Published online Jun 21, 2014. doi: 10.3748/wjg.v20.i23.7181
Revised: December 14, 2013
Accepted: January 8, 2014
Published online: June 21, 2014
Processing time: 237 Days and 13.8 Hours
Hepatitis B virus (HBV) infection is the leading cause of severe chronic liver disease. This article provides a critical view of the importance of genomic medicine for the study of HBV infection and its clinical outcomes in Latin America. Three levels of evolutionary adaptation may correlate with the clinical outcomes of HBV infection. Infections in Latin America are predominantly of genotype H in Mexico and genotype F in Central and South America; these strains have historically circulated among the indigenous population. Both genotypes appear to be linked to a benign course of disease among the native and mestizo Mexicans and native South Americans. In contrast, genotypes F, A and D are common in acute and chronic infections among mestizos with Caucasian ancestry. Hepatocellular carcinoma is rare in Mexicans, but it has been associated with genotype F1b among Argentineans. This observation illustrates the significance of ascertaining the genetic and environmental factors involved in the development of HBV-related liver disease in Latin America, which contrast with those reported in other regions of the world.
Core tip: We explore the influence of genetic and environmental factors that may participate in the clinical outcome of hepatitis B virus (HBV) infection among the Latin American population. Such features may be of interest to clinicians and scientists in the field of hepatology because this population differs importantly from others worldwide. A novel genomic medicine approach is required to implement new strategies for the prevention, management and treatment of HBV infection.
- Citation: Roman S, Jose-Abrego A, Fierro NA, Escobedo-Melendez G, Ojeda-Granados C, Martinez-Lopez E, Panduro A. Hepatitis B virus infection in Latin America: A genomic medicine approach. World J Gastroenterol 2014; 20(23): 7181-7196
- URL: https://www.wjgnet.com/1007-9327/full/v20/i23/7181.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i23.7181
Hepatitis B virus (HBV) is a globally distributed human pathogen that can cause life-threatening liver disease such as chronic hepatitis, liver cirrhosis and hepatocellular cancer[1]. At least 350 million people worldwide contribute to the large reservoir of chronic carriers, especially in developing regions, which may now be facing epidemiological shifts due to immigration[1,2].
The study of the HBV reflects many of the chronological stages of scientific and technological development in the field of medical virology[3]. During the immunological phase, the discovery of the hepatitis B surface antigen protein (HBsAg)[4] supported worldwide serological testing for HBV infection; later, the use of DNA amplification and sequencing tools marked the era of molecular epidemiology to test for viral genomes and genotypes, respectively[3]. Most recently, the Bayesian coalescent and phylogeographic framework[5] coupled with bioinformatics and specialized software is rapidly contributing new data regarding the geographic origin and spread of HBV infection throughout different populations globally[6].
Significant progress has also been made in regards to understanding the epidemiology, virology, natural history and therapy of HBV infection[3]. Moreover, novel prevention, management and treatment strategies should now be studied with a genomic medicine approach. Herein, we consider that the interrelationship between genetic and environmental factors involved in the development of human diseases should be based on the features of each population[7,8]. Hence, HBV infection is a particularly suitable candidate to examine such an approach.
This paper provides a critical view of why the application of genomic medicine is required for the study of HBV among the Latin American (LA) population. This complex ethnic group, which arose from the mixture of Native American, Caucasian and African lineages, presents a combined distribution of HBV genotypes from the Old and New World, which may have an impact on the clinical outcomes of and treatment strategies for HBV infection.
It is plausible that viruses have accompanied humans since their emergence on planet earth[9,10]. Within their genetic information and replication cycles may lie the history of key events in the diversification of all living creatures[11]. When did viruses first originate? It is an exciting question that still inspires researchers to develop new methods for genetic sequence analysis[12-14]. For example, the hepadnaviridae family is divided into two genera, orthohepadnavirus and avihepadnavirus, based on their genomic similarities and hosts[15]. Additionally, the Hepadnaviruses have the ability to integrate their genome into the host’s genome[16,17]. This feature is useful for paleovirologists to identify ancient endogenous viral elements and estimate how long they have coexisted with their host[18-20]. Interestingly, rock and sediment fossils are crucial to date the age of any plant or animal species, while in some viruses, fossil information can be found in contemporary cells’ genomes. The genus Avihepadnavirus primarily infects birds, and it is estimated that it emerged in the Mesozoic era, 65 million years ago[20], when mammals lived in the shadow of the dinosaurs[21]. In contrast, the genus Orthohepadnavirus affects mammals[15]. This genus includes the human hepatitis B virus, one of the most important etiological agents of viral hepatitis in the world[22]. The HBV genome is approximately 3.2 kb, made of partially doubled stranded DNA confined within a fenestrated nucleocapsid (core protein) enveloped in a membrane containing three surface antigen proteins (large, medium and small HBsAg)[15,23]. HBV genome replication is carried out by a viral DNA polymerase without DNA proof-reading capacity[23], which contributes to the high genetic diversity of HBV.
Based on their genomic divergence, HBV is classified into eight genotypes, designated A through H. New information has been gathered that suggests the existence of genotypes I and J[24]. However, the .origin of HBV still remains unclear[25] due to the lack of consensus on the estimated evolutionary rates for HBV and the inconsistency between these data and archeological evidence[26]. Recently, Paraskevis et al[6] estimated that diversification among the different genotypes may have occurred 33000 years ago. Thus, by adapting these results to the hypothesis that HBV came from Africa[6,25,27], it is plausible that the most recent common ancestor (MRCA) of all HBV genotypes traveled together with humans from Africa to their arrival in Beringia. Archaeological and genetic evidence show that modern humans originated 200000 years ago[28]. In addition, it has been estimated that humans began to migrate out of Africa 100000 years ago, traveling through Israel, India, China, Australia, Europe and Russia before reaching the limits of the old continent[29]. Later, the expansion was detained by the glacier of the Beringia region for approximately 36000 years[30]. In this region, the pre-Amerindians faced an extremely hostile climate that reduced their population from 9000 to approximately 3200 people, who then crossed the strait to the Americas[30]. In the last 20000-13000 years, humans have dispersed to Alaska, the continental United States, Mexico and Central and South America[18,31-35]. The ancestral population of Greenland may have been the last to cross over to the new continent, roughly 5500 years ago[36].
From Africa to South America, man was exposed to diverse climates, geographic altitudes, foodstuffs and pathogens. Consequently, due to host-environment interactions, inhabitants may have undergone region-specific genotypic and phenotypic adaptations[37,38]. Examples include the anatomical structure of the Eskimos[39,40], the height and pigmentation of the African population[41], tolerance of hypoxic conditions in the Tibetans[42,43] and adaptations to varying diets[44] and infectious agents[45,46]; these adaptations allowed them to survive. Based on such features, it is likely that each new human settlement carried an HBV strain with its own MRCA, which may have developed specific adaptations to its host, allowing it to survive and spread efficiently through its autochthonous population. These changes are reflected at the genomic level, giving rise to what we now know as genotypes.
It is estimated that the first genotype to diversify was C, followed by B, D, A, F, E, H and G[6]. HBV genotypes A through E are predominant among Old World populations. Genotypes B and C are predominant in Asia; genotype D in Africa, Europe and India; and genotype A in sub-Saharan Africa, North Africa and Western Europe; genotype E is restricted to East Africa[47]. Furthermore, genotype G may be the most recent, with an estimated time of MRCA of 800 years[6]; however, because its genome contains fragments of other genotypes[48,49] and the number of complete sequences studied is limited, further research is required to determine its geographic origin[8]. Nonetheless, genotype G is restricted primarily to populations of men who have sex with men in America[8], suggesting that these patients play a key role in the spread of HBV to other parts of the world[50-53].
Genotype H is predominant in Mexico[7,8], while genotype F prevails in Central and South America[54,55]. Phylogeny tests performed by the Neighbor-Joining and Maximum Likelihood methods group these genotypes as “sisters”, near the root of many phylogenetic trees[56-60] due to their strong genetic similarities. Thus, by this methodology, genotypes F and H belong to a monophyletic clade and can be considered direct descendants of the ancestor of all human hepatitis B genotypes. Because they share common epidemiological and transmission routes, such features may be of medical relevance to the clinical outcome and response to treatment among human populations in Mexico and Central and South America.
Based on their historical background and types of infection[61], human populations have developed a wide spectrum of adaptations to HBV. From an evolutionary perspective, “adaptation” is defined as all changes that increase the success of the survival of an organism[62]. Applying this concept to HBV, we may consider three levels of the adaptation process: incomplete, semi-complete and complete; these classifications may be related to the clinical outcomes of HBV infection in humans.
Incomplete adaptation: Incomplete adaptation events may be exemplified in patients who develop fulminant hepatitis[63], wherein a hyperimmune response to viral antigens may lead to the deterioration of liver function, severely compromising the patient’s life[64,65]. This is an inefficient state for the survival of HBV, which depends on the host to exist. Viruses exhibiting incomplete adaptation may be those that recently crossed the species barrier[66]. Alternatively, they may arise in the circulating population in the form of core and pre-core gene mutants, which enhance the encapsidation of virions and in some cases may cause fulminant hepatitis B[67-69]. In general, the frequency of fulminant hepatitis is approximately 0.1% to 1%[70,71], suggesting that the majority of HBV carriers have other forms of adaptation.
Another mechanism of incomplete adaptation, which is beneficial to the host, may occur when HBV DNA genomes are eliminated by an efficient and coordinated immune response[72]. In this type of infection, Th1 cytokines (IFN-γ, IL-2, TNF-α) quickly clear HBV DNA by means of an optimal polyclonal response against the viral antigens[73]. Then, T cell (CD4+ and CD8+) activity fights infection by cytolytic and non-cytolytic mechanisms[73,74]. This type of response may annihilate HBV; however, if all human populations responded in the same manner, today’s HBV would not have achieved such a broad distribution.
Semi-complete adaptation: Semi-complete adaptation can be exemplified in patients with chronic hepatitis B infection. In this group, it is a common characteristic to detect HBV DNA and HBsAg after more than six months[75]. It is likely that an inadequate immune response and HBV evasion mechanisms are responsible for chronicity[74]. This type of infection does not immediately compromise the host’s life, allowing hepatitis B virions to fulfill their life cycle and transmit to the susceptible population. Nevertheless, prolonged exposure to chronic infection is associated with the development of fibrosis and cirrhosis[22,75]. Additionally, the HBx protein has been related to the development of hepatocellular carcinoma by interacting with different signaling pathways[76], preventing DNA repair[77] and modulating the cell cycle[78]. There are 350 million people suffering from chronic HBV infection[71], many of whom are part of the Asian population, which harbors genotypes B and C and has a higher risk for hepatocellular carcinoma (HCC)[79]. This situation contrasts with the clinical presentation of HBV genotypes F and H[7,8] and may indicate that an important majority of the human population presents semi-complete adaptation; thus, in the long run, some HBV genotypes may be more aggressive than others.
Complete adaptation: Finally, complete adaptation comprises what is known as occult hepatitis B (OHB). In this scenario, infections are characterized by low viral loads (< 200 IU/mL) and the absence of HBsAg[80]. It is likely that the ability to integrate the HBV genome into the host’s cell[79], the stability of the cccDNA and immune tolerance[81] achieve a balance between the rate of viral replication and tissue damage, conferring a homeostatic state between the human host and the specific HBV genotype. Under this hypothesis, complete adaptation may allow patients to be asymptomatic for many years[82]. However, other viral infections (i.e., HIV or HCV)[83], intravenous drug use, immunosuppression, antiviral therapy and even massive vaccination may break the equilibrium state, triggering the onset of symptoms. Additionally, these environmental “stress” factors may provoke genetic diversification, thus enhancing viral fitness and survival.
Finally, HBV could have accompanied humans from their origin to their arrival in South America, developing variable degrees of mutual adaptation depending on the degree of endemicity, HBV genotype, extent of exposure and genetic background of the human population. Thus, the clinical manifestations and outcomes of HBV infection may reflect the degree of adaptation between the human host and their specific viral genotype companion.
The Latin American and Caribbean region encloses the Spanish, Portuguese and French-speaking countries of the American continent and covers almost 22000000 km2. It includes Mexico, the islands of the Caribbean and Central and South America, which possess a rich cultural and natural heritage[84]. In this region, HBV endemicity ranges from low to high with at least 7 to 12 million people infected[85,86]. In the following section, we offer an overview of the relationship between the genetic backgrounds of LA hosts and HBV genotypes, both endemic and foreign, with an eye to the clinical outcomes of HBV infection.
The peopling of the Americas, and consequently the origin of the present-day LA population, is an active topic of research. In a recent population genetics study, Reich et al[87] proposed that Native Americans descend from at least three streams of Asian gene flow. One stream of ancestry comprises the Native American descendants that came from the homogenous “First Americans”, who crossed the Bering Strait more than 15000 years ago (range, approximately 40000-15000 years). Two additional Asian lineages were detected among the Eskimo-Aleut speakers and Na-Dene-speaking Chipewyan, with a subsequent admixture between First Americans and the following streams of aforementioned Asian migrations[87]. The nomadic lifestyle of these people and the climatic changes allowed for their widespread expansion throughout the continent in a southward direction[87].
The establishment of human populations in continental Latin America began mainly in the northern part of Mexico. This dry land region, known as Aridoamerica, was inhabited by small and isolated semi-nomadic groups[88], while Mesoamerica, in central-south and southeast Mexico, with its extraordinary natural resources, especially in the Mexican Basin, invited the nomadic hunter-gatherers to become sedentary societies[87,88]. Anthropological evidence dates the earliest settlers in Mexico as far back as 30000 years, during the Lithic Period[89]. The climatic conditions and environment of Mesoamerica allowed the domestication and earliest cultivation of plants (7000-5500 BC), which resulted in the eventual emergence of agriculture systems. The growth of sedentary societies during the Pre-classical period (2200 BC) flourished in the Classical (150-900 AC) and Post-classical (900-1519 AC) period with the subsequent increase in population density[89].
The size of the most developed population, in Tenochtitlan, the Aztec capital city, at the time of the Spanish conquest was 25000000, which declined drastically to 5000000[90] due to warfare, overwork and the presence of epidemic diseases, thereby allowing the wide expansion and settlement of Europeans that gave rise to the initial genetic and cultural admixture[89,90]. The Spaniards later brought slaves from several regions of Africa and further admixture occurred[88]. These historical processes, in conjunction with successive colonization and industrial development from the 16th to the 18th century, brought other foreign settlers; in recent years, ongoing migration has come to shape the present-day gene pool of the Mexican population.
Several genome-wide analyses[87,88,91,92] have demonstrated that the Mexican population still retains its Native American ancestry, with the degree of Amerindian ancestry increasing from north to south (38%-76%) and a proportional decrease in European ancestry (50%-8.5%); African ancestry remains relatively low throughout the population (9%-18%)[92,93]. Likewise, most of the population of Central and South America underwent the same pattern of socio-demographic transformation as the Amerindian ancestry, which led to the heterogeneous distribution of the admixture background of LA. For example, the population of Argentina displays predominantly European ancestry (78%), followed by Amerindian (19%) and African ancestry (3%), although the precise proportions may differ according to the studied subpopulations[94]. Brazil and Colombia show similar proportions of European (71%) followed by the Amerindian ancestry (18%-19%), and the African ancestry (10%-11%)[95].
HBV infection in Latin America has a heterogeneous distribution when estimated by the HBsAg serological marker. According to the World Health Organization (WHO), the majority of LA countries are considered low seroprevalence (< 2%) regions, including Mexico, Honduras, Nicaragua, Costa Rica, Panama, Cuba, Paraguay, Uruguay, Chile, Argentina, Peru and north Colombia. Regions with intermediate seroprevalence of HBV infection in Central America are Guatemala, Belize, El Salvador, Honduras, Haiti the Dominican Republic and Puerto Rico (> 2.0%-< 8.0%). In South America, some countries or regions are classified as having intermediate endemicity, such as Ecuador, Venezuela, Guyana, Surinam, French Guyana and the south of Brazil; whereas Peru, south Colombia, northern Bolivia and northern Brazil are known for their high seroprevalence (HBsAg > 8%)[2,22,96,97]. However, WHO reports are primarily estimates that require updated feedback obtained by national, large-scale epidemiological studies, which are not commonly carried out in developing countries, including those of Latin America. For example, the introduction of HBV vaccination campaigns among members of the WHO has diminished its prevalence in several regions of Latin America[86].
Interestingly, countries that have a pattern of low endemicity for HBsAg may have a higher prevalence of anti-HBc, a marker of past or ongoing infection[98,99], suggesting that exposure to HBV infection may be higher than previously estimated by HBsAg alone. Additionally, it is unlikely that a given prevalence reported by the WHO is applicable to all regions or risk groups in a given country (i.e., rural and urban areas, or native and mixed race populations). Furthermore, despite advancements in the sensitivity thresholds for HBsAg testing, many commercial kits have their limitations; thus, the surface antigen protein may be undetected[98,99]. Furthermore, among special populations, precautions should be taken in patients with comorbidities related to overt immunosuppression, such as cancer or HCV/HIV co-infections, as well as in cases of obesity and alcoholism, which may indeed mask HBV infection in the form of OHB[7,8]. Thus, given that Latin America is a diverse region and that the aforementioned situations are likely to be encountered, it may be advisable to proceed with further HBV testing by using the anti-HBc marker, as well as nucleic acid testing, because HBsAg provides only one view of the status of infection.
As previously mentioned, diagnostic tests based on molecular techniques, either manual or automated, to ascertain HBV genomes have revolutionized the clinical management of HBV infection worldwide. It is now a standard practice to use them to confirm serological testing, to decide who and when to treat, and during the follow-up of antiviral treatment. These guidelines are recommended by several liver associations worldwide. Paralleling these advancements is the gradual appearance of research in the field of molecular epidemiology[100,101] and phylogeography[58,102-104] of HBV genotypes in Latin America.
At the beginning of the molecular epidemiology era in Latin America, it was clear that the HBV genotypes F and H were the indigenous genotypes[54,57,105], while the incidence of the HBV genotypes A and D within this region was the result of admixture with European and African populations. Specifically, genotype F has been detected as the predominant genotype in Central America and occurs at a high frequency among the HBV-infected Amerindians in all countries of South America (i.e., Venezuela, Colombia, Peru, Bolivia, Argentina, and Brazil), as well as in Native Alaskan populations[106]. In general, its prevalence depends on the degree of admixture of the population with Amerindians[57] (see section Clinical Outcomes for further discussion of this topic). Moreover, F sequences isolated from sporadic cases among non-Amerindian populations appear as nested clades within the Amerindian genotype F radiation[6]. On the contrary, HBV genotype H has been predominantly isolated from both Amerindians and mestizos in Mexico with a frequency that ranges from 60% to 100% depending upon the ethnicity and geographic location of the sample population[7,107].
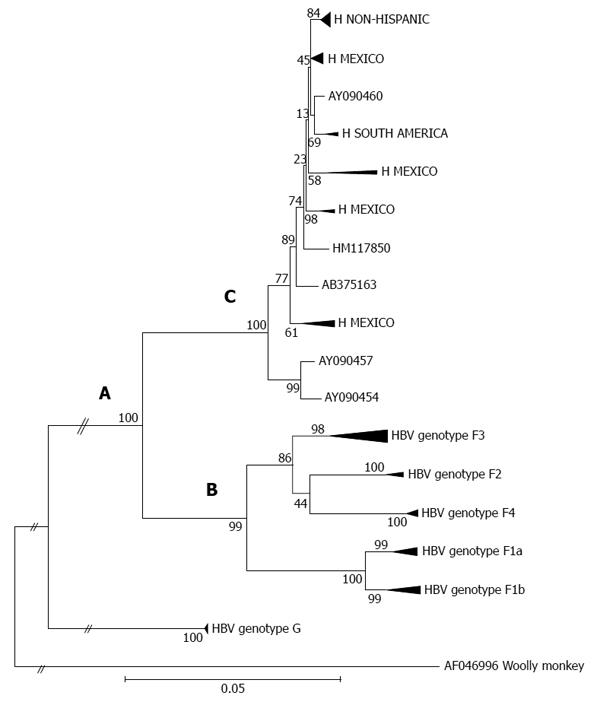
Genotypes F and H display a close phylogenetic relationship (Figure 1, section A), which suggests an introduction of the F/H ancestral strains to the Americas before the recent European colonization[54]. It has recently been proposed that the estimated time of the MRCA for genotypes F and H within the New World was approximately 8900 years ago[6]. It appears that both the ancestral and distinct F/H lineages (i.e., clusters or subgenotypes) emerged under appropriate conditions for human settlement, development of agricultural systems and an increase in population size throughout the American continent[103]. However, genotype F presents greater genetic diversity than genotype H. Phylogenetic tree topology and genetic distances built using maximum likelihood and maximum parsimony methods show the deep clusters and geographical structure typical of genotype F[103]. Notably, the phylogeography of genotype F denotes the presence of subgenotypes designated F1-F4; some are further classified in subdivisions “a” or “b” suggesting a high level of isolation of the Amerindian populations carrying HBV[6] (Figure 1, section B). The estimated times of the MRCAs for the F1-F4 subgenotypes are, in chronological order (i.e., oldest to newest), 6.4 ky, 3.3 ky, 3.2 ky and 2.4 ky, respectively[6]. Additionally, characteristic amino acid positions of complete genomes of HBV genotype F confirm the existence of the four subgenotypes[58,104].
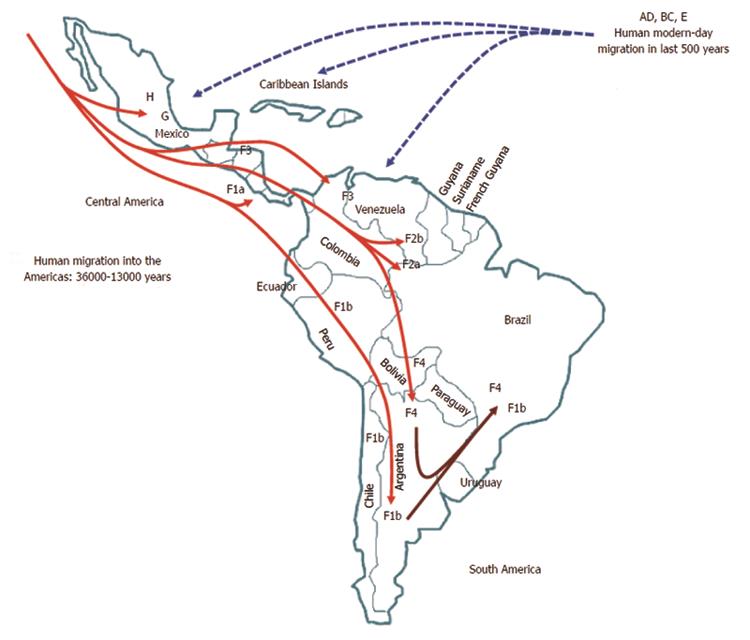
Within genotype H, at least four nested sub-clusters (Figure 1, section C) are noticeable, though they are not yet definite clades or subgenotypes. So far, these correspond to the geographic region of origin of the isolates[7]. Accordingly, genotype H isolates depict an intragenotypic divergence of 0.032%-3.82% (data not shown). In Table 1, the genetic distance by pair-wise analysis among the four F subgenotypes and the HMEX, HUSA, HSA and HNON-HISPANIC subsets range from 7.17% to 10.40% on the basis of complete genomes. Interestingly, the highest divergence is within the H subsets and F1 genotypes, followed by F2 and F4, while F3 had the lowest (Table 1). These data are consistent with the fact that F3 and H share amino acid positions[58,104] and with the similarity in the estimated times of MRCA, 3.2 ky and 4.1 ky, respectively[6]. Furthermore, recent studies based on coalescence and phylogeographic methods have provided new insights into the introduction and spread of F genotypes (F1-F4) among the LA populations through human migration, especially in Colombia[58], Brazil[102] and Argentina[103] (Figure 2).
| Cluster | HMEX | HUSA | HSA | Hnon-Hispanic |
| F1a | 8.63 | 8.89 | 8.70 | 8.51 |
| (8.11-9.9) | (8.16-9.5) | (8.5-8.8) | (8.22-8.91) | |
| F1b | 8.63 | 8.88 | 8.59 | 8.49 |
| (8.22-10.1) | (8.04-9.86) | (8.19-9.39) | (8.04-9.31) | |
| F2 | 8.07 | 8.83 | 8.77 | 7.9 |
| (7.59-9.66) | (8.04-9.86) | (8.09-8.24) | (7.66-8.48) | |
| F3 | 7.84 | 7.76 | 7.94 | 7.69 |
| (7.17-9.91) | (7.28-8.94) | (7.24-8.97) | (7.24-8.82) | |
| F4 | 8.36 | 8.66 | 8.50 | 8.29 |
| (7.77-10.40) | (7.84-9.98) | (8.06-9.34) | (7.91-9.33) |
In regards to genotypes D and A, they have maximum frequencies of 35% and 5%, respectively, among urban populations in Guadalajara, Jalisco, Mexico[7], while in various cities in Argentina in South America, they occur at frequencies of 22% and 45%, respectively (see references cited in Table 2). Both cases are in accordance with the ethnic demographic shift that took place during the European colonization. Likewise, genotypes B and C are dispersed among the LA populations due to Asian immigrants (Tables 2, 3).
| Ref. | Country | Study population | n | HBsAg | Anti-HBc | HBV genotypes1 | Diagnosis | ||||||
| Positive | A | B | C | D | F | G | H | ||||||
| Cortes-Mancera et al[130], 2011 | Colombia | Patients with end-stage liver disease | 131 | 14 (11) | 6 (5) | - | - | - | - | 7 (100) | - | - | Cirrhosis in 71% |
| F1a, F3 | HCC in 12%; | ||||||||||||
| Cirrhosis and HCC in 17% | |||||||||||||
| Devesa et al[131], 2008 | HBV-infected patients | 100 | NA | NA | 2 (2) | - | 2 (2) | 8 (8) | 86 (86) | 2 (2) | - | NA | |
| F3 | |||||||||||||
| Alvarado Mora et al[58], 2011 | Blood donors | 143 | 143 (100) | NA | 8 (15) | - | - | - | 40 (77) | 8 (15) | - | NA | |
| Nakano et al[134], 2001 | Venezuela | Amerindian | 12 | 12 (100) | NA | - | - | - | - | 12 (100) | - | - | CH in 33% |
| Quintero et al[132], 2002 | Afro-Venezuelan | 222 | 8 (4) | NA | 3 (50) | - | - | - | 3 (50) | - | - | CH in 100% | |
| Gutierrez et al[138], 2004 | Blood donors | 258 | 2 (0.8) | 258 (100) | 1 (9) | - | - | 7 (64) | 3 (27) | - | - | OHB in 4.3%; Rare severe liver disease | |
| Kato et al[123], 2005 | HBV-infected patients | 2 | NA | NA | - | - | - | - | 2 (100) | - | - | AH in 100% | |
| F1 | |||||||||||||
| Devesa et al[131], 2008 | Amerindian | 89 | NA | NA | 3 (3) | - | - | 5 (6) | 81 (91) | - | - | NA | |
| F1; F2a; F2b; F3 | |||||||||||||
| Cardona et al[135], 2011 | Amerindian | 70 | 2 (3) | 25 (36) | - | - | - | - | 25 (100) | - | - | OHB in 34%; | |
| F3 | |||||||||||||
| Palumbo et al[133], 2007 | Ecuador | Immigrants in Italy | 2 | 2 (100) | 0 (0) | 2 (100) | - | - | - | - | - | - | CH in 100% |
| Casey et al[139], 1996 | Peru | Military personnel | 84 | 77 (88) | 84 (95) | - | - | - | - | 15 (100) | - | - | CH in 68% |
| Von Meltzer et al[140], 2008 | HBV-infected patients | 9 | 9 (100) | NA | - | - | - | - | 9 (9) | - | - | CH in 100% | |
| F1b | |||||||||||||
| Sitnik et al[143], 2004 | Brazil | HBV-infected patients | 103 | 103 (100) | NA | 51 (49) | 3 (3) | 14 (14) | 25 (24) | 10 (10) | - | - | CH in 100% |
| Palumbo et al[133], 2007 | Immigrants in Italy | 12 | 12 (100) | 0 (0) | 3 (25) | - | - | 9 (75) | - | - | - | CH in 100% | |
| Bertolini et al[144], 2012 | Blood donors | 228 | 228 (100) | NA | 32 (14) | - | 3 (1) | 189 (83) | 3 (1) | - | 1 (0.4) | NA | |
| A1, A2 | F2a, F4 | ||||||||||||
| Mello et al[102], 2013 | HBV-infected patients | 12 | NA | NA | - | - | - | - | 12 (100) F2a, F1b, F4 | - | - | NA | |
| Eloy et al[145], 2013 | HBV-infected patients | 119 | 80 (100) | 80 (100) | 74 (92) | - | 4 (5) | 1 (1) | 1 (1) | - | - | Asymp in 70; AH in 2%; CH in 28% | |
| Araujo et al[146], 2013 | HBV/HIV-coinfected patients | 1 | 1 (100) | 1 (100) | - | - | - | - | 1 (100) | 1 | - | CH in 100% | |
| F4 | -100 | ||||||||||||
| Khan et al[147], 2008 | Bolivia | Japanese immigrants | 287 | 10 (8) | NA | - | 1 (1) | 5 (50) | - | 4 (4) | - | - | NA |
| Ba | C2 | ||||||||||||
| Khan et al[147], 2008 | Native population | 200 | 12 (6) | NA | - | 1 (8) | 3 (25) | - | 8 (68); F4 | - | - | NA | |
| Di Lello et al[141], 2009, | Chile | HBV-infected patients | 40 | NA | NA | 3 (7) | 2 (5) | 3 (7) | - | 27 (67) | - | - | NA |
| Venegas et al[142],2011 | HBV-infected patients | 21 | 21 (100) | 21 (100) | - | - | - | - | 21 (100) | - | - | CH in 100% | |
| F1b | |||||||||||||
| Solari et al[149], 2009 | Argentina | HBV-infected patients | 21 | 21 (100) | - | 8 (38) | 3 (14) | - | 3 (14) | 7 (33) | - | - | CH in 100% |
| Pezzano et al[150], 2011 | HBV-infected patients | 139 | 128 (100) | 128 (100) | 22 (28) | 1 (0.8) | 3 (2) | 28 (22) | 60 (47) | - | - | AH in 37%; CH in 63% | |
| Trinks et al[148], 2012 | HBV-infected patients | 33 | 33 (100) | 33 (100) | 15 (45) A1, A2 | - | - | 2 (6) | 13 (39) | - | - | CH in 100% | |
| D1 | F1b, F4 | ||||||||||||
| Barbini et al[101], 2013 | HBV-infected patients | 29 | 29 (100) | 29 (100) | 4 (14) A2 | - | - | 2 (7) D2, D3 | 20 (62) | 2 (7) | - | CH in 100% | |
| F1b, F4 | |||||||||||||
| Araujo et al[146], 2013 | HBV/HIV-coinfected patients | 1 | 1 (100) | 1 (100) | - | - | - | - | 1 (100); F1b | 1 | - | CH in 100% | |
| -100 | |||||||||||||
HBV genotype G is a minor strain that exists throughout the Americas in special populations with blood-borne infectious diseases, however due to the high frequency of this genotype in Mexico and the common practice of sexual relationships among native men, it may be associated with an Amerindian ancestry[7,8,98]. Finally, an uncommon genotype E was reported in Colombia[108], as well as an F3/A1 recombinant among an Afro-Colombian community[109].
Thus, with the advancement of molecular epidemiology and phylogeography, future progress can allow a better understanding of the relationship between the evolutionary history of the HBV genotypes and its impact on the clinical outcomes of HBV infection among the LA populations.
Mexico: HBV endemicity and clinical outcomes among the native or mestizo Mexican population are associated with the predominance of genotype H, followed by genotypes A, D and G[7,8,50,98,107] (Table 3). Two outstanding features of HBV infection are OHB and low viral load regardless of the degree of endemicity[7]. HBV genotype A is most likely to be detected in acute infections and associated with mixed infections and high viral loads; in contrast, genotype D manifests at very low or undetectable viral loads or in mixed infections. The progression to chronic infection occurs primarily among mestizo adults through horizontal transmission and, to a lesser degree, in children by vertical transmission[7,99,110]. OHB is a key feature in native populations with high rates of endemic HBV infection, although surprisingly few clinical manifestations occur. Recently, in an analysis of native Mexican groups, we reported differences in cytokine levels in the serum that can distinguish OHB genotype H-infected patients from patients that resolved HBV infection. This result suggests that cytokine expression, along with specific immune responses, can influence the severity of OHB disease. Differences in immune response may be responsible for viral transcription repression, which in turn results in both HBsAg and minimal detectable levels of HBV DNA[111].
| Ref. | Country | Study population | n | HBsAg | Anti-HBc | HBV genotypes1 | Diagnosis | |||||||
| Positive | A | B | C | D | E | F | G | H | ||||||
| Sanchez et al[107], 2002 | Mexico | HBV-infected adults | 15 | 13 (87) | 6 (40) | 3 (20) | - | - | 1 (7) | - | 10 (67) | 1 (7) | - | AH in 30%; |
| CH in 27%; | ||||||||||||||
| cirrhosis in 7%. | ||||||||||||||
| Sanchez et al[50], 2007 | HBV-infected patients | 67 | 67 (100) | NA | 10 (15) | - | - | 4 (6) | 4 (6) | 44 (66) | AH in 15%; | |||
| CH in 48% | ||||||||||||||
| Ruiz-Tachiquin et al[151], 2007 | Blood donors and HBV-infected patients | 33 | 33 (100) | NA | - | - | 4 (12) | 1 (3) | - | 26 (79) | Asymp in 64%; CH in 36% | |||
| Roman et al[98], 2010 | Native adults | 306 | 17 (5.6) | 100 (32.7) | 6 (24) | 1 (4) | 2 (8) | 4 (16) | - | 1 (4) | 11 (44) | OHB in 14.2%; asymp in 100% | ||
| Arauz-Ruiz et al[115], 1997 | Central America: | HBV-infected patients, blood donors and pregnant woman | 330 | 330 (100) | 71 (21) | 13 (14) | - | 1 (1) | 5 (6) | 71 (79) | - | CH in 21%; | ||
| Costa Rica, Nicaragua, | F1a; F1b; F2 | AH in 16% | ||||||||||||
| Honduras, El Salvador, Guatemala | ||||||||||||||
| Leon et al[122], 2005 | Costa Rica | HBV-infected patients | 50 | 32 (64) | 18 (36) | - | - | - | 2 (4) | - | 48 (96) | - | CH in 47%; | |
| HCC in 16% | ||||||||||||||
| Kato et al[123], 2005 | Panama | HBV-infected patients | 2 | NA | NA | - | - | - | - | 2 (100) | - | - | NA | |
| F1, F3 | ||||||||||||||
| Martinez et al[124], 2013 | Chinese residents | 320 | 42 (13) | - | 11 (61) | 7 (39) | - | - | - | - | - | No evident liver disease | ||
| B2 | C1 | |||||||||||||
| Andernach et al[125], 2009 | Haití | Pregnant woman | 320 | 320 (100) | 128 (71) | - | - | 40 (22) | 11 (6) | - | - | - | NA | |
| A1, A2, A5 | D3, D4 | |||||||||||||
| Couto et al[126], 2013 | Haitian’s in South Florida | 27 | NA | NA | - | - | - | 19 (79) | - | - | - | - | NA | |
Moreover, it is plausible that the course of liver disease and immune response in native populations in Mexico may be different from those described in other areas of the world. In general, liver cirrhosis and HCC associated with HBV infection do not occur frequently in native populations in Mexico[112,113], even in comparison with the rest of Latin America[113]. However, it is noteworthy that Mexico ranks first in mortality due to alcoholic liver disease[114] and ranks lowest for mortality due to HCC in general[112]. This epidemiological profile contrasts with what occurs in Asia, where countries such as China are exposed to HBV genotypes B and C at earlier ages through vertical transmission and have a higher prevalence of HCC and different responses to antiviral therapy, suggesting that genetic and environmental factors may modulate the degree of adaptation to HBV infection, as previously mentioned.
Central America: In Central America, data regarding the HBV genotype distribution in countries such as Costa Rica, Nicaragua, Honduras, El Salvador and Guatemala were reported in the late nineties (Table 3). HBV genotype F (specifically, F1a, F1b and F2) in patients with acute and chronic infection was associated with seropositivity for HBsAg and anti-HBc, as well as pre-core stop mutations[115].
In Guatemala, HBsAg prevalence was reported to be as low as 0.5% in a group of 77 pregnant women[116] and 1.3% (i.e., 6 cases) among 484 female sex workers from the Mexican-Guatemalan border[117]; in both populations, the infected individuals were asymptomatic carriers of HBsAg. However, among a group of Guatemalan refugees near the same border, the percentage of asymptomatic carriers of HBsAg increased to 17%[118]. In this country, the HBV genotype F1a was found, and the infection with this subgenotype apparently does not produce important hepatic inflammation. Likewise, in Belize, 35% of a studied population with acute hepatitis infection was indigenous[119], and chronic hepatitis B has not been reported in this region.
In Honduras and Nicaragua, HBsAg and anti-HBc seroprevalence have been reported in high-risk groups, such as multi-transfused adult patients[120] and children with cancer[121]. In addition, genotype F was reported in 96% of HBV cases in Costa Rica, followed by genotype D in 4%[122]; in contrast, genotype F (specifically, F1 and F3) was found in two patients in Panama[123]. All of these cases were in HBV-infected patients who presented with chronic hepatitis B and HCC associated with HBx gene mutations in genotype F strains. Therefore, the genetic characteristics of mutant HBV may increase the rate of HBV-related liver damage and HCC in Central America. On the other hand, the presence of HBV genotypes B and C among Chinese residents living in Panama without liver disease[124] provides the opportunity to study the natural history of these genotypes in a new environment.
Central America is an important gateway for immigration in which further serological and molecular epidemiological studies may provide new evidence concerning shifts in the genotype distribution and its effect on the clinical manifestations of HBV infection.
The Caribbean region: In general, HBV genotypes have rarely been reported in the Caribbean, but it seems that the HBV genotype distribution and progression of liver damage could be different from those observed in Central America. HBV genotype A (specifically, A1, A2 and A5) was the dominant strain reported in Haiti, followed by genotype D (specifically, D3 and D4) among 320 pregnant women without liver inflammation[125]. It is noteworthy that despite the fact that this country has significant African ancestry, HBV genotype E has rarely been found, suggesting that this genotype emerged after the slave trade in the Americas[125]. Additionally, in another study in Haitians living in Florida, United States, genotype D was found with spontaneous pre-core region mutations[126], whereas OHB was reported in Cuba in HIV-infected patients[127] in whom these HBV genotypes were unknown. In a Jamaican study, the HBsAg prevalence was reported at 3.2% among patients with sexually transmitted diseases, but liver damage was not studied[128]. Further, HCC was found to be associated with HBV infection in 5.3% of 114 veterans in Puerto Rico[129] (Table 3).
South America: Countries in this region, including Colombia, Venezuela, Ecuador, Peru, Brazil, Bolivia, Chile and Argentina, have defined the predominant circulating HBV genotypes, although the numbers of HBV strains in some regions are limited. Thus, in Colombia, a connecting country between Central and South America, HBV genotype F (specifically, F1a and F3) is more frequent (77% and 86%, respectively) than genotypes A and G (15% and 2%, respectively)[58,130,131]. In these studies, HBV-infected patients were at end-stage disease due to cirrhosis and HCC[130], whereas in other studies, the type of liver damage in HBV-infected patients and blood donors was not reported. Various studies report HBV genotype F as the most frequent in Venezuela, Peru and Chile with differences in the frequencies of the subgenotypes (specifically, F1a, F2a, F2b and F3) among the Afro-Venezuelan[132], Amerindian[133-137] and mestizo[138-142] populations. In these groups, chronic hepatitis B is common and OHB has been found in blood donors and Amerindians[139-142] (Table 2).
Although genotype F (specifically, F4) has been reported in Brazil, genotype A (specifically, A1 and A2) has been found to be dominant, followed by genotype D, in patients with chronic hepatitis B; coinfection with genotypes F and G was reported in one HIV-infected patient[102,132,143,144-146]. Interestingly, genotype F4 was reported in Bolivia[147] and Argentina[101], which have populations of mixed ethnic backgrounds, and the characteristics of the HBV genotypes could have changed over time in these regions. In fact, this population had chronic liver disease, HCC was associated with HBV infection, HBsAg was positive in all cases, and the viral load was high. On the other hand, HBV genotypes B and C (specifically, Ba and C2) were found in Japanese immigrants living in Bolivia[147], which reflects immigration events into South America. Regarding HBV genotypes A and D in patients in Argentina[148-151], a chronic progression of the infection with these genotypes was reported, contrasting with OHB and the asymptomatic clinical outcome reported in Mexico[151] (Tables 2 and 3).
Therefore, research on HBV strains in LA should be clinically associated with the natural evolution of liver disease, hepatic complications and the response to different treatments. These issues may have a strong impact on the prevention and control of HBV transmission because they may influence the prognosis and response to the hepatitis B vaccine.
This overview has illustrated the influence of genetic and environmental risk factors in the onset and development of HBV-related liver disease. The people of Latin America share a similar genetic ancestry with varying degrees of admixture from three distinct lineages. The closely related HBV genotypes F and H have been in contact with the native population of the Americas and tend to cause mild liver disease with no further complications. In contrast, the mestizo population of South America presents acute and chronic liver disease with a tendency toward HCC. Based on these features, personalized medicine strategies provide a novel framework for the prevention, management and treatment of HBV-related liver disease in Latin America. Future genomic medicine research will have an important impact on the clinical approach to liver diseases. An integrated approach requires that genetic and environmental factors be taken into account, as both are involved in the endemicity and clinical outcome of HBV infection. Among the human host factors, the susceptibility to liver damage and the response to antiviral treatment, which in turn are modulated by the immune system, are linked to genetic polymorphisms that are associated with specific ethnic backgrounds. Genes involved in various metabolic pathways are influenced by changes in environmental lifestyle factors, such as nutrition, physical activity and emotional stress. Other comorbidities that enter the picture are the worldwide obesity epidemic and increased consumption of alcohol, which impose a greater burden on liver health.
P- Reviewers: Dash S, Xiao YT S- Editor: Wen LL L- Editor: A E- Editor: Liu XM
| 1. | Ioannou GN. Chronic hepatitis B infection: a global disease requiring global strategies. Hepatology. 2013;58:839-843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 2. | Lavanchy D. Worldwide epidemiology of HBV infection, disease burden, and vaccine prevention. J Clin Virol. 2005;34 Suppl 1:S1-S3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 319] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 3. | Gerlich WH. Medical virology of hepatitis B: how it began and where we are now. Virol J. 2013;10:239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 220] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 4. | Blumberg BS. Hepatitis B: The Hunt for a Killer Virus. Princeton: University Press 2002; 1-164. |
| 5. | Holmes EC. Evolutionary history and phylogeography of human viruses. Annu Rev Microbiol. 2008;62:307-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 145] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 6. | Paraskevis D, Magiorkinis G, Magiorkinis E, Ho SY, Belshaw R, Allain JP, Hatzakis A. Dating the origin and dispersal of hepatitis B virus infection in humans and primates. Hepatology. 2013;57:908-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 117] [Article Influence: 9.8] [Reference Citation Analysis (2)] |
| 7. | Panduro A, Maldonado-Gonzalez M, Fierro NA, Roman S. Distribution of HBV genotypes F and H in Mexico and Central America. Antivir Ther. 2013;18:475-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (1)] |
| 8. | Roman S, Panduro A. HBV endemicity in Mexico is associated with HBV genotypes H and G. World J Gastroenterol. 2013;19:5446-5453. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 44] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | Forterre P. Defining life: the virus viewpoint. Orig Life Evol Biosph. 2010;40:151-160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 77] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 10. | Holmes EC. What does virus evolution tell us about virus origins? J Virol. 2011;85:5247-5251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 89] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 11. | Forterre P. The two ages of the RNA world, and the transition to the DNA world: a story of viruses and cells. Biochimie. 2005;87:793-803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 141] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 12. | Pybus OG, Rambaut A. Evolutionary analysis of the dynamics of viral infectious disease. Nat Rev Genet. 2009;10:540-550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 454] [Cited by in RCA: 445] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 13. | Li WL, Drummond AJ. Model averaging and Bayes factor calculation of relaxed molecular clocks in Bayesian phylogenetics. Mol Biol Evol. 2012;29:751-761. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 84] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 14. | Rosenberg NA, Nordborg M. Genealogical trees, coalescent theory and the analysis of genetic polymorphisms. Nat Rev Genet. 2002;3:380-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 446] [Cited by in RCA: 372] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 15. | King AMQ, Adams MJ, Lefkowitz E, Carstens EB. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. United States: Elsevier 2012; 442-455. |
| 16. | Yokosuka O, Omata M, Zhou YZ, Imazeki F, Okuda K. Duck hepatitis B virus DNA in liver and serum of Chinese ducks: integration of viral DNA in a hepatocellular carcinoma. Proc Natl Acad Sci USA. 1985;82:5180-5184. [PubMed] |
| 17. | Yaginuma K, Kobayashi M, Yoshida E, Koike K. Hepatitis B virus integration in hepatocellular carcinoma DNA: duplication of cellular flanking sequences at the integration site. Proc Natl Acad Sci USA. 1985;82:4458-4462. [PubMed] |
| 18. | Patel MR, Emerman M, Malik HS. Paleovirology - ghosts and gifts of viruses past. Curr Opin Virol. 2011;1:304-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 89] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 19. | Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6:e1001191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 480] [Cited by in RCA: 471] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 20. | Suh A, Brosius J, Schmitz J, Kriegs JO. The genome of a Mesozoic paleovirus reveals the evolution of hepatitis B viruses. Nat Commun. 2013;4:1791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 21. | Martin T. Paleontology. Early mammalian evolutionary experiments. Science. 2006;311:1109-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 22. | Dienstag JL. Hepatitis B virus infection. N Engl J Med. 2008;359:1486-1500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 689] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 23. | Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1094] [Cited by in RCA: 1111] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 24. | Kurbanov F, Tanaka Y, Mizokami M. Geographical and genetic diversity of the human hepatitis B virus. Hepatol Res. 2010;40:14-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 158] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 25. | Simmonds P. The origin and evolution of hepatitis viruses in humans. J Gen Virol. 2001;82:693-712. [PubMed] |
| 26. | Bouckaert R, Alvarado-Mora MV, Pinho JR. Evolutionary rates and HBV: issues of rate estimation with Bayesian molecular methods. Antivir Ther. 2013;18:497-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 27. | Magnius LO, Norder H. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology. 1995;38:24-34. [PubMed] |
| 28. | Hedges SB. Human evolution. A start for population genomics. Nature. 2000;408:652-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 23] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 29. | Lewin R. Human Evolution: An Illustrated Introduction. 5th ed. United Kingdom: Wiley 2009; 187-190. |
| 30. | Kitchen A, Miyamoto MM, Mulligan CJ. A three-stage colonization model for the peopling of the Americas. PLoS One. 2008;3:e1596. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 98] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 31. | Wang S, Lewis CM, Jakobsson M, Ramachandran S, Ray N, Bedoya G, Rojas W, Parra MV, Molina JA, Gallo C. Genetic variation and population structure in native Americans. PLoS Genet. 2007;3:e185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 393] [Cited by in RCA: 355] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 32. | Hamilton MJ, Buchanan B. Archaeological support for the three-stage expansion of modern humans across northeastern Eurasia and into the Americas. PLoS One. 2010;5:e12472. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 33. | Lell JT, Sukernik RI, Starikovskaya YB, Su B, Jin L, Schurr TG, Underhill PA, Wallace DC. The dual origin and Siberian affinities of Native American Y chromosomes. Am J Hum Genet. 2002;70:192-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 104] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 34. | O'Rourke DH, Raff JA. The human genetic history of the Americas: the final frontier. Curr Biol. 2010;20:R202-R207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 93] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 35. | Goebel T, Waters MR, O’Rourke DH. The late Pleistocene dispersal of modern humans in the Americas. Science. 2008;319:1497-1502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 522] [Cited by in RCA: 259] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 36. | Rasmussen M, Li Y, Lindgreen S, Pedersen JS, Albrechtsen A, Moltke I, Metspalu M, Metspalu E, Kivisild T, Gupta R. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757-762. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 684] [Cited by in RCA: 482] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 37. | Hancock AM, Alkorta-Aranburu G, Witonsky DB, Di Rienzo A. Adaptations to new environments in humans: the role of subtle allele frequency shifts. Philos Trans R Soc Lond B Biol Sci. 2010;365:2459-2468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 101] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 38. | Summerhayes GR, Leavesley M, Fairbairn A, Mandui H, Field J, Ford A, Fullagar R. Human adaptation and plant use in highland New Guinea 49,000 to 44,000 years ago. Science. 2010;330:78-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 39. | Mäkinen TM. Nordic Society for Arctic Medicine. The effects of cold adaptation on human performance. Int J Circumpolar Health. 2003;62:445. [PubMed] |
| 40. | Lebedev MD, Bobrov NI, Keerig IuIu. [Various indicators of human adaptation to the extreme conditions of the Arctic]. Gig Sanit. 1987;18-21. [PubMed] |
| 41. | Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao X, Humphreville VR, Humbert JE. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782-1786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 756] [Cited by in RCA: 750] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 42. | Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 755] [Cited by in RCA: 860] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 43. | Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1069] [Cited by in RCA: 1120] [Article Influence: 74.7] [Reference Citation Analysis (0)] |
| 44. | Teaford MF, Ungar PS. Diet and the evolution of the earliest human ancestors. Proc Natl Acad Sci USA. 2000;97:13506-13511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 212] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 45. | Casanova JL, Abel L. Genetic dissection of immunity to mycobacteria: the human model. Annu Rev Immunol. 2002;20:581-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 743] [Cited by in RCA: 727] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 46. | Stephens JC, Reich DE, Goldstein DB, Shin HD, Smith MW, Carrington M, Winkler C, Huttley GA, Allikmets R, Schriml L. Dating the origin of the CCR5-Delta32 AIDS-resistance allele by the coalescence of haplotypes. Am J Hum Genet. 1998;62:1507-1515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 382] [Cited by in RCA: 344] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 47. | Kao JH. Molecular epidemiology of hepatitis B virus. Korean J Intern Med. 2011;26:255-261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 48. | Simmonds P, Midgley S. Recombination in the genesis and evolution of hepatitis B virus genotypes. J Virol. 2005;79:15467-15476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 189] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 49. | Lindh M. HBV genotype G-an odd genotype of unknown origin. J Clin Virol. 2005;34:315-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 50. | Sánchez LV, Tanaka Y, Maldonado M, Mizokami M, Panduro A. Difference of hepatitis B virus genotype distribution in two groups of mexican patients with different risk factors. High prevalence of genotype H and G. Intervirology. 2007;50:9-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 51. | van Houdt R, Bruisten SM, Geskus RB, Bakker M, Wolthers KC, Prins M, Coutinho RA. Ongoing transmission of a single hepatitis B virus strain among men having sex with men in Amsterdam. J Viral Hepat. 2010;17:108-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 52. | Lama JR, Agurto HS, Guanira JV, Ganoza C, Casapia M, Ojeda N, Ortiz A, Zamalloa V, Suarez-Ognio L, Cabezas C. Hepatitis B infection and association with other sexually transmitted infections among men who have sex with men in Peru. Am J Trop Med Hyg. 2010;83:194-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 53. | Remis RS, Dufour A, Alary M, Vincelette J, Otis J, Mâsse B, Turmel B, LeClerc R, Parent R, Lavoie R. Association of hepatitis B virus infection with other sexually transmitted infections in homosexual men. Omega Study Group. Am J Public Health. 2000;90:1570-1574. [PubMed] |
| 54. | Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J Gen Virol. 2002;83:2059-2073. [PubMed] |
| 55. | Alvarado-Mora MV, Pinho JR. Distribution of HBV genotypes in Latin America. Antivir Ther. 2013;18:459-465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 56. | Norder H, Couroucé AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, Robertson BH, Locarnini S, Magnius LO. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 630] [Cited by in RCA: 649] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 57. | Devesa M, Pujol FH. Hepatitis B virus genetic diversity in Latin America. Virus Res. 2007;127:177-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 45] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 58. | Alvarado Mora MV, Romano CM, Gomes-Gouvêa MS, Gutierrez MF, Botelho L, Carrilho FJ, Pinho JR. Molecular characterization of the Hepatitis B virus genotypes in Colombia: a Bayesian inference on the genotype F. Infect Genet Evol. 2011;11:103-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 59. | Starkman SE, MacDonald DM, Lewis JC, Holmes EC, Simmonds P. Geographic and species association of hepatitis B virus genotypes in non-human primates. Virology. 2003;314:381-393. [PubMed] |
| 60. | Godoy BA, Alvarado-Mora MV, Gomes-Gouvêa MS, Pinho JR, Fagundes N. Origin of HBV and its arrival in the Americas--the importance of natural selection on time estimates. Antivir Ther. 2013;18:505-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 61. | Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1700] [Cited by in RCA: 1712] [Article Influence: 81.5] [Reference Citation Analysis (0)] |
| 62. | Solomon EP, Berg LR, Martin DW. Biology. 9th ed. Canada: Brook & Cole 2010; 481-708. |
| 63. | De Cock KM, Govindarajan S, Valinluck B, Redeker AG. Hepatitis B virus DNA in fulminant hepatitis B. Ann Intern Med. 1986;105:546-547. [PubMed] |
| 64. | Ando K, Moriyama T, Guidotti LG, Wirth S, Schreiber RD, Schlicht HJ, Huang SN, Chisari FV. Mechanisms of class I restricted immunopathology. A transgenic mouse model of fulminant hepatitis. J Exp Med. 1993;178:1541-1554. [PubMed] |
| 65. | Kanada A, Takehara T, Ohkawa K, Tatsumi T, Sakamori R, Yamaguchi S, Uemura A, Kohga K, Sasakawa A, Hikita H. Type B fulminant hepatitis is closely associated with a highly mutated hepatitis B virus strain. Intervirology. 2007;50:394-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 66. | Taubenberger JK. The origin and virulence of the 1918 “Spanish” influenza virus. Proc Am Philos Soc. 2006;150:86-112. [PubMed] |
| 67. | Liang TJ, Hasegawa K, Munoz SJ, Shapiro CN, Yoffe B, McMahon BJ, Feng C, Bei H, Alter MJ, Dienstag JL. Hepatitis B virus precore mutation and fulminant hepatitis in the United States. A polymerase chain reaction-based assay for the detection of specific mutation. J Clin Invest. 1994;93:550-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 64] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 68. | Omata M, Ehata T, Yokosuka O, Hosoda K, Ohto M. Mutations in the precore region of hepatitis B virus DNA in patients with fulminant and severe hepatitis. N Engl J Med. 1991;324:1699-1704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 377] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 69. | Baumert TF, Yang C, Schürmann P, Köck J, Ziegler C, Grüllich C, Nassal M, Liang TJ, Blum HE, von Weizsäcker F. Hepatitis B virus mutations associated with fulminant hepatitis induce apoptosis in primary Tupaia hepatocytes. Hepatology. 2005;41:247-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 70. | Garfein RS, Bower WA, Loney CM, Hutin YJ, Xia GL, Jawanda J, Groom AV, Nainan OV, Murphy JS, Bell BP. Factors associated with fulminant liver failure during an outbreak among injection drug users with acute hepatitis B. Hepatology. 2004;40:865-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 71. | Elgouhari HM, Abu-Rajab Tamimi TI, Carey WD. Hepatitis B virus infection: understanding its epidemiology, course, and diagnosis. Cleve Clin J Med. 2008;75:881-889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 72. | Wai CT, Fontana RJ. Cytokine gene polymorphisms in chronic hepatitis B: a step up the immunology ladder. Am J Gastroenterol. 2003;98:6-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 73. | Larrubia JR, Benito-Martínez S, Miquel-Plaza J, Sanz-de-Villalobos E, González-Mateos F, Parra T. Cytokines - their pathogenic and therapeutic role in chronic viral hepatitis. Rev Esp Enferm Dig. 2009;101:343-351. [PubMed] |
| 74. | Wieland SF, Chisari FV. Stealth and cunning: hepatitis B and hepatitis C viruses. J Virol. 2005;79:9369-9380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 346] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 75. | Liaw YF, Chu CM. Hepatitis B virus infection. Lancet. 2009;373:582-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 929] [Cited by in RCA: 992] [Article Influence: 62.0] [Reference Citation Analysis (1)] |
| 76. | Wu ZJ, Zhu Y, Huang DR, Wang ZQ. Constructing the HBV-human protein interaction network to understand the relationship between HBV and hepatocellular carcinoma. J Exp Clin Cancer Res. 2010;29:146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 77. | Qadri I, Fatima K, AbdeL-Hafiz H. Hepatitis B virus X protein impedes the DNA repair via its association with transcription factor, TFIIH. BMC Microbiol. 2011;11:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 78. | Wang T, Zhao R, Wu Y, Kong D, Zhang L, Wu D, Li C, Zhang C, Yu Z, Jin X. Hepatitis B virus induces G1 phase arrest by regulating cell cycle genes in HepG2.2.15 cells. Virol J. 2011;8:231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 79. | Wong GL, Chan HL, Yiu KK, Lai JW, Chan VK, Cheung KK, Wong EW, Wong VW. Meta-analysis: The association of hepatitis B virus genotypes and hepatocellular carcinoma. Aliment Pharmacol Ther. 2013;37:517-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 80. | Habibollahi P, Safari S, Daryani NE, Alavian SM. Occult hepatitis B infection and its possible impact on chronic hepatitis C virus infection. Saudi J Gastroenterol. 2009;15:220-224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 81. | Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5:215-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1202] [Cited by in RCA: 1212] [Article Influence: 60.6] [Reference Citation Analysis (0)] |
| 82. | van Hemert FJ, Zaaijer HL, Berkhout B, Lukashov VV. Occult hepatitis B infection: an evolutionary scenario. Virol J. 2008;5:146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 83. | Altınbaş A, Ergünay K, Calık Başaran N, Alp A, Turgut D, Hasçelik G, Uzun Ö, Unal S. [Investigation of occult hepatitis B in HIV infected patients]. Mikrobiyol Bul. 2011;45:353-358. [PubMed] |
| 84. | World Heritage Convention. Latin America and the Caribbean. Available from: http://whc.unesco.org/en/lac/. |
| 85. | Te HS, Jensen DM. Epidemiology of hepatitis B and C viruses: a global overview. Clin Liver Dis. 2010;14:1-21, vii. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 292] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 86. | Alvarado-Mora MV, Pinho JR. Epidemiological update of hepatitis B, C and delta in Latin America. Antivir Ther. 2013;18:429-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 87. | Reich D, Patterson N, Campbell D, Tandon A, Mazieres S, Ray N, Parra MV, Rojas W, Duque C, Mesa N. Reconstructing Native American population history. Nature. 2012;488:370-374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 526] [Cited by in RCA: 476] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
| 88. | Martínez-Cortés G, Salazar-Flores J, Fernández-Rodríguez LG, Rubi-Castellanos R, Rodríguez-Loya C, Velarde-Félix JS, Muñoz-Valle JF, Parra-Rojas I, Rangel-Villalobos H. Admixture and population structure in Mexican-Mestizos based on paternal lineages. J Hum Genet. 2012;57:568-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 89. | García-Bárcena J. La cuenca de Mexico. Etapa Lítica (30,000- 2000 a.C). Los primeros pobladores. Spanish. Revista de Arqueología Mexicana. 2007;15: 30-33. |
| 90. | Sanchez-Albornoz N. The Population of Latin America: A History. Berkeley: University of California Press 1974; 1-299. |
| 91. | Rubi-Castellanos R, Martínez-Cortés G, Muñoz-Valle JF, González-Martín A, Cerda-Flores RM, Anaya-Palafox M, Rangel-Villalobos H. Pre-Hispanic Mesoamerican demography approximates the present-day ancestry of Mestizos throughout the territory of Mexico. Am J Phys Anthropol. 2009;139:284-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 136] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 92. | Galanter JM, Fernandez-Lopez JC, Gignoux CR, Barnholtz-Sloan J, Fernandez-Rozadilla C, Via M, Hidalgo-Miranda A, Contreras AV, Figueroa LU, Raska P. Development of a panel of genome-wide ancestry informative markers to study admixture throughout the Americas. PLoS Genet. 2012;8:e1002554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 182] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 93. | Rangel-Villalobos H, Salazar-Flores J, Dondiego R, Anaya-Palafox M, Nuño-Arana I, Canseco-Ávila LM, Rubi-Castellanos R. South to North increasing gradient of paternal European ancestry throughout the Mexican territory: Evidence of Y-linked short tandem repeats. Forensic Sci Int Genet. 2009;2:448-450. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 94. | Seldin MF, Tian C, Shigeta R, Scherbarth HR, Silva G, Belmont JW, Kittles R, Gamron S, Allevi A, Palatnik SA. Argentine population genetic structure: large variance in Amerindian contribution. Am J Phys Anthropol. 2007;132:455-462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 61] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 95. | Price AL, Patterson N, Yu F, Cox DR, Waliszewska A, McDonald GJ, Tandon A, Schirmer C, Neubauer J, Bedoya G. A genomewide admixture map for Latino populations. Am J Hum Genet. 2007;80:1024-1036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 96. | Tanaka J. Hepatitis B epidemiology in Latin America. Vaccine. 2000;18 Suppl 1:S17-S19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 70] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 97. | World Health Organization. International travel and health. Hepatitis B. Available from: http://www.who.int/ith/diseases/hepatitisB/en/. |
| 98. | Roman S, Tanaka Y, Khan A, Kurbanov F, Kato H, Mizokami M, Panduro A. Occult hepatitis B in the genotype H-infected Nahuas and Huichol native Mexican population. J Med Virol. 2010;82:1527-1536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 99. | Roman S, Panduro A, Aguilar-Gutierrez Y, Maldonado M, Vazquez-Vandyck M, Martinez-Lopez E, Ruiz-Madrigal B, Hernandez-Nazara Z. A low steady HBsAg seroprevalence is associated with a low incidence of HBV-related liver cirrhosis and hepatocellular carcinoma in Mexico: a systematic review. Hepatol Int. 2009;3:343-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 100. | Campos RH, Mbayed VA, Pineiro Y Leone FG. Molecular epidemiology of hepatitis B virus in Latin America. J Clin Virol. 2005;34 Suppl 2:S8-S13. [PubMed] |
| 101. | Barbini L, Elizalde M, Torres C, Campos R. Molecular epidemiology and genetic diversity of hepatitis B virus in Mar del Plata city, Argentina. Infect Genet Evol. 2013;19:152-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 102. | Mello FC, Araujo OC, Lago BV, Motta-Castro AR, Moraes MT, Gomes SA, Bello G, Araujo NM. Phylogeography and evolutionary history of hepatitis B virus genotype F in Brazil. Virol J. 2013;10:236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 103. | Torres C, Piñeiro y Leone FG, Pezzano SC, Mbayed VA, Campos RH. New perspectives on the evolutionary history of hepatitis B virus genotype F. Mol Phylogenet Evol. 2011;59:114-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 104. | Piñeiro y Leone FG, Mbayed VA, Campos RH. Evolutionary history of Hepatitis B virus genotype F: an in-depth analysis of Argentine isolates. Virus Genes. 2003;27:103-110. [PubMed] |
| 105. | Norder H, Hammas B, Lee SD, Bile K, Couroucé AM, Mushahwar IK, Magnius LO. Genetic relatedness of hepatitis B viral strains of diverse geographical origin and natural variations in the primary structure of the surface antigen. J Gen Virol. 1993;74:1341-1348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 255] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 106. | Livingston SE, Simonetti JP, McMahon BJ, Bulkow LR, Hurlburt KJ, Homan CE, Snowball MM, Cagle HH, Williams JL, Chulanov VP. Hepatitis B virus genotypes in Alaska Native people with hepatocellular carcinoma: preponderance of genotype F. J Infect Dis. 2007;195:5-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 193] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 107. | Sánchez LV, Maldonado M, Bastidas-Ramírez BE, Norder H, Panduro A. Genotypes and S-gene variability of Mexican hepatitis B virus strains. J Med Virol. 2002;68:24-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 67] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 108. | Alvarado Mora MV, Romano CM, Gomes-Gouvêa MS, Gutierrez MF, Carrilho FJ, Pinho JR. Molecular epidemiology and genetic diversity of hepatitis B virus genotype E in an isolated Afro-Colombian community. J Gen Virol. 2010;91:501-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 109. | Alvarado-Mora MV, Romano CM, Gomes-Gouvêa MS, Gutierrez MF, Carrilho FJ, Pinho JR. Phylogenetic analysis of complete genome sequences of hepatitis B virus from an Afro-Colombian community: presence of HBV F3/A1 recombinant strain. Virol J. 2012;9:244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 110. | Escobedo-Meléndez G, Fierro NA, Roman S, Maldonado-González M, Zepeda-Carrillo E, Panduro A. Prevalence of hepatitis A, B and C serological markers in children from western Mexico. Ann Hepatol. 2012;11:194-201. [PubMed] |
| 111. | Fierro NA, Roman S, Realpe M, Hernandez-Nazara Z, Zepeda-Carrillo EA, Panduro A. Multiple cytokine expression profiles reveal immune-based differences in occult hepatitis B genotype H-infected Mexican Nahua patients. Mem Inst Oswaldo Cruz. 2011;106:1007-1013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 112. | Roman S, Fierro NA, Moreno-Luna L, Panduro A. Hepatitis B virus genotype H and environmental factors associated to the low prevalence of hepatocellular carcinoma in Mexico. J Cancer Ther. 2013;2A:367-376. [RCA] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 113. | Pujol FH, Roman S, Panduro A, Navas MC, Lampe E. Hepatocellular carcinoma in Latin America. Isabelle Chemin, Ed In: Hepatocellular Carcinoma: A Global Challenge. New York: Nova Science Publishers Inc 2012; 55-68. |
| 114. | Roman S, Zepeda-Carrillo EA, Moreno-Luna LE, Panduro A. Alcoholism and liver disease in Mexico: genetic and environmental factors. World J Gastroenterol. 2013;19:7972-7982. [PubMed] [DOI] [Full Text] |
| 115. | Arauz-Ruiz P, Norder H, Visoná KA, Magnius LO. Genotype F prevails in HBV infected patients of hispanic origin in Central America and may carry the precore stop mutant. J Med Virol. 1997;51:305-312. [PubMed] |
| 116. | Samayoa B, Anderson MR, Alonso Pacheco KP, Lee C, Pittard A, Soltren A, Barrios Matos I, Arathoon E. Seroprevalence of HIV, hepatitis B, and syphilis among pregnant women at the general hospital, Guatemala City, 2005-2009. J Int Assoc Physicians AIDS Care (Chic). 2010;9:313-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 117. | Uribe-Salas F, Conde-Glez CJ, Juárez-Figueroa L, Hernández-Castellanos A. Sociodemographic dynamics and sexually transmitted infections in female sex workers at the Mexican-Guatemalan border. Sex Transm Dis. 2003;30:266-271. [PubMed] |
| 118. | Alvarez-Muñoz T, Bustamante-Calvillo E, Martínez-García C, Moreno-Altamirando L, Guiscafre-Gallardo H, Guiscafre JP, Muñoz O. Seroepidemiology of the hepatitis B and delta in the southeast of Chiapas, Mexico. Arch Invest Med (Mex). 1989;20:189-195. [PubMed] |
| 119. | Bryan JP, Reyes L, Hakre S, Gloria R, Kishore GM, Tillett W, Engle R, Tsarev S, Cruess D, Purcell RH. Epidemiology of acute hepatitis in the Stann Creek District of Belize, Central America. Am J Trop Med Hyg. 2001;65:318-324. [PubMed] |
| 120. | Vinelli E, Lorenzana I. Transfusion-transmitted infections inmulti-transfused patients in Honduras. J Clin Virol. 2005;34 Suppl 2:S53-S60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 121. | Visoná K, Baez F, Taylor L, Berríos R, León B, Pacheco C, Jirón R, Luftig RB, Somarriba MM. Impact of hepatitis B and hepatitis C virus infections in a hematology-oncology unit at a children’s hospital in Nicaragua, 1997 to 1999. Clin Diagn Lab Immunol. 2002;9:622-626. [PubMed] |
| 122. | León B, Taylor L, Vargas M, Luftig RB, Albertazzi F, Herrero L, Visona K. HBx M130K and V131I (T-A) mutations in HBV genotype F during a follow-up study in chronic carriers. Virol J. 2005;2:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 123. | Kato H, Fujiwara K, Gish RG, Sakugawa H, Yoshizawa H, Sugauchi F, Orito E, Ueda R, Tanaka Y, Kato T. Classifying genotype F of hepatitis B virus into F1 and F2 subtypes. World J Gastroenterol. 2005;11:6295-6304. [PubMed] |
| 124. | Martinez AA, Zaldivar Y, Hong CCh, Alvarado-Mora MV, Smith R, Ortiz AY, Pinho JR, Cristina J, Pascale JM. Molecular characterisation of hepatitis B virus in the resident Chinese population in Panama City. Mem Inst Oswaldo Cruz. 2013;108:541-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 125. | Andernach IE, Nolte C, Pape JW, Muller CP. Slave trade and hepatitis B virus genotypes and subgenotypes in Haiti and Africa. Emerg Infect Dis. 2009;15:1222-1228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 97] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 126. | Couto CA, Levy C, Morris CJ, Hill M, de Medina M, Sanborn MR, Cloherty GA, Schiff ER, Martin P. High frequency of genotype D and spontaneous hepatitis B virus genomic mutations among Haitians in a multiethnic North American population. J Clin Gastroenterol. 2013;47:83-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 127. | Marité B, Montalvo MC, Rodríguez Lde L, Sariego S, Verdasquera D, Vincent M, Gutiérrez A, Sánchez M. Occult hepatitis B in Cuban HIV patients. MEDICC Rev. 2011;13:32-37. [PubMed] |
| 128. | Smikle M, Dowe G, Hylton-Kong T, Williams E. Hepatitis B and C viruses and sexually transmitted disease patients in Jamaica. Sex Transm Infect. 2001;77:295-296. [PubMed] |
| 129. | López-García LJ, Toro DH, Mayol H, Martínez-Souss J, Dueño MI, Rodríguez-Pérez F. Hepatocellular carcinoma: ten years experience among veterans in Puerto Rico. P R Health Sci J. 2007;26:103-107. [PubMed] |
| 130. | Cortes-Mancera F, Loureiro CL, Hoyos S, Restrepo JC, Correa G, Jaramillo S, Norder H, Pujol FH, Navas MC. Etiology and Viral Genotype in Patients with End-Stage Liver Diseases admitted to a Hepatology Unit in Colombia. Hepat Res Treat. 2011;2011:363205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 131. | Devesa M, Loureiro CL, Rivas Y, Monsalve F, Cardona N, Duarte MC, Poblete F, Gutierrez MF, Botto C, Pujol FH. Subgenotype diversity of hepatitis B virus American genotype F in Amerindians from Venezuela and the general population of Colombia. J Med Virol. 2008;80:20-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 69] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 132. | Quintero A, Martínez D, Alarcón De Noya B, Costagliola A, Urbina L, González N, Liprandi F, Castro De Guerra D, Pujol FH. Molecular epidemiology of hepatitis B virus in Afro-Venezuelan populations. Arch Virol. 2002;147:1829-1836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 133. | Palumbo E, Scotto G, Faleo G, Cibelli DC, Angarano G. Prevalence of HBV genotypes in South American immigrants affected by HBV-related chronic active hepatitis. Braz J Infect Dis. 2007;11:311-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 134. | Nakano T, Lu L, Hu X, Mizokami M, Orito E, Shapiro C, Hadler S, Robertson B. Characterization of hepatitis B virus genotypes among Yucpa Indians in Venezuela. J Gen Virol. 2001;82:359-365. [PubMed] |
| 135. | Cardona NE, Loureiro CL, Garzaro DJ, Duarte MC, García DM, Pacheco MC, Chemin I, Pujol FH. Unusual presentation of hepatitis B serological markers in an Amerindian community of Venezuela with a majority of occult cases. Virol J. 2011;8:527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 136. | Pujol FH, Navas MC, Hainaut P, Chemin I. Worldwide genetic diversity of HBV genotypes and risk of hepatocellular carcinoma. Cancer Lett. 2009;286:80-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 57] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 137. | Duarte MC, Cardona N, Poblete F, González K, García M, Pacheco M, Botto C, Pujol FH, Williams JR. A comparative epidemiological study of hepatitis B and hepatitis D virus infections in Yanomami and Piaroa Amerindians of Amazonas State, Venezuela. Trop Med Int Health. 2010;15:924-933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 138. | Gutiérrez C, Devesa M, Loureiro CL, León G, Liprandi F, Pujol FH. Molecular and serological evaluation of surface antigen negative hepatitis B virus infection in blood donors from Venezuela. J Med Virol. 2004;73:200-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 139. | Casey JL, Niro GA, Engle RE, Vega A, Gomez H, McCarthy M, Watts DM, Hyams KC, Gerin JL. Hepatitis B virus (HBV)/hepatitis D virus (HDV) coinfection in outbreaks of acute hepatitis in the Peruvian Amazon basin: the roles of HDV genotype III and HBV genotype F. J Infect Dis. 1996;174:920-926. [PubMed] |
| 140. | von Meltzer M, Vásquez S, Sun J, Wendt UC, May A, Gerlich WH, Radtke M, Schaefer S. A new clade of hepatitis B virus subgenotype F1 from Peru with unusual properties. Virus Genes. 2008;37:225-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 141. | Di Lello FA, Piñeiro Y Leone FG, Muñoz G, Campos RH. Diversity of hepatitis B and C viruses in Chile. J Med Virol. 2009;81:1887-1894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 142. | Venegas M, Alvarado-Mora MV, Villanueva RA, Rebello Pinho JR, Carrilho FJ, Locarnini S, Yuen L, Brahm J. Phylogenetic analysis of hepatitis B virus genotype F complete genome sequences from Chilean patients with chronic infection. J Med Virol. 2011;83:1530-1536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 143. | Sitnik R, Pinho JR, Bertolini DA, Bernardini AP, Da Silva LC, Carrilho FJ. Hepatitis B virus genotypes and precore and core mutants in Brazilian patients. J Clin Microbiol. 2004;42:2455-2460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 99] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 144. | Bertolini DA, Gomes-Gouvêa MS, Guedes de Carvalho-Mello IM, Saraceni CP, Sitnik R, Grazziotin FG, Laurino JP, Fagundes NJ, Carrilho FJ, Pinho JR. Hepatitis B virus genotypes from European origin explains the high endemicity found in some areas from southern Brazil. Infect Genet Evol. 2012;12:1295-1304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 145. | Eloy AM, Moreira RC, Lemos MF, de Almeida Silva JL, Coêlho MR. Hepatitis B virus in the State of Alagoas, Brazil: genotypes characterization and mutations of the precore and basal core promoter regions. Braz J Infect Dis. 2013;17:704-706. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 146. | Araujo NM, Araujo OC, Silva EM, Villela-Nogueira CA, Nabuco LC, Parana R, Bessone F, Gomes SA, Trepo C, Kay A. Identification of novel recombinants of hepatitis B virus genotypes F and G in human immunodeficiency virus-positive patients from Argentina and Brazil. J Gen Virol. 2013;94:150-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 147. | Khan A, Tanaka Y, Saito H, Ebinuma H, Sekiguchi H, Iwama H, Wakabayashi G, Kamiya T, Kurbanov F, Elkady A. Transmission of hepatitis B virus (HBV) genotypes among Japanese immigrants and natives in Bolivia. Virus Res. 2008;132:174-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 148. | Trinks J, Frías S, Frider B, Alessio A, Pozzati M, Daleoso G, León L, Batalla VM, Díaz A, Ameigeiras B. Genotypes B and C hepatocellular carcinoma-associated hepatitis B virus pre-S mutants: their detection among F1b and A2 - but not F4 - isolates from Argentina. J Viral Hepat. 2012;19:823-828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 149. | Solari J, Galdame O, Rezzonico L, Frider B, Villamil A, Casciato P, Reig M, Bandi JC, Alessio A, Gadano A. Twenty-four weeks therapy with peginterferon alfa-2a could be similar to 48 weeks therapy in patients with HBeAg positive chronic hepatitis B and good predictors of response: results of a pilot study. Acta Gastroenterol Latinoam. 2009;39:254-260. [PubMed] |
| 150. | Pezzano SC, Torres C, Fainboim HA, Bouzas MB, Schroder T, Giuliano SF, Paz S, Alvarez E, Campos RH, Mbayed VA. Hepatitis B virus in Buenos Aires, Argentina: genotypes, virological characteristics and clinical outcomes. Clin Microbiol Infect. 2011;17:223-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 151. | Ruiz-Tachiquín ME, Valdez-Salazar HA, Juárez-Barreto V, Dehesa-Violante M, Torres J, Muñoz-Hernández O, Alvarez-Muñoz MT. Molecular analysis of hepatitis B virus “a” determinant in asymptomatic and symptomatic Mexican carriers. Virol J. 2007;4:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |