in ESCC.
Recently, metabolomics-based techniques have been developed to identify cancer-related metabolic signatures for early cancer detection. Among metabolites, profiling of plasma free amino acids (PFAAs) is a promising approach because PFAAs link all organ systems and have important roles in metabolism. However, several investigators have reported that PFAA profiles are not the same in different cancers, including esophageal squamous cell carcinoma (ESCC). Therefore, this study aimed to identify the PFAA profile of human plasma as a means to identify an ESCC-related metabolic signature, which may be helpful for improving non-invasive ESCC screening or diagnosis and for providing novel insights about ESCC metabolism.
PFAA profiles have been shown to be influenced by metabolic variations in different solid tumors. Regarding ESCC, few studies have focused on the quantitation of PFAA profiles. In this study, authors used high-performance liquid chromatography to determine the PFAA profiles of patients with ESCC and compared the alterations between ESCC patients and controls using univariate analyses.
This is the first study to quantitatively detect PFAA profiles of ESCC patients and healthy people, and systematically analyze the discriminating metabolites in the different pathological stages of ESCC. The results showed that most of the amino acids were differentially expressed in ESCC patients and control subjects, and were related to the ESCC clinicopathological characteristics. The partial least squares-discriminant analysis model identified patients with ESCC and healthy controls, suggesting that PFAA profiling is useful for screening or diagnosis of ESCC.
PFAA profiles may be helpful for improving non-invasive ESCC screening or diagnosis and for providing novel insights about ESCC metabolism.
Metabolomics is a quantitative measurement of the dynamic multiparametric metabolic response of living systems to pathophysiological stimuli or genetic modification. Amino acids and their metabolites, which play important roles as both basic substrates and regulators in many metabolic pathways. Amino acid metabolism in cancer patients differs from that in healthy people. Some studies indicated that free amino acid profiling is of potential value for screening or diagnosing cancer.
The authors performed PFAA profile of ESCC and its association with clinicopathological characteristics. It revealed that most of the amino acids were differentially expressed in ESCC patients and controls, and were related to the ESCC clinicopathological characteristics. The results derived from this study are well-presented and has potential clinical value for the screening and diagnosis of esophageal cancer.
P- Reviewer: Chen GS S- Editor: Gou SX L- Editor: Cant MR E- Editor: Zhang DN
| 1. | Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137-2150. [PubMed] |
| 2. | Ruol A, Castoro C, Portale G, Cavallin F, Sileni VC, Cagol M, Alfieri R, Corti L, Boso C, Zaninotto G. Trends in management and prognosis for esophageal cancer surgery: twenty-five years of experience at a single institution. Arch Surg. 2009;144:247-254; discussion 254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 89] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 3. | Zhang YM. The distribution of esophageal cancer in Xinjiang. Xinjiang Yike Daxue Xuebao. 1988;11:139-144. |
| 4. | Koshy M, Esiashvilli N, Landry JC, Thomas CR, Matthews RH. Multiple management modalities in esophageal cancer: combined modality management approaches. Oncologist. 2004;9:147-159. [PubMed] |
| 5. | Lambert R, Hainaut P. The multidisciplinary management of gastrointestinal cancer. Epidemiology of oesophagogastric cancer. Best Pract Res Clin Gastroenterol. 2007;21:921-945. [PubMed] |
| 6. | Altman BJ, Dang CV. Normal and cancer cell metabolism: lymphocytes and lymphoma. FEBS J. 2012;279:2598-2609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 7. | Lai HS, Lee JC, Lee PH, Wang ST, Chen WJ. Plasma free amino acid profile in cancer patients. Semin Cancer Biol. 2005;15:267-276. [PubMed] |
| 8. | Miyagi Y, Higashiyama M, Gochi A, Akaike M, Ishikawa T, Miura T, Saruki N, Bando E, Kimura H, Imamura F. Plasma free amino acid profiling of five types of cancer patients and its application for early detection. PLoS One. 2011;6:e24143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 339] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 9. | Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181-1189. [PubMed] |
| 10. | Fiehn O. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comp Funct Genomics. 2001;2:155-168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 644] [Cited by in RCA: 555] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 11. | Chadeau-Hyam M, Ebbels TM, Brown IJ, Chan Q, Stamler J, Huang CC, Daviglus ML, Ueshima H, Zhao L, Holmes E. Metabolic profiling and the metabolome-wide association study: significance level for biomarker identification. J Proteome Res. 2010;9:4620-4627. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 92] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 12. | Hasim A, Ali M, Mamtimin B, Ma JQ, Li QZ, Abudula A. Metabonomic signature analysis of cervical carcinoma and precancerous lesions in women by (1)H NMR spectroscopy. Exp Ther Med. 2012;3:945-951. [PubMed] |
| 13. | Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res. 2009;15:431-440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 582] [Cited by in RCA: 539] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 14. | Mamas M, Dunn WB, Neyses L, Goodacre R. The role of metabolites and metabolomics in clinically applicable biomarkers of disease. Arch Toxicol. 2011;85:5-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 254] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 15. | Shimbo K, Kubo S, Harada Y, Oonuki T, Yokokura T, Yoshida H, Amao M, Nakamura M, Kageyama N, Yamazaki J. Automated precolumn derivatization system for analyzing physiological amino acids by liquid chromatography/mass spectrometry. Biomed Chromatogr. 2010;24:683-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 16. | Hasim A, Ma H, Mamtimin B, Abudula A, Niyaz M, Zhang LW, Anwer J, Sheyhidin I. Revealing the metabonomic variation of EC using ¹H-NMR spectroscopy and its association with the clinicopathological characteristics. Mol Biol Rep. 2012;39:8955-8964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 17. | Kubota A, Meguid MM, Hitch DC. Amino acid profiles correlate diagnostically with organ site in three kinds of malignant tumors. Cancer. 1992;69:2343-2348. [PubMed] |
| 18. | Shingyoji M, Iizasa T, Higashiyama M, Imamura F, Saruki N, Imaizumi A, Yamamoto H, Daimon T, Tochikubo O, Mitsushima T. The significance and robustness of a plasma free amino acid (PFAA) profile-based multiplex function for detecting lung cancer. BMC Cancer. 2013;13:77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 19. | Wang L, Chen J, Chen L, Deng P, Bu Q, Xiang P, Li M, Lu W, Xu Y, Lin H. 1H-NMR based metabonomic profiling of human esophageal cancer tissue. Mol Cancer. 2013;12:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 20. | Garber K. Energy boost: the Warburg effect returns in a new theory of cancer. J Natl Cancer Inst. 2004;96:1805-1806. [PubMed] |
| 21. | Phang JM, Liu W, Hancock C. Bridging epigenetics and metabolism: role of non-essential amino acids. Epigenetics. 2013;8:231-236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 22. | Ong ES, Zou L, Li S, Cheah PY, Eu KW, Ong CN. Metabolic profiling in colorectal cancer reveals signature metabolic shifts during tumorigenesis. Mol Cell Proteomics. 2010;Epub ahead of print. [PubMed] |
| 23. | de Aguiar Picanço E, Lopes-Paulo F, Marques RG, Diestel CF, Caetano CE, de Souza MV, Moscoso GM, Pazos HM. L-arginine and glycine supplementation in the repair of the irradiated colonic wall of rats. Int J Colorectal Dis. 2011;26:561-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 24. | Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040-1044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1023] [Cited by in RCA: 1118] [Article Influence: 86.0] [Reference Citation Analysis (0)] |
| 25. | Stumvoll M, Meyer C, Perriello G, Kreider M, Welle S, Gerich J. Human kidney and liver gluconeogenesis: evidence for organ substrate selectivity. Am J Physiol. 1998;274:E817-E826. [PubMed] |
| 26. | Tessem MB, Swanson MG, Keshari KR, Albers MJ, Joun D, Tabatabai ZL, Simko JP, Shinohara K, Nelson SJ, Vigneron DB. Evaluation of lactate and alanine as metabolic biomarkers of prostate cancer using 1H HR-MAS spectroscopy of biopsy tissues. Magn Reson Med. 2008;60:510-516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 153] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 27. | Moreadith RW, Lehninger AL. The pathways of glutamate and glutamine oxidation by tumor cell mitochondria. Role of mitochondrial NAD(P)+-dependent malic enzyme. J Biol Chem. 1984;259:6215-6221. [PubMed] |
| 28. | Leichtle AB, Nuoffer JM, Ceglarek U, Kase J, Conrad T, Witzigmann H, Thiery J, Fiedler GM. Serum amino acid profiles and their alterations in colorectal cancer. Metabolomics. 2012;8:643-653. [PubMed] |
| 29. | Tajiri K, Shimizu Y. Branched-chain amino acids in liver diseases. World J Gastroenterol. 2013;19:7620-7629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 146] [Cited by in RCA: 141] [Article Influence: 11.8] [Reference Citation Analysis (0)] |