Published online Jun 7, 2014. doi: 10.3748/wjg.v20.i21.6523
Revised: January 14, 2014
Accepted: February 20, 2014
Published online: June 7, 2014
Processing time: 206 Days and 22.1 Hours
AIM: To investigate the role and mechanism of insulin-like growth factor binding protein-related protein 1 (IGFBPrP1) in the development of liver fibrosis.
METHODS: An in vitro model using hepatic stellate cell (HSC)-T6 cells and an in vivo model of rat liver overexpressing IGFBPrP1 were established using an IGFBPrP1-expressing recombinant adenovirus. The expression of IGFBPrP1 was examined by immunofluorescence, and the expression of collagen I and fibronectin was measured by real-time reverse transcription-polymerase chain reaction and Western blot analysis. The expression of Smad2/3 and p-Smad2/3 was examined by Western blot and immunohistochemistry. A shSmad3-expressing recombinant adenovirus (AdshSmad3) was designed and used to knockdown the Smad3 gene in HSC-T6 cells and rat liver fibrosis transfected with IGFBPrP1. The expression of collagen I, fibronectin, and α-smooth muscle actin (α-SMA) was determined by Western blot analysis and immunohistochemistry. Hepatocyte apoptosis was assessed using TUNEL assay.
RESULTS: IGFBPrP1 overexpression induced collagen deposition and up-regulated the expression of α-SMA and p-Smad2/3, and AdshSmad3 inhibited IGFBPrP1-stimulated p-Smad2/3 activation and the expression of α-SMA, collagen I and fibronectin in HSC-T6 cells. Similarly, increased hepatocyte apoptosis (38.56% ± 3.42% vs 0.24% ± 0.03%, P < 0.05), α-SMA positive stained cells (29.84% ± 1.36% vs 5.83% ± 1.47%, P < 0.05), and increased numbers of Smad3 (35.88% ± 2.15% vs 10.24% ± 1.31%, P < 0.05) and p-Smad2/3 positive cells (28.87% ± 2.73% vs 8.23% ± 0.98%, P < 0.05) were detected in the livers of IGFBPrP1-overexpressing rats compared with the control group. Moreover, AdshSmad3 reduced IGFBPrP1-stimulated Smad3 expression and attenuated α-SMA expression (29.84% ± 1.36% vs 8.23% ± 1.28%, P < 0.05), hepatocyte apoptosis (38.56% ± 3.42% vs 6.75% ± 0.52%, P < 0.05), and both collagen I and fibronectin deposition in the livers of AdIGFBPrP1-treated rats.
CONCLUSION: IGFBPrP1 induces liver fibrosis by mediating the activation of hepatic stellate cells and hepatocyte apoptosis in a Smad3-dependent mechanism.
Core tip: This study investigated the role and mechanism of insulin-like growth factor binding protein-related protein 1 (IGFBPrP1) in liver fibrosis using an adenovirus vector carrying IGFBPrP1 or a small interfering RNA targeting Smad3. We found that overexpression of IGFBPrP1 induced liver fibrosis by mediating hepatocyte apoptosis and hepatic stellate cells activation. We also identified the important role of the IGFBPrP1-Smad pathway in the regulation of IGFBPrP1 action in the development of liver fibrosis, and this pathway is a potential therapeutic target for liver fibrosis.
-
Citation: Zhang Y, Zhang QQ, Guo XH, Zhang HY, Liu LX. IGFBPrP1 induces liver fibrosis by inducing hepatic stellate cell activation and hepatocyte apoptosis
via Smad2/3 signaling. World J Gastroenterol 2014; 20(21): 6523-6533 - URL: https://www.wjgnet.com/1007-9327/full/v20/i21/6523.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i21.6523
Hepatic fibrosis is characterized by excessive production and deposition of extracellular matrix (ECM) components including collagen and fibronectin, and often results in hepatic cirrhosis and carcinoma[1]. During the process of liver fibrosis, hepatic stellate cells (HSCs) transform into myofibroblasts and are responsible for progressive ECM accumulation[2-4]. Several cytokines and growth factors have been shown to regulate HSC activation, proliferation and ECM production[5]. In addition, hepatocyte apoptosis may also contribute to HSC activation and the development of liver fibrosis. To date, there are no reports that a single molecule leads to hepatocyte apoptosis and HSC activation.
Insulin-like growth factor binding protein-related protein 1 (IGFBPrP1) has been shown to be a tumor suppressor by regulating cell proliferation, senescence and apoptosis. We previously reported that IGFBPrP1 was up-regulated in the liver of patients with hepatic cirrhosis and in mice with thioacetamide (TAA)-induced hepatic cirrhosis[6]. Most importantly, we demonstrated that recombinant IGFBPrP1 was capable of triggering HSC activation[7,8]. These findings suggest that IGFBPrP1 plays an important role in liver fibrosis. However, the mechanism has not been described.
Recent studies found that IGFBPrP1 stimulated glioma growth or fibroblast activation by binding activin A, a transforming growth factor (TGF)-β superfamily member, to regulate TGF-β signaling. TGF-β combines with transmembrane type I and type II serine/threonine kinase receptors (TβRI and TβRII) to form a complex, which will activate the downstream Smad pathway[9,10] or non-Smad pathway[11-14], such as the p44/p42 mitogen-activated protein kinase pathway and phosphoinositide 3-kinase-Akt-mTOR regulating ECM production. The TGF-β-Smad signaling pathway is one of the most important pathways responsible for regulating ECM production and liver fibrosis[15]. Since IGFBPrP1 can regulate the TGF-β pathway, it is not surprising that IGFBPrP1 may contribute to liver fibrosis via the Smad signaling pathway.
The aim of this study was to identify the role and mechanism of IGFBPrP1 in liver fibrosis using an adenovirus vector carrying IGFBPrP1 (AdIGFBPrP1) or a small interfering RNA targeting Smad3 (AdshSmad3). We found that overexpression of IGFBPrP1 induced liver fibrosis by mediating hepatocyte apoptosis and HSC activation. We also identified the important role of the IGFBPrP1-Smad pathway in the regulation of IGFBPrP1 action in the development of liver fibrosis, and this pathway is a potential therapeutic target for liver fibrosis.
The recombinant replication deficient adenovirus 5 expressing EGFP was constructed as previously described. The full-length cDNA of rat IGFBPrP1 was obtained from the cDNA library using the PCR method, then subcloned into the shuttle vector AdMax for preparation of replication-deficient adenovirus type 5 expressing IGFBPrP1 (AdIGFBPrP1) or no cDNA (cAd) at the GenePharma Company (Shanghai, China). Both AdIGFBPrP1 and cAd contained an EGFP marker, which was used to determine the transduction efficiency and to optimize viral infection in HSCs.
Four shRNAs targeting rat Smad3 mRNA (nt553-572, 906-925, 958-977, and 1054-1073) and a scrambled shRNA used as a negative control (shNC) were designed using software found on the Ambio website and synthesized by the GenePharma Company (Shanghai, China). The most effective shSmad3 (1054-1073) or shNC was then used to construct the adenoviral vectors containing shSmad3 (AdshSmad3) or shNC (AdshNC). Both AdshSmad3 and AdshNC contained an RFP marker, which was used to determine the transduction efficiency.
The HSC-T6 cell line was a gift from Scott L. Friedman of the Mount Sinai School of Medicine (NY, United States) and was cultured in RPMI 1640 medium (Gibco, United States) supplemented with 10% fetal calf serum, 100 U/mL penicillin and 100 g/mL streptomycin. After 24 h, HSC-T6 cells were transiently infected with AdshSmad3 or AdshNC in the presence of cAd or AdIGFBPrP1 at a multiplicity of infection (MOI) of 25, 50 and 100. The transfection efficiency was expressed as a percentage of the number of EGFP or RFP positive cells to the total cells.
Male wild-type Sprague-Dawley rats weighing 125-150 g were obtained from Shanxi Medical University Laboratory Animal Center (Shanxi, China). All procedures were approved by the Shanxi Medical University Animal Care and Use Committee. All rats were injected with 2 × 109 PFU of AdshNC or AdshSmad3 in the presence of PBS or 2 × 109 PFU of cAd or AdIGFBPrP1 administered via the tail vein. Ten rats were included in each experimental group. Rats were sacrificed 14 and 28 d after adenovirus administration. Blood and liver tissues were harvested.
Total RNA was extracted from the cells or tissues with Trizol reagent (Invitrogen Life Technology, CA, United States). cDNA was obtained using the Reverse Transcription reagent kit (Fermentas Life Sciences, CA, United States). Quantitative real-time PCR was performed using the SYBR Green PCR kit (Fermentas Life Sciences, CA, United States). The primer sequences were as follows: (1) IGFBPrP1 forward primer (5’-GCGAGCAAGGTCCTTCC AT-3’) and reverse primer (5’-CGGTCACCAGGCAGGAGTT-3’); (2) Collagen I forward primer (5’-AGCCAGCAGATCGAGAACAT-3’) and reverse primer (5’-TCT TGTCCTTGGGGTTCTTG-3’); (3) Smad3 forward primer (5’-GGGAGACATTCCACGCTTCA-3’) and reverse primer (5’-TAAGCTCCACGGCTGCATT-3’); (4) α-smooth muscle actin (α-SMA) forward primer (5’-TTCGTTACTACTGCTGAGCGTGAGA-3’) and reverse primer (5’ -AAAGATGGCTGGAAGAGGGTC-3’); (5) fibronectin forward primer (5’-CCAGGCACTGACTACAAGAT-3’) and reverse primer (5’-CATGATACCAGCAAGGACTT -3’); and (6) β-actin forward primer (5’-CTGGCACCACACCTTCTACA-3’) and reverse primer (5’-AGCACA GCCTGGATAGCAAC-3’). β-actin was used as an internal control. Experiments were performed at least 3 times with similar results. The mRNA results were expressed as number of folds relative to the control group.
Western blot was performed as previously described with antibodies to (1) IGFBPrP1 (1:300, Santa Cruz Biotechnology, United States); (2) α-SMA (1:500, Abcam, United Kingdom); (3) collagen I (1:300, Santa Cruz Biotechnology, United States); (4) fibronectin (1:300, Santa Cruz Biotechnology, United States); (5) TGF-β1 (1:200, Santa Cruz Biotechnology, United States); (6) Smad2/3 (1:300, Santa Cruz Biotechnology, United States); and (7) p-Smad2/3 (1:500, Abcam, United Kingdom). Immunoreactive blots were visualized using the Super ECL detection kit (Amersham Pharmacia Biotech, NJ, United States) according to the manufacturer’s instructions. Specific signals were scanned using scanning densitometry and quantified with Quantity One Image software.
All paraffin-embedded liver tissues were cut into 4 μm thick sections, and stained with hematoxylin and eosin or Sirius Red stain. Immunohistochemical staining was performed to examine the expression of α-SMA, Smad3 and p-Smad3. The results were analyzed with Image-Pro Plus 7.0 software and expressed as a percentage of the area occupied by the signal.
Hepatocyte apoptosis in liver sections was measured by TUNEL assay, which was performed according to the manufacturer’s instructions (In Situ Cell Death Detection Kit; Boehringer Mannheim, Indianapolis, IN, United States). The data were expressed as a percentage of the area of TUNEL-positive cells in 10 random fields.
Hydroxyproline content in whole liver specimens was quantified colorimetrically, which evaluated the total amount of collagen in the liver. The Hydroxyproline Assay Kit was purchased from Nanjing Jiancheng Bioengineering (Nanjing, China). In brief, liver specimens were hydrolyzed, lyophilized and the absorbance of each sample at 550 nm was assayed for hydroxyproline content using a spectrophotometer.
All data are expressed as mean ± SD. Statistical significance was determined using the Student’s t test as appropriate.
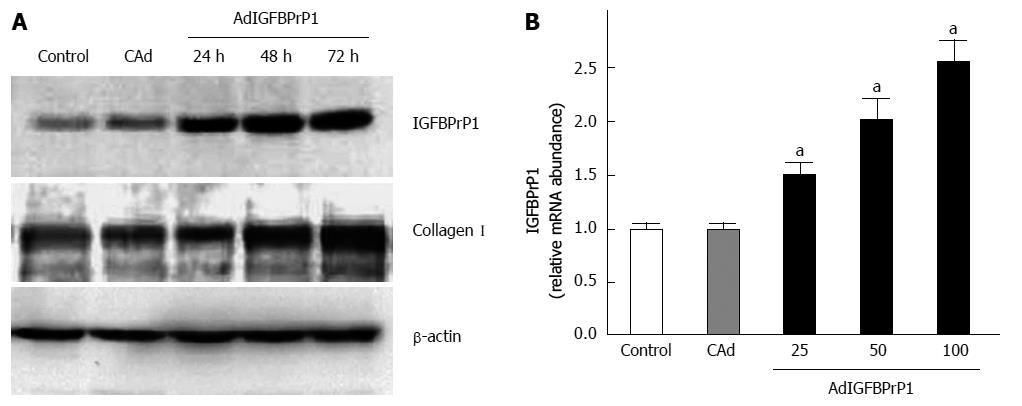
Having shown that rIGFBPrP1 induces HSC activation, we sought to determine whether endogenously expressed IGFBPrP1 exerts similar effects. We first established IGFBPrP1 overexpression using the adenovirus in HSC-T6 cells, a rat HSC cell line. As shown in Figure 1A and B, adenoviral gene transfer of IGFBPrP1 (AdIGFBPrP1) increased IGFBPrP1 mRNA and protein expression in HSC-T6 cells in a time-dependent manner compared to cAd (0.254 ± 0.072, 0.689 ± 0.023, 0.856 ± 0.034 vs 0.038 ± 0.062, P < 0.05). IGFBPrP1 overexpression similarly increased collagen I expression after transfection (Figure 1A, 0.614 ± 0.021, 0.986 ± 0.027, 1.294 ± 0.062 vs 0.596 ± 0.014, P < 0.05).
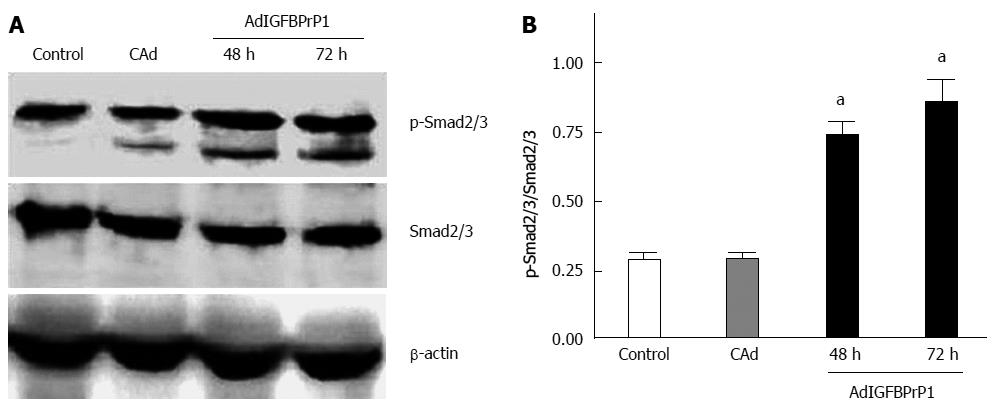
IGFBPrP1 has been shown to activate the TGF-β pathway in osteosarcoma cells. The TGF-β-Smad signaling pathway plays an important role in liver fibrosis. Smad2 and Smad3 are the main downstream mediators of TGF-β signaling in regulating ECM production. To determine the role of the Smad signaling pathway in mediating ECM up-regulation in response to elevated IGFBPrP1 levels, we measured Smad2/3 activation in HSC-T6 cells treated with AdIGFBPrP1. As shown in Figure 2A and B, Western blot analysis revealed that phosphorylation of Smad2/3 was 0.6-fold higher at 48 h and 1.5-fold higher at 72 h in HSC-T6 cells transduced with AdIGFBPrP1 than in the cAd group, suggesting that IGFBPrP1 overexpression activated the Smad2/3 pathway (P < 0.05).
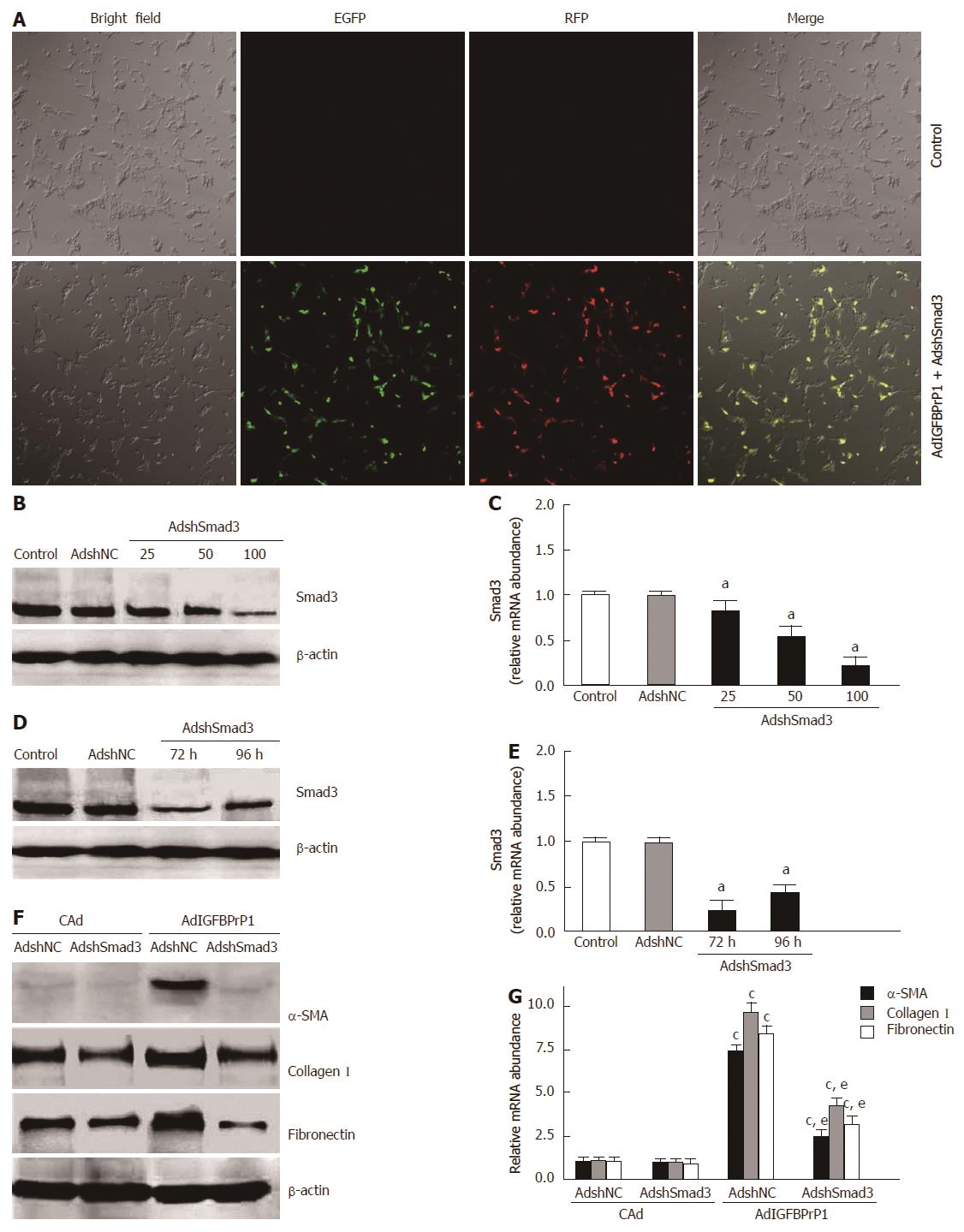
To further investigate whether activation of the Smad2/3 pathway contributes to IGFBPrP1-stimulated ECM production, we designed an adenovirus harboring an shRNA targeting Smad3 (AdshSmad3) to knock down the expression of Smad3 gene. HSC-T6 cells were co-transfected with AdshSmad3 or negative control (AdshNC) and AdIGFBPrP1 at three different MOI (25, 50 and 100). As shown in Figure 3A, transfection efficiency in HSC-T6 cells was approximately 85.23% ± 10.2% at an MOI of 100 (P < 0.05). As expected, real-time RT-PCR and Western blot results revealed that AdshSmad3 significantly reduced Smad3 protein by 68.45% ± 12.6% (P < 0.05) and mRNA by 76.45% ± 14.3% (P < 0.05) at 72 h at an MOI of 100 (Figure 3B and C). Our results also showed that AdshSmad3 inhibited Smad3 protein by 68.6% ± 12.6% and 58.6% ± 9.8% at 72 and 96 h, respectively, in HSC-T6 cells compared with AdshNC at an MOI of 100 (P < 0.05, Figure 3D). In addition, AdshSmad3 inhibited Smad3 mRNA by 74.3% ± 11.2% and 63.2% ± 10.4% (P < 0.05, Figure 3E). Importantly, knockdown of Smad3 significantly abrogated IGFBPrP1-stimulated induction of α-SMA (0.196 ± 0.012 vs 0.723 ± 0.015, P < 0.05), collagen I (0.482 ± 0.019 vs 1.268 ± 0.027, P < 0.05) and fibronectin expression (0.334 ± 0.024 vs 1.146 ± 0.015, P < 0.05) (Figure 3F). Similarly, up-regulation of α-SMA, collagen I and fibronectin mRNA in response to IGFBPrP1 overexpression was suppressed by AdshSmad3 (P < 0.05, Figure 3G). Taken together, these data indicated that IGFBPrP1-induced ECM expression in HSC-T6 cells was Smad dependent.
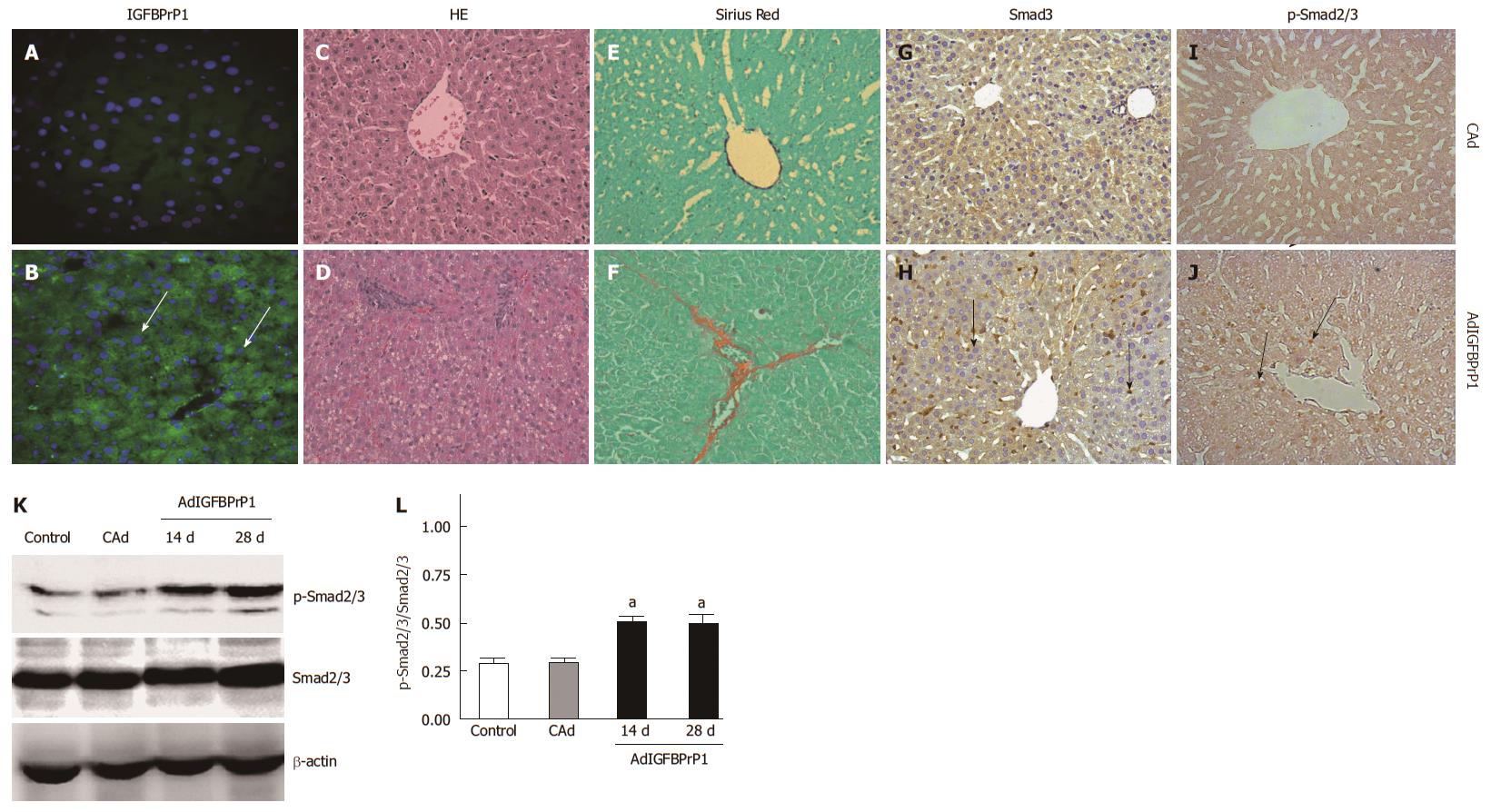
Based on these in vitro data, we hypothesized that IGFBPrP1 leads to the development of liver fibrosis via the Smad pathway. Rats were injected with 2 × 109 PFU of cAd or AdIGFBPrP1 via the tail vein. Expression of IGFBPrP1 in AdIGFBPrP1 or cAd-treated rat livers was examined by immunohistochemistry. As shown in Figure 4A and B, IGFBPrP1 was expressed mainly in hepatocytes and sinusoidal cells 2 d after adenovirus injection. Hepatocyte steatosis, cellular infiltration and excessive collagen deposition were observed at 28 d in IGFBPrP1-treated rats compared with cAd-treated rats (Figure 4C and D). Collagen content, quantified by Sirius Red staining, was markedly increased at 28 d in the liver of AdIGFBPrP1-injected rats compared with cAd-injected rats (Figure 4E and F).
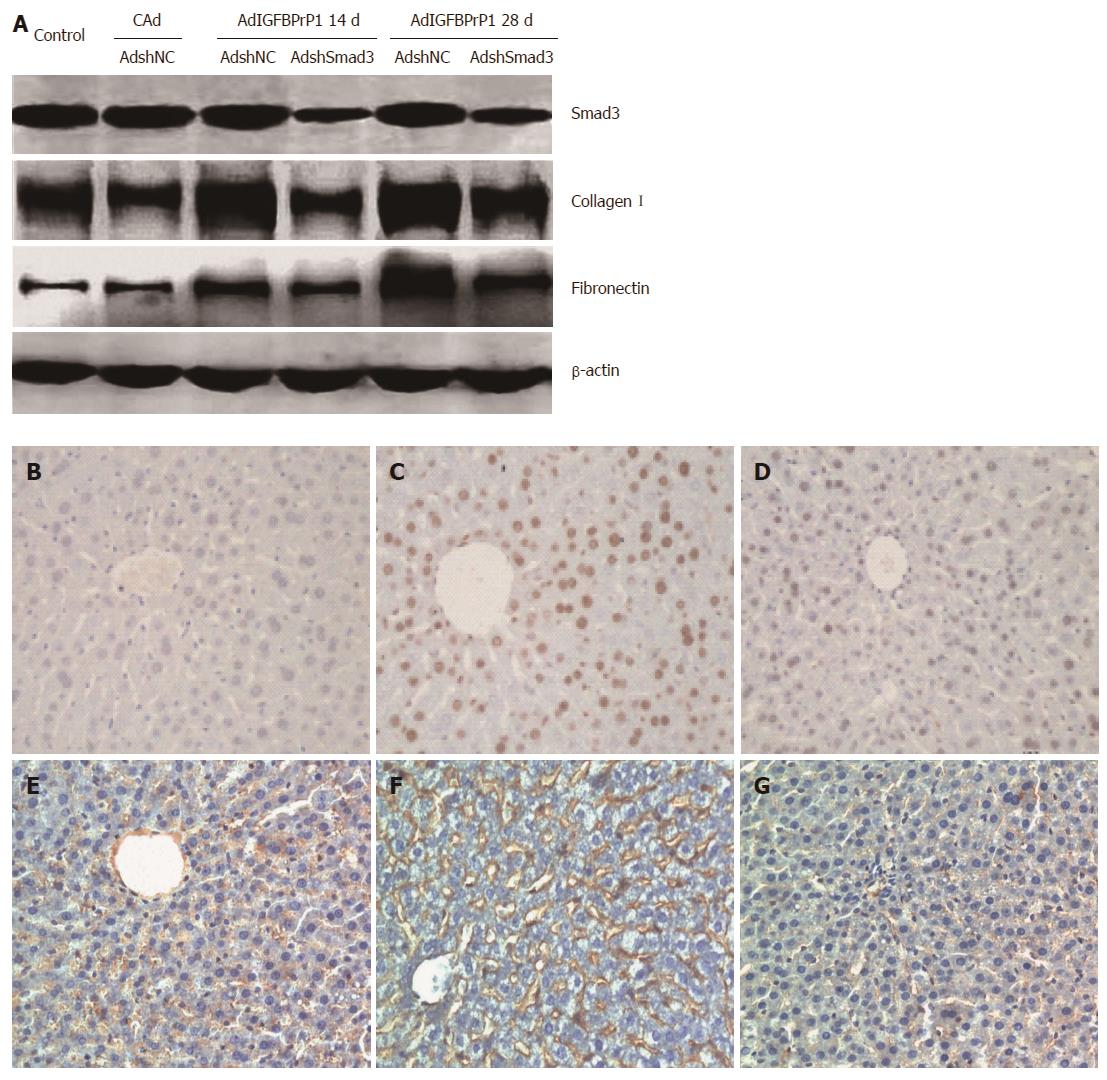
We then examined Smad2/3 expression. Immunohistochemistry revealed faint expression of Smad3 and phosphorylated Smad2/3 (p-Smad2/3) in the liver of normal rats. Moreover, Smad3 and p-Smad2/3 were strongly expressed in the IGFBPrP1-induced fibrotic liver (Figure 4G-J). The positive areas in IGFBPrP1-induced fibrotic liver were larger than those in the cAd group (Smad3, 35.88% ± 2.15% vs 10.24% ± 1.31%, P < 0.05; p-Smad2/3, 28.87% ± 2.73% vs 8.23% ± 0.98%, P < 0.05). Consistent with the immunohistochemistry staining results, Western blot results also showed that the expression of Smad2/3 and p-Smad2/3 protein was increased 14 and 28 d after IGFBPrP1 administration (Smad3, 1.342 ± 0.075, 1.586 ± 0.116 vs 0.657 ± 0.032, P < 0.05; p-Smad2/3, 0.682 ± 0.043, 0.856 ± 0.064 vs 0.189 ± 0.007, P < 0.05) (Figure 4K) and the ratio of p-Smad2/3 to total Smad2/3 was significantly up-regulated in the AdIGFBPrP1 group compared with the normal and cAd groups (P < 0.05) (Figure 4L).
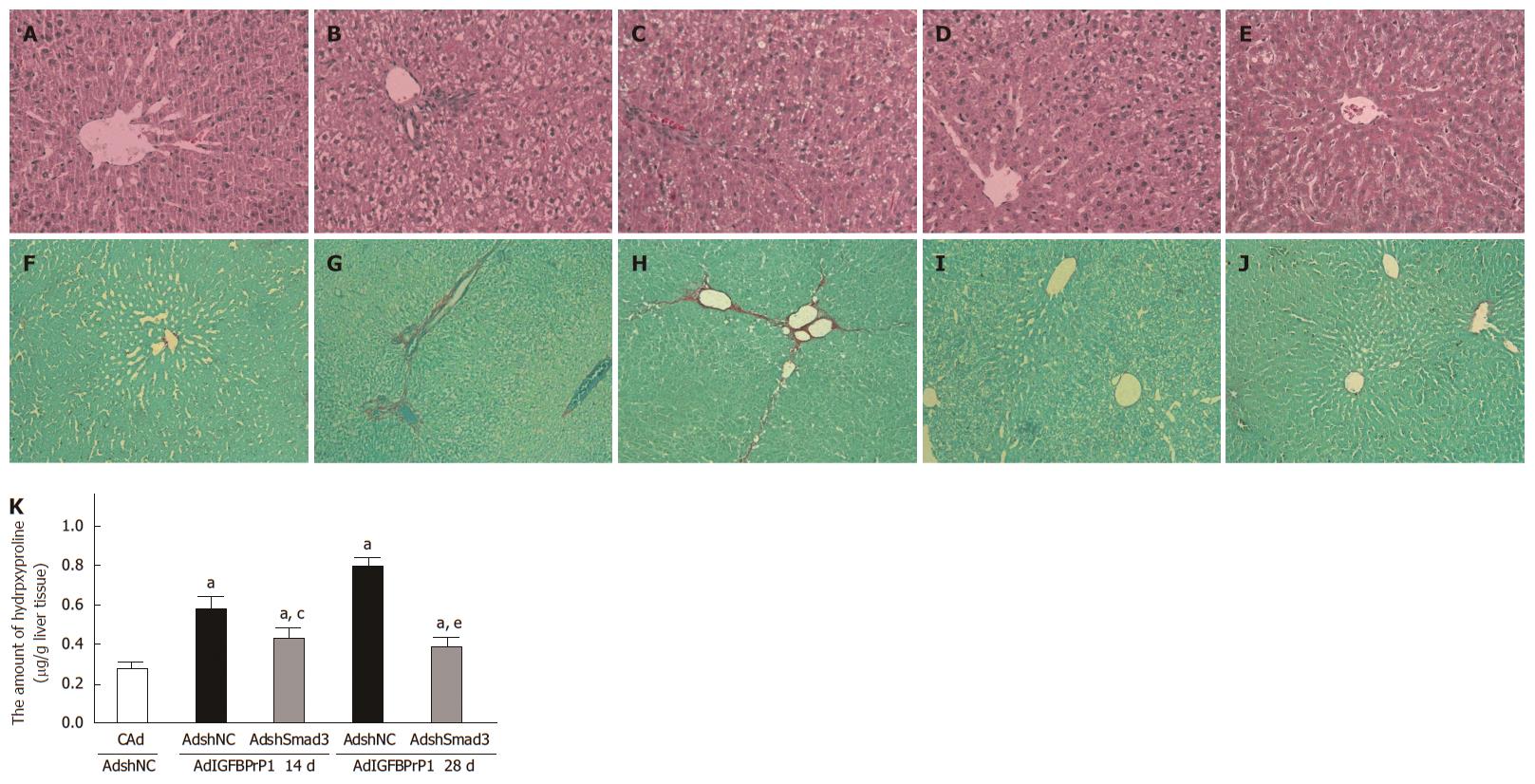
To further elucidate the effect of the Smad2/3 pathway on IGFBPrP1-induced liver fibrosis, we injected SD rats with 2 × 109 PFU of AdshNC or AdshSmad3 in the presence of 2 × 109 PFU of AdIGFBPrP1 into the tail vein. The levels of serum ALT and AST increased in AdIGFBPrP1-treated rats compared with the control group and decreased in AdshSmad3-treated rats compared with AdIGFBPrP1-treated rats. As shown in Figure 5A-J, AdshSmad3 ameliorated liver fibrosis induced by IGFBPrP1 as demonstrated by both hematoxylin and eosin and Sirius red staining. Hydroxyproline content in the AdshSmad3 group was down-regulated by 48.5% ± 12.6% as compared with the AdshNC group and IGFBPrP1 group (P < 0.05, Figure 5K). Moreover, the expression of Smad3, collagen I and fibronectin proteins was significantly up-regulated at 14 and 28 d in fibrotic livers induced by IGFBPrP1 (Smad3, 1.128 ± 0.164, 1.345 ± 0.156 vs 0.626 ± 0.021, P < 0.05; collagen I, 0.832 ± 0.031, 1.324 ± 0.076 vs 0.534 ± 0.018, P < 0.05; fibronectin, 0.647 ± 0.037, 1.225 ± 0.039 vs 0.324 ± 0.022, P < 0.05) and was markedly down-regulated 14 and 28 d after AdshSmad3 treatment as demonstrated by Western blot analysis (Smad3, 0.594 ± 0.147 vs 1.128 ± 0.164, 0.742 ± 0.189 vs 1.345 ± 0.156, P < 0.05; collagen I, 0.626 ± 0.025 vs 0.832 ± 0.031, 0.728 ± 0.014 vs 1.324 ± 0.076, P < 0.05; fibronectin, 0.428 ± 0.018 vs 0.647 ± 0.037, 0.532 ± 0.024 vs 1.225 ± 0.039, P < 0.05, Figure 6A).
The TGF-β/Smad pathway is not only associated with HSC activation, but also participates in hepatocyte apoptosis. In light of the mechanism of the Smad pathway in the development of liver fibrosis induced by IGFBPrP1, hepatocyte apoptosis and HSC activation in rat livers were evaluated 28 d after co-infection with AdIGFBPrP1 and AdShmad3. As shown in Figure 6B-D, no TUNEL-positive cells were identified in normal liver, whereas scattered TUNEL-positive cells were observed in AdIGFBPrP1-treated rat liver (38.56% ± 3.42% vs 0.24% ± 0.03%, P < 0.05). Interestingly, AdshSmad3 reduced AdIGFBPrP1-induced TUNEL-positive cells (6.75% ± 0.52% vs 38.56% ± 3.42%, P < 0.05). We then examined α-SMA expression, a marker of HSC activation, by immunohistochemistry. α-SMA-positive cells were more abundant in the liver of IGFBPrP1-treated rats compared with normal rats (29.84% ± 1.36% vs 5.83% ± 1.47%, P < 0.05). More importantly, AdshSmad3 reduced AdIGFBPrP1-induced α-SMA-positive cells (8.23% ± 1.28% vs 29.84% ± 1.36%, P < 0.05, Figure 6E-G).
Liver fibrosis is thought to be a reversible disease. HSCs have been recognized to play an important role in the development of liver fibrosis. Thus, many effective therapeutic approaches have intensified interest in regulating HSC activation and proliferation[16-19]. Recently, the IGFBPrP1 gene was found to be significantly increased during HSC activation[6]. Therefore, we examined the role of IGFBPrP1 in liver fibrosis. We found that anti-IGFBPrP1 antibody can attenuate TAA-induced hepatic fibrosis[7]. Moreover, siRNA targeting IGFBPrP1 reduced HSC activation and ECM production stimulated by TGF-β. Most importantly, we previously reported that recombinant IGFBPrP1 induces HSC activation in vitro[8]. However, the molecular mechanism underlying this process and the in vivo effect of IGFBPrP1 have not been elucidated. In this study, we demonstrated that overexpression of IGFBPrP1 induced liver fibrosis by stimulating hepatocyte apoptosis and HSC activation, and the underlying mechanism involved the Smad2/3 pathway.
IGFBPrP1, also known as Mac25 or IGFBP7, is a member of the IGFBP superfamily. It appears to be involved in diverse biological functions, such as regulation of cell growth, stimulation of prostacyclin production, and binding of type IV collagen, IGFs and insulin[20-23]. Interestingly, IGFBPrP1 demonstrated positive and negative roles in tumor progression by mediating fibroblast activation or epithelial cell senescence[24,25]. However, the relationship between IGFBPrP1 and liver fibrosis has not been investigated. We established an in vitro and an in vivo model in which we transiently overexpressed IGFBPrP1 in HSC-T6 cells and in rat liver by adenoviral-mediated IGFBPrP1 gene transfer, respectively, as the replication-deficient recombinant adenovirus has very high efficient delivery into target cells and was reported to be suitable for liver fibrosis[26,27]. With this approach, we showed that overexpression of IGFBPrP1 caused activation of HSCs and ECM production in HSC-T6 cells, which resulted in liver fibrosis, and AdIGFBPrP1-treated rats developed liver steatosis and fibrosis.
HSCs are known to have an important role in liver fibrosis, however, hepatocyte apoptosis is now emerging as a critical event in the progression of liver fibrosis[28,29]. Engulfment of apoptotic bodies by HSCs stimulates the activation of HSCs and ECM production. Hepatocyte apoptosis may also be responsible for the generation of inflammatory mediators leading to liver inflammation and fibrosis. We observed increased hepatocyte apoptosis and HSC activation in IGFBPrP1-treated fibrotic liver.
The TGFβ-Smad signaling pathway is the main pathway regulating ECM production and liver fibrosis. Recent studies found that IGFBPrP1 stimulated glioma growth or fibroblast activation by binding activin A to regulate the TGF-β pathway. Activin A belongs to the TGF-β superfamily and activates the Smad pathway in systemic sclerosis[30,31]. Therefore, it is not surprising that IGFBPrP1 may induce liver fibrosis via the activin A-Smads pathway. Kitamura et al[15] reported that Smad expression increased in the nucleus of HSCs in liver fibrosis both in vivo and in vitro. We found that overexpression of IGFBPrP1 up-regulated p-Smad2/3 expression in cultured HSC-T6 cells and IGFBPrP1-induced liver fibrosis. Our results were consistent with stimulation by TGF-β1[32], suggesting that IGFBPrP1 activated the Smad2/3 pathway in activated HSCs both in vivo and in vitro. Furthermore, our results also showed that strong p-Smad2/3 expression was observed in the nucleus of hepatocytes in IGFBPrP1-induced liver fibrosis. Taken together, our data suggest that the Smad pathway participated in IGFBPrP1-induced liver fibrosis.
Latella et al[33] previously demonstrated that targeted disruption of Smad3 inhibits the development of TAA-induced hepatic fibrosis in mice. In order to further evaluate the effect of the Smad pathway on IGFBPrP1-induced liver fibrosis, we successfully used AdshSmad3 to knockdown the Smad3 gene in AdIGFBPrP1-treated HSC-T6 cells and rat liver as demonstrated by real-time RT-PCR and Western blot analysis. Furthermore, AdshSmad3 attenuated AdIGFBPrP1-induced liver fibrosis and reduced the expression of α-SMA, collagen I and fibronectin both in vivo and in vitro. More importantly, AdshSmad3 attenuated AdIGFBPrP1-induced hepatocyte apoptosis. It was reported that the Smad2/3 pathway not only stimulated HSC activation, but also induced hepatocyte apoptosis[34,35]. Taken together, these data demonstrated that IGFBPrP1 may contribute to liver fibrosis by inducing HSC activation and hepatocyte apoptosis in a Smad-dependent manner.
In summary, we have shown that adenovirus-mediated IGFBPrP1 overexpression induced HSC activation and ECM production in vitro via the Smad pathway. More importantly, overexpression of IGFBPrP1 induced hepatocyte apoptosis and HSC activation in vivo in a Smad-dependent manner. These data suggest a novel potential therapeutic target for liver fibrosis.
Hepatic stellate cells (HSCs) play an important role in the development of liver fibrosis. Insulin-like growth factor binding protein-related protein 1 (IGFBPrP1) is a novel protein, which is involved in the activation of HSCs and regulates cell proliferation, senescence and apoptosis.
Significant IGFBPrP1 expression has been detected during fibrogenesis and our previous study demonstrated that rIGFBPrP1 activated HSCs and increased extracellular matrix production in vitro. In this study, the authors focused on the in vivo effects and the mechanism of endogenous IGFBPrP1 expression by administration of a recombinant adenovirus expressing IGFBPrP1 or shSmad3.
Recent studies have highlighted the importance of IGFBP5 and IGFBPrP1 in the activation of HSCs. The authors provide the first evidence that overexpression of IGFBPrP1 induces liver fibrosis by inducing HSC activation and hepatocyte apoptosis in a Smad3-dependent manner.
By understanding the effect and mechanism of IGFBPrP1 in the development of liver fibrosis, IGFBPrP1 could be a novel potential therapeutic target for liver fibrosis.
HSCs are responsible for extracellular matrix deposition during liver fibrosis. The activation of HSCs is regulated by many cytokines. Hepatocyte apoptosis may also contribute to HSC activation. IGFBPrP1 can induce hepatocyte apoptosis and HSC activation.
The authors demonstrated that overexpression of IGFBPrP1 induced liver fibrosis by mediating hepatocyte apoptosis and HSC activation and identified the IGFBPrP1-mediated pathway involved in liver fibrosis development. The interesting conclusion of the study was that this pathway could be a potential therapeutic target for liver fibrosis.
P- Reviewers: Gorrell MD, Leardkamolkarn V, Wong GLH, Zocco MA S- Editor: Gou SX L- Editor: Wang TQ E- Editor: Wang CH
| 1. | Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209-218. [PubMed] |
| 2. | Leask A, Denton CP, Abraham DJ. Insights into the molecular mechanism of chronic fibrosis: the role of connective tissue growth factor in scleroderma. J Invest Dermatol. 2004;122:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 103] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 3. | Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655-1669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2139] [Cited by in RCA: 2159] [Article Influence: 127.0] [Reference Citation Analysis (0)] |
| 4. | Pinzani M. Novel insights into the biology and physiology of the Ito cell. Pharmacol Ther. 1995;66:387-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 124] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 5. | Wong L, Yamasaki G, Johnson RJ, Friedman SL. Induction of beta-platelet-derived growth factor receptor in rat hepatic lipocytes during cellular activation in vivo and in culture. J Clin Invest. 1994;94:1563-1569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 242] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 6. | Liu LX, Zhang HY, Zhang QQ, Guo XH. Effects of insulin-like growth factor binding protein-related protein 1 in mice with hepatic fibrosis induced by thioacetamide. Chin Med J (Engl). 2010;123:2521-2526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 7. | Boers W, Aarrass S, Linthorst C, Pinzani M, Elferink RO, Bosma P. Transcriptional profiling reveals novel markers of liver fibrogenesis: gremlin and insulin-like growth factor-binding proteins. J Biol Chem. 2006;281:16289-16295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 86] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 8. | Liu LX, Huang S, Zhang QQ, Liu Y, Zhang DM, Guo XH, Han DW. Insulin-like growth factor binding protein-7 induces activation and transdifferentiation of hepatic stellate cells in vitro. World J Gastroenterol. 2009;15:3246-3253. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 22] [Cited by in RCA: 30] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 9. | Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2875] [Cited by in RCA: 2918] [Article Influence: 104.2] [Reference Citation Analysis (0)] |
| 10. | Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin CH, Miyazono K. TGF-beta receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16:5353-5362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 806] [Cited by in RCA: 854] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 11. | Mu Y, Gudey SK, Landström M. Non-Smad signaling pathways. Cell Tissue Res. 2012;347:11-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 432] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 12. | Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128-139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1451] [Cited by in RCA: 1427] [Article Influence: 89.2] [Reference Citation Analysis (0)] |
| 13. | Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3797] [Cited by in RCA: 4292] [Article Influence: 195.1] [Reference Citation Analysis (0)] |
| 14. | ten Dijke P, Hill CS. New insights into TGF-beta-Smad signalling. Trends Biochem Sci. 2004;29:265-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 914] [Cited by in RCA: 932] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 15. | Kitamura Y, Ninomiya H. Smad expression of hepatic stellate cells in liver cirrhosis in vivo and hepatic stellate cell line in vitro. Pathol Int. 2003;53:18-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 24] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 16. | Ji J, Zhang J, Huang G, Qian J, Wang X, Mei S. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 2009;583:759-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 252] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 17. | Adachi M, Brenner DA. High molecular weight adiponectin inhibits proliferation of hepatic stellate cells via activation of adenosine monophosphate-activated protein kinase. Hepatology. 2008;47:677-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 148] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 18. | Georgescu EF, Ionescu R, Niculescu M, Mogoanta L, Vancica L. Angiotensin-receptor blockers as therapy for mild-to-moderate hypertension-associated non-alcoholic steatohepatitis. World J Gastroenterol. 2009;15:942-954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 135] [Cited by in RCA: 136] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 19. | Jeong WI, Park O, Radaeva S, Gao B. STAT1 inhibits liver fibrosis in mice by inhibiting stellate cell proliferation and stimulating NK cell cytotoxicity. Hepatology. 2006;44:1441-1451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 218] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 20. | Rodgers BD, Roalson EH, Thompson C. Phylogenetic analysis of the insulin-like growth factor binding protein (IGFBP) and IGFBP-related protein gene families. Gen Comp Endocrinol. 2008;155:201-207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 21. | Oh Y, Nagalla SR, Yamanaka Y, Kim HS, Wilson E, Rosenfeld RG. Synthesis and characterization of insulin-like growth factor-binding protein (IGFBP)-7. Recombinant human mac25 protein specifically binds IGF-I and -II. J Biol Chem. 1996;271:30322-30325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 256] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 22. | Wolchinsky Z, Shivtiel S, Kouwenhoven EN, Putin D, Sprecher E, Zhou H, Rouleau M, Aberdam D. Angiomodulin is required for cardiogenesis of embryonic stem cells and is maintained by a feedback loop network of p63 and Activin-A. Stem Cell Res. 2014;12:49-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Yamanaka Y, Wilson EM, Rosenfeld RG, Oh Y. Inhibition of insulin receptor activation by insulin-like growth factor binding proteins. J Biol Chem. 1997;272:30729-30734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 158] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 24. | Komiya E, Furuya M, Watanabe N, Miyagi Y, Higashi S, Miyazaki K. Elevated expression of angiomodulin (AGM/IGFBP-rP1) in tumor stroma and its roles in fibroblast activation. Cancer Sci. 2012;103:691-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 25. | Jiang W, Xiang C, Cazacu S, Brodie C, Mikkelsen T. Insulin-like growth factor binding protein 7 mediates glioma cell growth and migration. Neoplasia. 2008;10:1335-1342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 62] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 26. | Nakatani T, Kuriyama S, Tominaga K, Tsujimoto T, Mitoro A, Yamazaki M, Tsujinoue H, Yoshiji H, Nagao S, Fukui H. Assessment of efficiency and safety of adenovirus mediated gene transfer into normal and damaged murine livers. Gut. 2000;47:563-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 27. | Hu PF, Chen H, Zhong W, Lin Y, Zhang X, Chen YX, Xie WF. Adenovirus-mediated transfer of siRNA against PAI-1 mRNA ameliorates hepatic fibrosis in rats. J Hepatol. 2009;51:102-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 28. | Canbay A, Taimr P, Torok N, Higuchi H, Friedman S, Gores GJ. Apoptotic body engulfment by a human stellate cell line is profibrogenic. Lab Invest. 2003;83:655-663. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 311] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 29. | Feldstein AE, Canbay A, Angulo P, Taniai M, Burgart LJ, Lindor KD, Gores GJ. Hepatocyte apoptosis and fas expression are prominent features of human nonalcoholic steatohepatitis. Gastroenterology. 2003;125:437-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 775] [Cited by in RCA: 795] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 30. | Takagi K, Kawaguchi Y, Kawamoto M, Ota Y, Tochimoto A, Gono T, Katsumata Y, Takagi M, Hara M, Yamanaka H. Activation of the activin A-ALK-Smad pathway in systemic sclerosis. J Autoimmun. 2011;36:181-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 31. | Ohga E, Matsuse T, Teramoto S, Ouchi Y. Activin receptors are expressed on human lung fibroblast and activin A facilitates fibroblast-mediated collagen gel contraction. Life Sci. 2000;66:1603-1613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 32. | Liu C, Gaça MD, Swenson ES, Vellucci VF, Reiss M, Wells RG. Smads 2 and 3 are differentially activated by transforming growth factor-beta (TGF-beta ) in quiescent and activated hepatic stellate cells. Constitutive nuclear localization of Smads in activated cells is TGF-beta-independent. J Biol Chem. 2003;278:11721-11728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 154] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 33. | Latella G, Vetuschi A, Sferra R, Catitti V, D’Angelo A, Zanninelli G, Flanders KC, Gaudio E. Targeted disruption of Smad3 confers resistance to the development of dimethylnitrosamine-induced hepatic fibrosis in mice. Liver Int. 2009;29:997-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 85] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 34. | Yoo J, Ghiassi M, Jirmanova L, Balliet AG, Hoffman B, Fornace AJ, Liebermann DA, Bottinger EP, Roberts AB. Transforming growth factor-beta-induced apoptosis is mediated by Smad-dependent expression of GADD45b through p38 activation. J Biol Chem. 2003;278:43001-43007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 207] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 35. | Yang YA, Zhang GM, Feigenbaum L, Zhang YE. Smad3 reduces susceptibility to hepatocarcinoma by sensitizing hepatocytes to apoptosis through downregulation of Bcl-2. Cancer Cell. 2006;9:445-457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 121] [Article Influence: 6.4] [Reference Citation Analysis (0)] |