Published online Jun 14, 2014. doi: 10.3748/wjg.v20.i22.6701
Revised: January 16, 2014
Accepted: March 12, 2014
Published online: June 14, 2014
Processing time: 260 Days and 10.3 Hours
While hepatitis B virus (HBV) screening relies on hepatitis B surface antigen to confirm HBV infection since the early days of hepatitis B disease management, hepatitis C virus (HCV) infection screening is based on anti-HCV testing which does not discriminate active from past infection. Thus to confirm infection HCV RNA testing has been required; recently a HCV core antigen assay became widely commercially available which could serve to confirm infection. That assay is less sensitive than current HCV RNA assays, but as more than 50% of anti-HCV positive persons will be HCV core antigen positive, HCV core antigen testing can be a cost effective and reflex test to confirm HCV infection in anti-HCV positive individuals and will be easier as it can be applied on the same platform. For treatment monitoring, more data need to be generated, but the early data available at present suggest that HCV core antigen may be an alternative to HCV RNA monitoring. With direct antivirals, HCV core antigen could even be superior to HCV RNA testing, as direct antivirals might already prevent virus formation when HCV core antigen is still produced and thereby correlates better with eventual viral clearance.
Core tip: Hepatitis C virus (HCV) core antigen can be a cost effective alternative to confirm HCV infection, though patients with low HCV RNA (< 1000 IU/mL) have a chance of being false negative. It could have a role in therapy monitoring, but that is currently not well enough studied.
- Citation: Tillmann HL. Hepatitis C virus core antigen testing: Role in diagnosis, disease monitoring and treatment. World J Gastroenterol 2014; 20(22): 6701-6706
- URL: https://www.wjgnet.com/1007-9327/full/v20/i22/6701.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i22.6701
The first report on identification of the non-A, non-B hepatitis virus now known as hepatitis C virus (HCV) came as a press release by the Chiron Corporation in May 1988[1]. The first scientific report on HCV was published in 1989 together with the first generation diagnostic assay to test for antibodies against HCV[2,3]. Subsequent assays were developed to reduce the rate of false positive in anti-HCV testing. A positive antibody reaction should be confirmed with either a confirmatory antibody assay such as recombinant immunoblot assay (RIBA) to confirm true presence of anti-HCV antibodies or preferentially by proof of viremia and thereby infection. Confirmation of infection was initially depending on “in house” polymerase chain reaction (PCR) assays until the first commercially available assay for HCV RNA detection was released based on bDNA technology. Later also qualitative and quantitative commercial PCR based assays to confirm infection were released and continuously improved in sensitivity and linear range accuracy. While earlier versions showed only moderate correlations in the range of r-values of 0.6-0.8 among each other[4], current assays show good correlations with each other. Still for HCV RNA level monitoring during antiviral therapy, it is recommended to use the same diagnostic assay to limit variability[5], though this might change with potent antiviral therapy, where monitoring during treatment becomes obsolete as its result are clinically irrelevant with everyone becoming undetectable.
As a rule of thumb, molecular assays are nowadays frequently more sensitive than serological assays such as in human immunodeficiency virus (HIV, HIV RNA vs p-24), but even though sensitivity is lower, such lower sensitivity assays have a role in diagnosis and management of disease. For CMV, there are studies indicating even higher sensitivity with an viral antigen targeting assay (pp65) compared to PCR[6], while others did not find same results[7]. Importantly concerning their clinical relevance, it seems that they may be interchangeable in several scenarios, as a very low level may be clinically less relevant[8].
For HCV as reviewed here, HCV RNA detection assays are more sensitive, still HCV antigen detection can serve as an alternative. HCV antigen detection might be the first next step following a positive antibody test. As anti-HCV and HCV core antigen testing can be done on the same platform, a reflex test for anti-HCV positive samples can be done to confirm HCV infection within 40 min of the positive anti-HCV result. There might be a small number of HCV core antigen negative individuals who would be positive for HCV RNA. However, HCV core antigen negative individuals can be confirmed to have either no or only low level HCV viremia, which may translate to less significant clinical disease. Thus, if accepting to miss low level HCV viremic patients, HCV core antigen could be the principal screening assay as with hepatitis B, where hepatitis surface antigen (HBsAg) is the principal screening assay though some patients may be HBsAg negative but HBV DNA positive.
A first HCV core antigen test was developed around 2000, but did not really take off, in part due to cost concerns. Now a newer and more sensitive HCV core antigen assay has become available, which is about 25 times more sensitive and licensed in several countries. Importantly, as mentioned in some papers, in addition to a faster turnaround time compared to molecular tests, it is cheaper and thereby very attractive[9]. The currently available assay is a Chemiluminescent Microparticle Immunoassay and allows for a quantitative determination of HCV core antigen in human serum and plasma.
The purpose of this paper is to review the current knowledge on this newer assay with a sensitivity of 3 fmol/L in different scenarios and reflect on it utility. In Japan there are additional 4 assays with slightly reduced sensitivity marketed[10]. General considerations on the data presently available for HCV core antigen are also discussed.
In principal, a big advantage of HCV core antigen testing is that the same testing platform and sample used for anti-HCV testing, can be used for HCV core antigen testing allowing for reflex HCV core antigen testing in anti-HCV positive samples, thereby enhancing speed of clinically meaningful release of important results[11]. Rather than testing for anti-HCV and releasing that result, and awaiting a new sample for confirmation of infection or transfer of the anti-HCV positive sample to the molecular testing facility, physicians could get the results of both anti-HCV and HCV core antigen within the same hour.
One limitation of the earlier and but still also of the more recent HCV core antigen assay is the lower sensitivity compared to HCV RNA assays. Current HCV RNA assays have a lower level of detection between about 5-15 IU/mL.
The sensitivity for the currently available HCV core antigen assay by Abbott was improved to about 3.00 fmol/L (0.0 6 pg/mL), which is about 25 times more sensitive than the Trac-C assay, which was in development by Ortho Diagnostic and had a sensitivity of 1.5 pm/mL. Importantly, the intra-run and between-run precision is now well under 10%. Therefore, samples tested for HCV core antigen do not need to be tested in duplicates anymore; only samples with a value between the lower limit of detection for HCV core antigen of 3 fmol/L and 10 fmol/L should be re-tested in duplicate, but otherwise samples can be run in singleton.
Ross et al[12] estimated that the sensitivity of 3 fmol/L should approximately be equivalent to 507 IU/mL for subtype 1a, 405 IU/mL for subtype 1b, 600 IU/mL for genotype 2 and 771 IU/mL for genotype 3. However, they also found that the analytic sensitivity to detect 95% of samples at the respective level was slightly higher than the 3 fmol, varying from 3.9 fmol/L (equivalent to about 1002 IU/mL HCV RNA) for genotype 3a to 13.5 fmol/L (equivalent to about 2700 IU/mL HCV RNA) for genotype 2a[12]. However, these differences are not likely to be clinically relevant.
In general, it seems that about 90% of HCV RNA positive samples are positive with a viral load above 10.000 IU/mL[13], and thus well in the sensitivity range of the HCV core antigen assay.
Specificity in HCV RNA negative samples has been determined as high as 99.98% in 5394 anti-HCV-negative samples[14]. In smaller studies of 420 and 100 persons, none was found to be false positive[12,15]. However, some studies found that individual patients with detectable HCV core antigen were negative for HCV RNA, i.e., Miedouge et al[16] tested for HCV RNA in mini-pools, but the assay had an insufficient sensitivity to exclude HCV RNA. Overall, it appears that the assay is highly specific.
HCV core antigen assay performance was also assessed in seroconversion panels and its performance was compared to HCV RNA tests. Several such studies explored the option to shorten the window of HCV seroconversion when HCV core antigen would be used vs anti-HCV testing and compared the performance of the HCV core antigen to that of HCV RNA testing, and in common HCV core antigen leads to earlier detection compared to anti-HCV. For the setting of blood transfusion the modern sensitive HCV RNA assays are the most sensitive option[17], but where costs are a constrain, testing with HCV core antigen might be a better option than not testing for HCV virema at all.
Numerous studies have explored the correlation between HCV RNA values and HCV core antigens also in relation to genotypes, and overwhelmingly high correlations were reported between 0.7 to > 0.9 for r-values[10,16,17]. This would indicate that HCV core antigen might be a substitute for HCV RNA testing. Interestingly, the fluctuations in individual patients during a time course without any antiviral treatment were less pronounced with HCV core antigen vs the HCV RNA[18].
Furthermore, a significant lower correlation between HCV core antigen levels and HCV RNA levels is found in HBV coinfection patients with a “r-value” of 0.04[19], indicating that HCV core antigen actually may reveal additional information compared to HCV RNA levels, though this aspect has not been sufficiently explored yet.
The ability to commercialize an HCV antigen assay and need for a quantitative vs a qualitative assay will depend on the scenario, each of which will be addressed below in regard to where we stand and what we still need now: (1) role in a diagnostic algorithm; (2) role in a blood bank/organ donor setting; (3) monitoring during natural history; (4) monitoring during and after therapy; and (5) predicting histological chances.
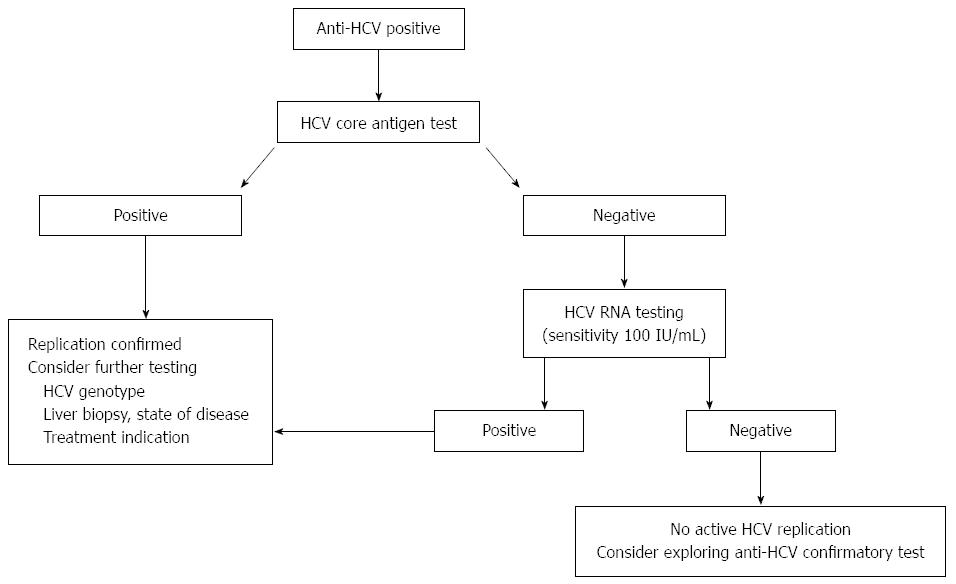
An HCV core antigen assay can be useful in an diagnostic algorithm (as previously already suggested for the less sensitive assay[20]), but such use is even greater with a newer more sensitive assay (Figure 1)[11,19].
As HCV RNA is still more sensitive, utility of HCV core antigen in such diagnostic algorithm will depend on potential cost saving for diagnosing HCV infection in a population setting. A big advantage is the ease to allowing reflex testing when the same testing-instrument is used for anti-HCV and HCV core antigen testing.
Thus anti-HCV positive results would be reflexed to HCV core antigen testing, and additional HCV RNA testing would only be required if HCV core antigen would be negative and potential confirmation of anti-HCV with RIBA (Recombinant ImmunoBlot Assay) only when both HCV core antigen and HCV RNA were negative.
In a setting where samples need to be shipped to a laboratory, HCV core antigen could prove to be more stable than HCV RNA in a setting where blood samples cannot be assessed soon after blood draw[16]. But this would likely require some more data.
The principal usefulness for population screening was indeed confirmed by some recent papers[21]. With a special focus on dialysis patients in some studies, a number of anti-HCV negative patients could be identified to be infected by HCV by a positive HCV core antigen assay[16,22]. Concerning the sensitivity in dialysis patients, it might be worth mentioning that in one study where all dialysis patients were individually tested for HCV RNA, only one HCV RNA positive patient was found to be HCV core antigen negative, but that patient had a low HCV viral load of HCV RNA < 100 IU/mL[22].
As outlined above, the sensitivity of HCV core antigen is inferior to HCV RNA, but superior to no testing. Thus, in a resource limited setting, where HCV RNA testing might be impossible due to cost restrains at present, HCV core antigen testing would allow for a compromise.
Given the cost of organ transplantation, HCV RNA testing cost is unlikely to be a rate limiting step, and therefore not likely to be relevant to organ donation. Still if currently no HCV RNA testing is performed, HCV core antigen might be better than no testing for infectivity.
There currently is no role in monitoring the viral load of patients not undergoing antiviral therapy, but patients frequently want to know that their viral load has not significantly changed. For these circumstances HCV core antigen might be an alternative to quantitative HCV RNA testing.
Depending on country, there are or will soon be regimens available with a cure rate close to 100%. In such a setting it might well be that all patients who tested HCV viremic (with either HCV core antigen or HCV RNA) would get treated in an attempt to eradicate HCV. In such situation monitoring of untreated patients would become obsolete. However, likely the cost of such newer regimens might be prohibitive to treat all infected individuals in all countries.
Monitoring HCV viral load with HCV core antigen during antiviral treatment might be an attractive tool for the future. Unfortunately data are too limited for strong recommendations thereof (Table 1). Especially, there are no data with the newer antivirals available at present. For Pegylated Interferon plus Ribavirin regimens, there have been some studies suggesting that one can predict response as early as day 3[23], week 1[23-26] or week 2[9,23,26-28].
| Ref. | n | Genotype | Time point | Threshold | PPV | NPV |
| Wada et al[25] | 64 | gt 2 | 1 wk | 100 fmol/L | 96% (SVR) | 100% (SVR) |
| Tedder et al[9] | 41 | u.n. | week 12 | HCV RNA > 2log reduction | 70% (EOTR) | 74% (EOTR) |
| HCV core Ag negative | 85% (EOTR) | 93% (EOTR) | ||||
| within 2 wk | HCV RNA > 2log reduction | 86% (EOTR) | 72% (EOTR) | |||
| HCV core 1log reduction | 75% (EOTR) | 76% (EOTR) | ||||
| Vermehren et al[26] | 160 | 1 | 1 | Undetectable | 90.9% (RVR) | 92.8% (RVR) |
| 2 | Undetectable | 79.4% (RVR) | 97.6% (RVR) | |||
| 4 | Undetectable | 47.6% (RVR) | 100% (RVR) | |||
| Tamai et al[28] | 106 | 1b | 2 | HCV RNA 1log reduction | 65% (SVR) | 90% (SVR) |
| 2 | HCV core Ag 1log reduction | 64% (SVR) | 97% (SVR) | |||
| 2 | HCV RNA 2log reduction | 86% (SVR) | 67% (SVR) | |||
| 2 | HCV core Ag 2log reduction | 93% (SVR) | 69% (SVR) | |||
| Ross et al[12] | 29 | 1/4 | week 12 | 2log reduction | 45% (SVR) | 100% (SVR) |
| 9 | 2/3 | week 12 | 2log reduction | 87.5% (SVR) | 1/1 (SVR) | |
| Fujino et al[23] | 60 | 1b | day 3 | 500 fmol/L | 96.7% (SVR) | 46.6% (SVR) |
| day 7 | 500 fmol/L | 90% (SVR) | 76.7% (SVR) | |||
| day 14 | 500 fmol/L | 96.7% (SVR) | 53.3% (SVR) | |||
| day 28 | 500 fmol/L | 100% (SVR) | 26.7% (SVR) |
When 2log rule is used to terminate treatment at week 12, Ross et al[12] suggest that failure of 2log HCV core antigen decline reflects a lack of eventual response in 10/10 patients. Furthermore, with the introduction of direct antiviral agents targeting HCV maturation and replication, HCV core antigen could even be superior to HCV RNA testing, as direct antivirals might already prevent virus formation when HCV core antigen is still produced and thereby correlates better with eventual viral clearance. No data on such therapy in relation to HCV core antigen and achieving SVR are currently available.
It has recently been shown that quantitative HBsAg is useful in excluding significant liver damage in HBeAg positive patients[29]. It would be unlikely that high HCV core antigen levels will translate into mild disease, but the ratio of HCV core antigen to HCV RNA might reveal differences in histology. In a study with a moderate size of 114 patients Durante-Mangoni et al[30] found some correlation with histology activity index and liver fibrosis, though both correlated slightly better with viral load determined by HCV RNA than by HCV core antigen. Only steatosis correlated better with the HCV core antigen level.
Interestingly, the ratio between HCV RNA and HCV core antigen was in an earlier version of an HCV core antigen assay found to potentially play a role in identifying patients who will have a long term biochemically inactive disease[31].
HCV core antigen has a relative strong role in a diagnostic algorithm for HCV infection, while it is too insensitive in its present form to substitute for HCV RNA testing in the blood bank setting. Its role in treatment monitoring remains to be determined, especially for the newer direct antiviral.
P- Reviewers: Gowans EJ, Triyatni M S- Editor: Zhai HH L- Editor: Wang TQ E- Editor: Ma S
| 2. | Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362. [PubMed] |
| 3. | Kuo G, Choo QL, Alter HJ, Gitnick GL, Redeker AG, Purcell RH, Miyamura T, Dienstag JL, Alter MJ, Stevens CE. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362-364. [PubMed] |
| 4. | Pradat P, Chossegros P, Bailly F, Pontisso P, Saracco G, Sauleda S, Thursz M, Tillmann H, Vlassopoulou H, Alberti A. Comparison between three quantitative assays in patients with chronic hepatitis C and their relevance in the prediction of response to therapy. J Viral Hepat. 2000;7:203-210. [PubMed] |
| 5. | Pas S, Molenkamp R, Schinkel J, Rebers S, Copra C, Seven-Deniz S, Thamke D, de Knegt RJ, Haagmans BL, Schutten M. Performance evaluation of the new Roche cobas AmpliPrep/cobas TaqMan HCV test, version 2.0, for detection and quantification of hepatitis C virus RNA. J Clin Microbiol. 2013;51:238-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Ji W, Kim DS, Jung SW, Yu YD, Suh SO. Pre-emptive therapy for the cytomegalovirus infection after liver transplantation in endemic areas and its optimal diagnostic method. Transplant Proc. 2013;45:3065-3068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 7. | Marchetti S, Santangelo R, Manzara S, D’onghia S, Fadda G, Cattani P. Comparison of real-time PCR and pp65 antigen assays for monitoring the development of Cytomegalovirus disease in recipients of solid organ and bone marrow transplants. New Microbiol. 2011;34:157-164. [PubMed] |
| 8. | Drew WL. Laboratory diagnosis of cytomegalovirus infection and disease in immunocompromised patients. Curr Opin Infect Dis. 2007;20:408-411. [PubMed] |
| 9. | Tedder RS, Tuke P, Wallis N, Wright M, Nicholson L, Grant PR. Therapy-induced clearance of HCV core antigen from plasma predicts an end of treatment viral response. J Viral Hepat. 2013;20:65-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 10. | Murayama A, Sugiyama N, Watashi K, Masaki T, Suzuki R, Aizaki H, Mizuochi T, Wakita T, Kato T. Japanese reference panel of blood specimens for evaluation of hepatitis C virus RNA and core antigen quantitative assays. J Clin Microbiol. 2012;50:1943-1949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 11. | Ottiger C, Gygli N, Huber AR. Detection limit of architect hepatitis C core antigen assay in correlation with HCV RNA, and renewed confirmation algorithm for reactive anti-HCV samples. J Clin Virol. 2013;58:535-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 12. | Ross RS, Viazov S, Salloum S, Hilgard P, Gerken G, Roggendorf M. Analytical performance characteristics and clinical utility of a novel assay for total hepatitis C virus core antigen quantification. J Clin Microbiol. 2010;48:1161-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 75] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 13. | Fytili P, Tiemann C, Wang C, Schulz S, Schaffer S, Manns MP, Wedemeyer H. Frequency of very low HCV viremia detected by a highly sensitive HCV-RNA assay. J Clin Virol. 2007;39:308-311. [PubMed] |
| 14. | Morota K, Fujinami R, Kinukawa H, Machida T, Ohno K, Saegusa H, Takeda K. A new sensitive and automated chemiluminescent microparticle immunoassay for quantitative determination of hepatitis C virus core antigen. J Virol Methods. 2009;157:8-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 98] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 15. | Medici MC, Furlini G, Rodella A, Fuertes A, Monachetti A, Calderaro A, Galli S, Terlenghi L, Olivares M, Bagnarelli P. Hepatitis C virus core antigen: analytical performances, correlation with viremia and potential applications of a quantitative, automated immunoassay. J Clin Virol. 2011;51:264-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 68] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 16. | Miedouge M, Saune K, Kamar N, Rieu M, Rostaing L, Izopet J. Analytical evaluation of HCV core antigen and interest for HCV screening in haemodialysis patients. J Clin Virol. 2010;48:18-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 17. | Waldenström J, Konar J, Ekermo B, Norder H, Lagging M. Neonatal transfusion-transmitted hepatitis C virus infection following a pre-seroconversion window-phase donation in Sweden. Scand J Infect Dis. 2013;45:796-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 18. | Mederacke I, Wedemeyer H, Ciesek S, Steinmann E, Raupach R, Wursthorn K, Manns MP, Tillmann HL. Performance and clinical utility of a novel fully automated quantitative HCV-core antigen assay. J Clin Virol. 2009;46:210-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 68] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 19. | Mederacke I, Potthoff A, Meyer-Olson D, Meier M, Raupach R, Manns MP, Wedemeyer H, Tillmann HL. HCV core antigen testing in HIV- and HBV-coinfected patients, and in HCV-infected patients on hemodialysis. J Clin Virol. 2012;53:110-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 20. | Tillmann HL, Wiegand J, Glomb I, Jelineck A, Picchio G, Wedemeyer H, Manns MP. Diagnostic algorithm for chronic hepatitis C virus infection: role of the new HCV-core antigen assay. Z Gastroenterol. 2005;43:11-16. [PubMed] |
| 21. | Kuo YH, Chang KC, Wang JH, Tsai PS, Hung SF, Hung CH, Chen CH, Lu SN. Is hepatitis C virus core antigen an adequate marker for community screening? J Clin Microbiol. 2012;50:1989-1993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 22. | Moini M, Ziyaeyan M, Aghaei S, Sagheb MM, Taghavi SA, Moeini M, Jamalidoust M, Hamidpour L. Hepatitis C virus (HCV) Infection Rate among Seronegative Hemodialysis Patients Screened by Two Methods; HCV Core Antigen and Polymerase Chain Reaction. Hepat Mon. 2013;13:e9147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 23. | Fujino T, Nakamuta M, Aoyagi Y, Fukuizumi K, Takemoto R, Yoshimoto T, Miyahara T, Harada N, Sakai H, Nakashima M. Early decline of the HCV core antigen can predict SVR in patients with HCV treated by Pegylated interferon plus ribavirin combination therapy. J Dig Dis. 2009;10:21-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 24. | Matsui K, Iwabuchi S, Shimizu H, Yoshida A, Fujikawa T, Takatsuka K. Two week induction of interferon-beta followed by pegylated interferon alpha-2b and ribavirin for chronic infection with hepatitis C. Hepatol Res. 2010;40:757-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 25. | Wada Y, Tamai H, Uno A, Kawashima A, Shingaki N, Mori Y, Moribata K, Miyata K, Higashi K, Deguchi H. Prediction of efficacy to pegylated interferon-α-2b plus ribavirin in patients with genotype 2 hepatitis C virus using viral response within 2 weeks. Hepatol Res. 2014;44:179-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 26. | Vermehren J, Susser S, Berger A, Perner D, Peiffer KH, Allwinn R, Zeuzem S, Sarrazin C. Clinical utility of the ARCHITECT HCV Ag assay for early treatment monitoring in patients with chronic hepatitis C genotype 1 infection. J Clin Virol. 2012;55:17-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 27. | Loggi E, Cursaro C, Scuteri A, Grandini E, Panno AM, Galli S, Furlini G, Bernardi M, Galli C, Andreone P. Patterns of HCV-RNA and HCV core antigen in the early monitoring of standard treatment for chronic hepatitis C. J Clin Virol. 2013;56:207-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 28. | Tamai H, Shingaki N, Shiraki T, Tukuda H, Mori Y, Moribata K, Enomoto S, Deguchi H, Ueda K, Inoue I. Prediction of sustained response to low-dose pegylated interferon alpha-2b plus ribavirin in patients with genotype 1b and high hepatitis C virus level using viral reduction within 2 weeks after therapy initiation. Hepatol Res. 2011;41:1137-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 29. | Shouval D. Focus: quantitative HBsAg measurement as a new surrogate marker for assessment of hepatic fibrosis in HBeAg+ chronic hepatitis B. J Hepatol. 2013;58:1063-1064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 30. | Durante-Mangoni E, Vallefuoco L, Sorrentino R, Iossa D, Perna E, Molaro R, Braschi U, Zampino R, Sodano G, Adinolfi LE. Clinico-pathological significance of hepatitis C virus core antigen levels in chronic infection. J Med Virol. 2013;85:1913-1918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 31. | Carabaich A, Ruvoletto M, Bernardinello E, Tono N, Cavalletto L, Chemello L, Gatta A, Pontisso P. Profiles of HCV core protein and viremia in chronic hepatitis C: possible protective role of core antigen in liver damage. J Med Virol. 2005;76:55-60. [PubMed] |