Published online Jun 7, 2014. doi: 10.3748/wjg.v20.i21.6448
Revised: February 16, 2014
Accepted: March 12, 2014
Published online: June 7, 2014
Processing time: 219 Days and 17.6 Hours
Epstein-Barr virus (EBV)-associated gastric carcinoma (EBVaGC) comprises nearly 10% of gastric carcinoma cases worldwide. Recently, it was recognised to have unique clinicopathologic characteristics, including male predominance, lower rates of lymph node involvement, and better prognosis. EBVaGC is further characterised by abnormal hypermethylation of tumour suppressor gene promoter regions, causing down-regulation of their expression. In the present review, we critically discuss the role of EBV in gastric carcinogenesis, summarising the role of viral proteins and microRNAs with respect to aberrant methylation in EBVaGC. Given the role of epigenetic dysregulation in tumourigenesis, epigenetic modifiers may represent a novel therapeutic strategy.
Core tip: Epstein-Barr virus (EBV)-associated gastric carcinoma (EBVaGC) comprises nearly 10% of gastric carcinoma cases worldwide. In the present review, we critically discuss the role of EBV in gastric carcinogenesis, summarising the role of viral proteins and microRNAs) with respect to aberrant methylation in EBVaGC. Given the role of epigenetic dysregulation in tumourigenesis, epigenetic modifiers may represent a novel therapeutic strategy.
- Citation: Yau TO, Tang CM, Yu J. Epigenetic dysregulation in Epstein-Barr virus-associated gastric carcinoma: Disease and treatments. World J Gastroenterol 2014; 20(21): 6448-6456
- URL: https://www.wjgnet.com/1007-9327/full/v20/i21/6448.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i21.6448
Epstein-Barr virus (EBV) infection is ubiquitous, and is accepted as a causative microorganism for various malignancies including nasopharyngeal carcinoma (NPC), Burkitt’s lymphoma, and gastric carcinoma (GC). EBV-associated GC (EBVaGC) accounts for approximately 10% of cases worldwide[1,2], and is characterised by unique clinicopathologic features including a relatively favourable prognosis (Table 1)[1-4]. In recent years, the molecular mechanisms underlying EBV-related carcinogenesis have become increasingly understood. EBV may contribute to tumourigenesis through the expression of viral proteins and microRNAs (miRNAs). Previous studies have also reported that promoter methylation was observed more frequently in EBVaGC. Hence another method by which EBV contributes to gastric carcinogenesis is through aberrant DNA methylation and histone modification. Thus EBVaGC is characterised by distinct variations on genomic, epigenomic, and transcriptomic levels[5]. Here, we review the mechanism by which EBV infection causes aberrant methylation, transformation, cancer development, and its associated therapeutic implications.
| Clinical and pathological features | |
| Age | Younger1 |
| Gender | Male predominance |
| Associations | Smoking |
| Prevalence | 10% of gastric carcinoma cases |
| Location | Gastric body/cardia |
| Remnant stomach | |
| Clinical | Multiple carcinomas1 |
| Thickening of gastric wall | |
| Ulcerated (saucer-like) neoplasm | |
| Lower rate of lymph node involvement1 | |
| Histology | Lymphoepithelioma-like |
| Lymphocytic infiltration in various degrees | |
| Atrophic gastritis | |
| Lace pattern within the mucosa | |
| Moderate to poorly differentiated adenocarcinoma | |
| Prognosis | Longer survival1 |
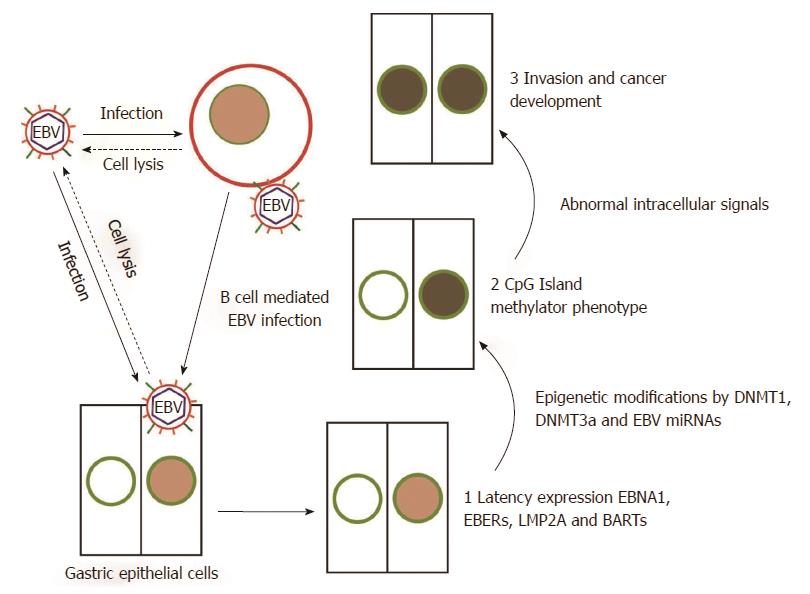
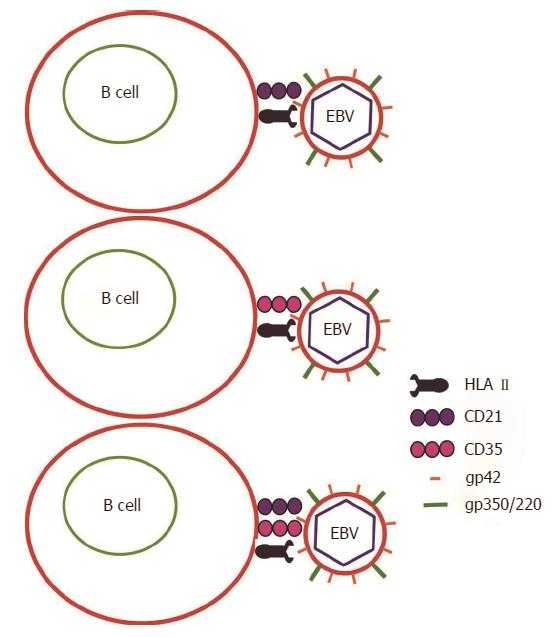
EBV may infect host gastric epithelial cells directly or indirectly (Figure 1). In direct infection, the viral envelope glycoprotein BMRF-2 interacts with cellular β1 integrins. Subsequently, viral protein gH/gL attaches to cellular αvβ6/8 integrins, and triggers fusion of the viral envelope with the epithelial cell membrane[6]. EBV preferentially infects B lymphocytes, which then mediates subsequent infection to epithelial cells[7]. In B cell invasion, EBV envelope glycoproteins gp350/220 bind to B cell receptors CD21 and/or CD35[8,9]. Simultaneously, viral glycoprotein gp42 interacts with Human Leukocyte Antigen (HLA) class II molecules on the B cell membrane to trigger the core fusion complex, enabling EBV entry into the B cell (Figure 2)[8,10]. Through direct cell-to-cell contact, EBV-infected B cells may subsequently infect epithelial cells[11]. The exact mechanism of epithelial cell invasion is unclear, but involves CD21-mediated co-capping of EBV and integrins on B cells, as well as conjugate formation between EBV-infected B cells and epithelial cells via the capped adhesion molecules[11]. Once EBV enters epithelial cells, the viral capsid dissolves and the viral genome is transported to the cell nucleus.
Following infection, EBV typically persists in a latent stage. During latency, the viral genome is largely silenced by host-driven methylation of CpG island motifs. Based on the subset of viral genes which are expressed, tumours may be classified into four types; latency Ia, Ib, II, and III (Table 2). EBVaGC belongs to latency type I, where the viral genes EBV nuclear antigen 1 (EBNA1), EBV-encoded small RNA (EBER1/2), BamHI-A rightward transcripts (BARTs), and latent membrane protein 2A (LMP2A) may be expressed[12,13]. Notably, the expression of latency genes is associated with malignancy. For example, EBER1 up-regulates the expression of insulin-growth factor-1, an autocrine growth factor which accelerates cell proliferation in EBVaGC[14].
| Genes | Latency Ia | Latency Ib | Latency II | Latency III |
| EBNA1 | + | + | + | + |
| EBNA2 | – | – | – | + |
| EBNA3a | – | – | – | + |
| EBNA3b | – | – | – | + |
| EBNA3c | – | – | – | + |
| EBNA-LP | – | – | + | + |
| LMP1 | – | – | + | + |
| LMP2A | – | + | + | + |
| LMP2B | – | – | + | + |
| EBER1 | + | + | + | + |
| EBER2 | + | + | + | + |
| BARTs | + | + | + | + |
| Disease | EBVaGC, Burkitt’s lymphoma | EBVaGC | NPC, Hodgkin’s lymphoma, NK/T-cell lymphoma | AIDS-associated B-cell lymphomas, Pyothorax-associated lymphoma |
Half of all EBVaGCs also express LMP2A. LMP2A plays a critical role in the oncogenic processes in EBVaGC, and thus EBV latency patterns should be further subdivided into Ia or Ib based on the absence or presence of LMP2A[12,15]. LMP2A not only inhibits apoptosis through up-regulation of the cellular survivin gene via the NF-κB pathway[16], but induces expression of phosphorylated signal transducer and activator of transcription 3 (pSTAT3), which causes up-regulation of DNA methyltransferase DNMT1[16] and DNMT3B[17] in EBV-infected GC cells. DNA methyltransferases play important roles in controlling DNA methylation. The subsequent overdrive of CpG methylation and silencing of tumour suppressor genes such as PTEN, p16, and p73 leads to the transformation of EBV-infected cells. Hence epigenetic dysregulation plays an important role in gastric carcinogenesis.
Epigenetics refer to functionally relevant and heritable changes in gene expression that occur without alteration of the underlying DNA sequence. The two primary mechanisms which may produce this change are DNA methylation, and histone modification. According to the epigenetic progenitor model, tumour-progenitor genes promote the polyclonal epigenetic disruption of stem cells as a first step in the development of cancer[18]. This epigenetic plasticity causes genomic instability, and collectively drives tumour progression[19].
The CpG island methylator phenotype was first observed in EBVaGC in 1999[20]. EBV infection was shown to induce extensive methylation and repression of tumour suppressor genes over 18 wk in MKN7, a low methylation GC cell line[21]. Subsequent studies confirmed that EBVaGC has higher rates of aberrant DNA methylation than EBV non-associated GC (EBVnGC)[21,22]. Nevertheless, the mechanisms by which EBV induces aberrant DNA methylation and histone modification remain poorly understood.
Viral encoded miRNAs play a pivotal role in alterations to DNA methylation status in host cells. The expression of EBV miRNAs vary under different latency programs (Table 3)[23]. For example, miR-BART1-5p, 6, and 17-5p suppresses LMP1 expression[24], whilst miR-BART-22 regulates expression of LMP2A[25]. EBV miRNAs further repress cellular proteins, including PUMA, DICER1, and BIM. EBV infection may also affect host cell miRNA expression. Specifically, miR-200a and miR-200b are down-regulated in EBVaGC compared to EBVnGC and adjacent mucosa. This down-regulation may be mediated by viral proteins such as BRAF0, EBER, and LMP2A, as well as by aberrant DNA methylation following EBV infection[26]. More recently, miRNA sequencing studies have revealed that EBV-infection mediates down-regulation of tumour suppressor miRNAs including the Let-7 family. Further research is required to elucidate their role in tumourigenesis[27].
| Gene name | Gene targets in EBV | Gene targets in host cell |
| miR-BHRF1-1 | - | GUF1[58], SCRN1[58] |
| miR-BART1-5p | LMP1[24] | CLEC2D[58,59], LY75[58,59], SP100[58,59], DICER1[58,59], MICB[58,59] |
| miR-BART1-3 | - | CXCL11[60] |
| miR-BART2-5p | BALF5[61] | MICB[62] |
| miR-BART3 | - | DICER1[58], MICB[58] |
| miR-BART3-3p | - | IPO7[63] |
| miR-BART5 | LMP1[59] | PUMA[66] |
| miR-BART6 | LMP1[24] | DICER1[65] |
| miR-BART10 | BHRF1[59] | - |
| miR-BART13 | - | CAPRIN2[59] |
| miR-BART16 | LMP1[24] | TOMM22[63] |
| miR-BART17-p | LMP1[24] | - |
| miR-BART19 | LMP1[59] | - |
| miR-BART22 | LMP2A[25] | - |
| miR-BARTs | - | BIM[66] |
Currently, GC is subdivided into three subtypes based on CpG-island methylator phenotype (CIMP). Defined as high (CIMP-H), low (CIMP-L), or none (CIMP-N), the classification is based on the number of methylated loci (≥ 4, 1-3, and 0 respectively) in the promoter regions of five genes (LOX, HRASLS, FLNC, HAND1, and THBD)[28]. It was previously shown that promoter methylation of cancer-related genes was seen more frequently in EBVaGC than EBVnGC. EBVaGC is thus classified as CIMP-H[29].
In a genome-wide study comparing promoter methylation between EBV-infected and EBV non-infected GC cell lines, hundreds of genes involved in cancer pathways such as cell adhesion molecules, wnt signalling pathway, and mitogen-activated protein kinase signalling were observed to be hypermethylated following EBV infection[17]. Further investigation through epigenomic and transcriptomic sequencing revealed that 216 genes were down-regulated by promoter hypermethylation. Significantly, hypermethylation of tumour suppressor genes, including p14, p15, p16, APC, E-cadherin, and PTEN were noted in EBVaGC, but not EBVnGC[30,31]. All studies unanimously agreed that p16 was significantly more hypermethylated in EBVaGC[29-35]. P16 is a tumour suppressor gene which acts in the G1 phase of the cell cycle to phosphorylate the retinoblastoma gene product (pRb). Loss of p16 leads to uncontrolled cell growth[36], and is thus commonly found in tumours[37,38].
Another important cellular abnormality in EBVaGC is its resistance to apoptosis. The frequency of apoptosis is significantly lower in EBVaGC than in EBVnGC[38]. It may be caused by hypermethylation of SSTR1 and GSTP1; both genes are frequently hypermethylated in NPC and GC infected EBV tissues, and regulate cell migration, proliferation, and apoptosis[30,31,39-42]. Notably, EBV infection also up-regulates expression of FAM3B and IHH[5]. FAM3B is associated with invasion[43], and Indian Hedgehog (IHH) with increased metastatic potential through angiogenesis and Snail protein expression, as well as a decrease in e-cadherin and tight junctions[44,45]. Table 4 shows a comprehensive list of hypermethylated genes and their role in carcinogenesis. Hence aberrant DNA methylation plays an important role in gastric carcinogenesis.
| Function | Hypermethylated genes |
| Apoptosis | DAPK[30], BNIP3[29], FAM3B[5], HRK[29], IL15RA[17], MINT31[32], p16[29-35], p73[30,32-34], PTEN[31,67], RASSF1A[31] |
| Cell adhesion | EPHB6[17], FLNc[30], FSD1[34], REC8[17], CSPG2[29] |
| Cell-cell interactions | MDGA2[17], THBS1[31] |
| Cell cycle regulation | APC[31], p15[30], p16[29-35], p57[29], p73[30,32-34] |
| Cell invasion | E-Cadherin[30,68,69] |
| Cell migration | EPHB6[17] |
| Cell proliferation | E-Cadherin[30,68,69], HRASLS[30], IL15RA[17], MINT31[32], NKX3.1[34], RUNX3[32], TIMP2[21], TIMP3[30] |
| Cell signalling | 14-3-3Sigma[31], CSPG2[29], MINT1[31], MINT2[31,32], PLXND1[21] |
| Differentiation | HAND1[30] |
| Dna repair | hMLH1[32,43,53], MGMT[31] |
| Exocytosis | SCRN1[34] |
| Metastasis | E-Cadherin[30,68,69], LOX[30] |
| Other | BCL7A[34], BLU[34], CHFR[29], CXXC4[21], GSTP1[30,31,40], HLTF[29], HOXA10[70], IHH[5], MARK1[34], MINT25[31], PAX5-β[29], SCARF2[17], SSTR1[17,39], THBD[30], WNT5A[71] |
Current treatment guidelines from the National Institute for Health and Clinical Excellence (NICE) for the management of GC depends on the stage of disease. Broadly, the mainstay for cure is surgical excision with clearance of adjacent lymph nodes. Radiotherapy, and chemotherapeutic agents including cisplatin, docetaxel, epirubicin, and 5-fluorouracil (5-FU) may be used as adjuvants or in the palliative setting. Notably, no differentiation is made between the distinct subtypes of GC in the treatment guidelines.
Research has established that EBVaGC represents a distinct entity of GC, characterised not only by unique genomic aberrations, but also by clinicopathologic features such as less lymph node involvement, and significantly better prognosis[2]. Naturally, there are associated therapeutic implications, as evidenced by resistance to docetaxel and 5-FU in EBV-positive, but not EBVnGC cell lines[46,47]. The chemoresistance is mediated by EBV-lytic gene expression, which induces expression of Bcl-2 and survivin whilst simultaneously suppressing p21 to inhibit apoptosis[48]. In support of this hypothesis, silencing of EBV-lytic gene LMP1 through specific small interfering RNA (siRNA) enhanced chemosensitivity of cancer cells to bleomycin and cisplatin[49]. Since epigenetic dysregulation is implicated in the expression of EBV-lytic genes and consequent tumour progression, we believe that epigenetic processes are a rational therapeutic target in EBVaGC.
Crucially, aberrant DNA methylation in cancer is reversible. Thus the enzymes which regulate epigenetic modifications are attractive targets for pharmacological intervention. Current epigenetic therapies may be classified into histone acetyltransferase (HAT), histone deacetylase (HDAC), and DNA methyltransferase (DNMT) inhibitors (Table 5). Broadly, they facilitate demethylation and re-expression of epigenetically silenced tumour suppressor genes, lowering the apoptotic threshold to sensitise tumour cells to chemotherapy and radiotherapy. Consequently, there has been an emphasis on investigating the clinical value of epigenetic therapies in combination with conventional cytotoxic agents and radiation.
| Target | Drug name | Status | Ref. |
| DNA methylation | Azacitine (5-Aza-CR) | Approved | [72] |
| Decitabine (5-Aza-CdR) | Approved | [72] | |
| Hydralazine | Phase II/III | [73] | |
| Epigallocatechin-3-gallate (EGCG) | Phase II | [74-76] | |
| 5-Fluoro-deoxycytidine (FdCyd/FDAC) | Phase I/II | [77] | |
| 5-fluoro-2′-deoxycytidine (FCdR) | Phase I/II | [78] | |
| Procainamide | Phase I | [79] | |
| Procaine | Phase I | [80] | |
| Psammaplin A | Phase 0 | [81,82] | |
| RG108 | Phase 0 | [83-86] | |
| Zebularine | Phase 0 | [87-89] | |
| Histone deacetylases | Vorinostat | Approved | [90] |
| Romidepsin | Approved | [90] | |
| Panobinostat | Phase II | [91] | |
| SEN196 | Phase II | [92] | |
| Phenyl butyrate | Phase I/II | [93-95] | |
| Valporic acid | Phase I | [93-95] | |
| Compound 6J (R = -C4H8) | Phase 0 | [96] |
It was previously reported that the combination of irradiation and 5-aza-CdR significantly decreased growth activity compared with irradiation alone in OCUM-2M, OCUM-12, and MKN-45 GC cell lines (P < 0.05). The cell cycle arrest and increased apoptotic rate may be partly mediated by enhanced expression of p53, RASSF1, and DAPK gene families by 5-aza-CdR[50]. The use of epigenetic therapies in conjunction with targeted therapies such as geftinib in lung cancer, imatinib in chronic myeloid leukemia, and trastuzumab in breast cancer cell lines and in vivo tumour models also had synergistic effects on the induction of apoptosis[51,52]. More recently, epigenetic modifiers and ZEB1 inhibitors have been used to induce lytic transformation of EBV-infected gastric cancer cells. Expressed only in the lytic form of infection, virally encoded kinases convert ganciclovir into its active form, potentiating its cytotoxic effects[53,54]. Hence epigenetic modifiers may be a useful therapeutic strategy in EBVaGC.
However, several problems must be considered. Firstly, methylation is reversible, so re-methylation and re-silencing after cessation of drug therapy may occur[55]. Moreover, there have been numerous concerns raised regarding the systemic effects of non-specific gene activation in non-cancerous cells by epigenetic therapies. Conflicting evidence exists in the literature regarding the effect of epigenetic therapies on normal cells. Some studies have demonstrated that 5-Aza and decitabine increases mutation frequency, causes chromosomal re-arrangements, and decreases fertility in mice. Conversely, no increase in chromosomal integrity was observed following administration of low dose 5-aza-CdR in patients with myelodysplastic syndrome[56]. Additionally, treatment of 41 leukemia patients with 5-aza-CdR showed only mild effects on global genomic de-methylation, as measured by changes in Alu methylation[57]. Few adverse effects were observed, and original methylation levels were regained within two weeks after therapy. No development of secondary malignancies were recorded. Consequently, further studies are needed to investigate the long term effects of epigenetic therapies.
EBVaGC is a unique type of GC. The characteristic global hypermethylation of the promoter region in tumour-suppressor genes may be due to overexpression of DNMTs by viral latent proteins, miRNAs, and various epigenomic changes. However, the precise role of EBV in the multifactorial etiology of GC is still not fully understood. Further studies are needed to elucidate the intricate relationship between EBV infection, environmental factors, genetic backgrounds, and aberrant DNA methylation in GC. A better understanding of the role of EBV in gastric carcinogenesis will enable discovery of novel therapeutic targets and strategies.
P- Reviewers: Engin AB, Jung YD S- Editor: Zhai HH L- Editor: A E- Editor: Wang CH
| 1. | Fukayama M, Hino R, Uozaki H. Epstein-Barr virus and gastric carcinoma: virus-host interactions leading to carcinoma. Cancer Sci. 2008;99:1726-1733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 126] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 2. | van Beek J, zur Hausen A, Klein Kranenbarg E, van de Velde CJ, Middeldorp JM, van den Brule AJ, Meijer CJ, Bloemena E. EBV-positive gastric adenocarcinomas: a distinct clinicopathologic entity with a low frequency of lymph node involvement. J Clin Oncol. 2004;22:664-670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 216] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 3. | Camargo MC, Kim WH, Chiaravalli AM, Kim KM, Corvalan AH, Matsuo K, Yu J, Sung JJ, Herrera-Goepfert R, Meneses-Gonzalez F. Improved survival of gastric cancer with tumour Epstein-Barr virus positivity: an international pooled analysis. Gut. 2014;63:236-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 293] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 4. | Camargo MC, Koriyama C, Matsuo K, Kim WH, Herrera-Goepfert R, Liao LM, Yu J, Carrasquilla G, Sung JJ, Alvarado-Cabrero I. Case-case comparison of smoking and alcohol risk associations with Epstein-Barr virus-positive gastric cancer. Int J Cancer. 2014;134:948-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 5. | Liang Q, Yao X, Tang S, Yau TO, Zhao J, Sung JJ, Yu J. Integrative Identification of EBV-Associated Variations At Genomic, Epigenomic and Transcriptomic Levels in Gastric Cancer. Gastroenterology. 2013;144:S525. [DOI] [Full Text] |
| 6. | Tugizov SM, Berline JW, Palefsky JM. Epstein-Barr virus infection of polarized tongue and nasopharyngeal epithelial cells. Nat Med. 2003;9:307-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 206] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 7. | Sixbey JW, Davis DS, Young LS, Hutt-Fletcher L, Tedder TF, Rickinson AB. Human Epithelial Cell Expression of an Epstein-barr Virus Receptor. J Gen Virol. 1987;68:805-811. [RCA] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 91] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 8. | Tanner J, Weis J, Fearon D, Whang Y, Kieff E. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell. 1987;50:203-213. [PubMed] |
| 9. | Ogembo JG, Kannan L, Ghiran I, Nicholson-Weller A, Finberg RW, Tsokos GC, Fingeroth JD. Human complement receptor type 1/CD35 is an Epstein-Barr Virus receptor. Cell Rep. 2013;3:371-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 105] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 10. | Tanner J, Whang Y, Sample J, Sears A, Kieff E. Soluble gp350/220 and deletion mutant glycoproteins block Epstein-Barr virus adsorption to lymphocytes. J Virol. 1988;62:4452-4464. [PubMed] |
| 11. | Shannon-Lowe C, Rowe M. Epstein-Barr virus infection of polarized epithelial cells via the basolateral surface by memory B cell-mediated transfer infection. PLoS Pathog. 2011;7:e1001338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 89] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 12. | Sugiura M, Imai S, Tokunaga M, Koizumi S, Uchizawa M, Okamoto K, Osato T. Transcriptional analysis of Epstein-Barr virus gene expression in EBV-positive gastric carcinoma: unique viral latency in the tumour cells. Br J Cancer. 1996;74:625-631. [PubMed] |
| 13. | Niedobitek G, Agathanggelou A, Herbst H, Whitehead L, Wright DH, Young LS. Epstein-Barr virus (EBV) infection in infectious mononucleosis: virus latency, replication and phenotype of EBV-infected cells. J Pathol. 1997;182:151-159. [PubMed] |
| 14. | Iwakiri D, Eizuru Y, Tokunaga M, Takada K. Autocrine growth of Epstein-Barr virus-positive gastric carcinoma cells mediated by an Epstein-Barr virus-encoded small RNA. Cancer Res. 2003;63:7062-7067. [PubMed] |
| 15. | Luo B, Wang Y, Wang XF, Liang H, Yan LP, Huang BH, Zhao P. Expression of Epstein-Barr virus genes in EBV-associated gastric carcinomas. World J Gastroenterol. 2005;11:629-633. [PubMed] |
| 16. | Hino R, Uozaki H, Inoue Y, Shintani Y, Ushiku T, Sakatani T, Takada K, Fukayama M. Survival advantage of EBV-associated gastric carcinoma: survivin up-regulation by viral latent membrane protein 2A. Cancer Res. 2008;68:1427-1435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 80] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 17. | Zhao J, Liang Q, Cheung KF, Kang W, Lung RW, Tong JH, To KF, Sung JJ, Yu J. Genome-wide identification of Epstein-Barr virus-driven promoter methylation profiles of human genes in gastric cancer cells. Cancer. 2013;119:304-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 119] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 18. | Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1431] [Cited by in RCA: 1266] [Article Influence: 66.6] [Reference Citation Analysis (0)] |
| 19. | Pfeifer GP, Tang M, Denissenko MF. Mutation hotspots and DNA methylation. Curr Top Microbiol Immunol. 2000;249:1-19. [PubMed] |
| 20. | Toyota M, Ahuja N, Suzuki H, Itoh F, Ohe-Toyota M, Imai K, Baylin SB, Issa JP. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999;59:5438-5442. [PubMed] |
| 21. | Matsusaka K, Kaneda A, Nagae G, Ushiku T, Kikuchi Y, Hino R, Uozaki H, Seto Y, Takada K, Aburatani H. Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res. 2011;71:7187-7197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 190] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 22. | Kaneda A, Matsusaka K, Aburatani H, Fukayama M. Epstein-Barr virus infection as an epigenetic driver of tumorigenesis. Cancer Res. 2012;72:3445-3450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 150] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 23. | Qiu J, Cosmopoulos K, Pegtel M, Hopmans E, Murray P, Middeldorp J, Shapiro M, Thorley-Lawson DA. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog. 2011;7:e1002193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 24. | Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, Hayward SD. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci USA. 2007;104:16164-16169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 283] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 25. | Lung RW, Tong JH, Sung YM, Leung PS, Ng DC, Chau SL, Chan AW, Ng EK, Lo KW, To KF. Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22. Neoplasia. 2009;11:1174-1184. [PubMed] |
| 26. | Shinozaki A, Sakatani T, Ushiku T, Hino R, Isogai M, Ishikawa S, Uozaki H, Takada K, Fukayama M. Downregulation of microRNA-200 in EBV-associated gastric carcinoma. Cancer Res. 2010;70:4719-4727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 128] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 27. | Marquitz AR, Mathur A, Chugh PE, Dittmer DP, Raab-Traub N. Expression profile of microRNAs in Epstein-Barr virus-infected AGS gastric carcinoma cells. J Virol. 2014;88:1389-1393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 28. | Kaneda A, Kaminishi M, Yanagihara K, Sugimura T, Ushijima T. Identification of silencing of nine genes in human gastric cancers. Cancer Res. 2002;62:6645-6650. [PubMed] |
| 29. | Kusano M, Toyota M, Suzuki H, Akino K, Aoki F, Fujita M, Hosokawa M, Shinomura Y, Imai K, Tokino T. Genetic, epigenetic, and clinicopathologic features of gastric carcinomas with the CpG island methylator phenotype and an association with Epstein-Barr virus. Cancer. 2006;106:1467-1479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 138] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 30. | Chang MS, Uozaki H, Chong JM, Ushiku T, Sakuma K, Ishikawa S, Hino R, Barua RR, Iwasaki Y, Arai K. CpG island methylation status in gastric carcinoma with and without infection of Epstein-Barr virus. Clin Cancer Res. 2006;12:2995-3002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 153] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 31. | Kang GH, Lee S, Kim WH, Lee HW, Kim JC, Rhyu MG, Ro JY. Epstein-barr virus-positive gastric carcinoma demonstrates frequent aberrant methylation of multiple genes and constitutes CpG island methylator phenotype-positive gastric carcinoma. Am J Pathol. 2002;160:787-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 259] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 32. | Saito M, Nishikawa J, Okada T, Morishige A, Sakai K, Nakamura M, Kiyotoki S, Hamabe K, Okamoto T, Oga A. Role of DNA methylation in the development of Epstein-Barr virus-associated gastric carcinoma. J Med Virol. 2013;85:121-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 33. | Ushiku T, Chong JM, Uozaki H, Hino R, Chang MS, Sudo M, Rani BR, Sakuma K, Nagai H, Fukayama M. p73 gene promoter methylation in Epstein-Barr virus-associated gastric carcinoma. Int J Cancer. 2007;120:60-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 34. | Okada T, Nakamura M, Nishikawa J, Sakai K, Zhang Y, Saito M, Morishige A, Oga A, Sasaki K, Suehiro Y. Identification of genes specifically methylated in Epstein-Barr virus-associated gastric carcinomas. Cancer Sci. 2013;104:1309-1314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 35. | Sakuma K, Chong JM, Sudo M, Ushiku T, Inoue Y, Shibahara J, Uozaki H, Nagai H, Fukayama M. High-density methylation of p14ARF and p16INK4A in Epstein-Barr virus-associated gastric carcinoma. Int J Cancer. 2004;112:273-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 37. | Jang TJ, Kim DI, Shin YM, Chang HK, Yang CH. p16(INK4a) Promoter hypermethylation of non-tumorous tissue adjacent to gastric cancer is correlated with glandular atrophy and chronic inflammation. Int J Cancer. 2001;93:629-634. [PubMed] |
| 38. | Ohfuji S, Osaki M, Tsujitani S, Ikeguchi M, Sairenji T, Ito H. Low frequency of apoptosis in Epstein-Barr virus-associated gastric carcinoma with lymphoid stroma. Int J Cancer. 1996;68:710-715. [PubMed] |
| 39. | Zhao J, Liang Q, Cheung KF, Kang W, Dong Y, Lung RW, Tong JH, To KF, Sung JJ, Yu J. Somatostatin receptor 1, a novel EBV-associated CpG hypermethylated gene, contributes to the pathogenesis of EBV-associated gastric cancer. Br J Cancer. 2013;108:2557-2564. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 40. | Kim J, Lee HS, Bae SI, Lee YM, Kim WH. Silencing and CpG island methylation of GSTP1 is rare in ordinary gastric carcinomas but common in Epstein-Barr virus-associated gastric carcinomas. Anticancer Res. 2005;25:4013-4019. [PubMed] |
| 41. | Challouf S, Ziadi S, Zaghdoudi R, Ksiaa F, Ben Gacem R, Trimeche M. Patterns of aberrant DNA hypermethylation in nasopharyngeal carcinoma in Tunisian patients. Clin Chim Acta. 2012;413:795-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 42. | Guo X, Zeng Y, Deng H, Liao J, Zheng Y, Li J, Kessing B, O’Brien SJ. Genetic Polymorphisms of CYP2E1, GSTP1, NQO1 and MPO and the Risk of Nasopharyngeal Carcinoma in a Han Chinese Population of Southern China. BMC Res Notes. 2010;3:212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 43. | Li Z, Mou H, Wang T, Xue J, Deng B, Qian L, Zhou Y, Gong W, Wang JM, Wu G. A non-secretory form of FAM3B promotes invasion and metastasis of human colon cancer cells by upregulating Slug expression. Cancer Lett. 2013;328:278-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 44. | Evangelista M, Tian H, de Sauvage FJ. The hedgehog signaling pathway in cancer. Clin Cancer Res. 2006;12:5924-5928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 181] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 45. | Fan H, Oro AE, Scott MP, Khavari PA. Induction of basal cell carcinoma features in transgenic human skin expressing Sonic Hedgehog. Nat Med. 1997;3:788-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 191] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 46. | Blenk S, Engelmann J, Weniger M, Schultz J, Dittrich M, Rosenwald A, Müller-Hermelink HK, Müller T, Dandekar T. Germinal center B cell-like (GCB) and activated B cell-like (ABC) type of diffuse large B cell lymphoma (DLBCL): analysis of molecular predictors, signatures, cell cycle state and patient survival. Cancer Inform. 2007;3:399-420. [PubMed] |
| 47. | Park SY, Kook MC, Kim YW, Cho NY, Jung N, Kwon HJ, Kim TY, Kang GH. CpG island hypermethylator phenotype in gastric carcinoma and its clinicopathological features. Virchows Arch. 2010;457:415-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 48. | Shin HJ, Kim do N, Lee SK. Association between Epstein-Barr virus infection and chemoresistance to docetaxel in gastric carcinoma. Mol Cells. 2011;32:173-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 49. | Mei YP, Zhou JM, Wang Y, Huang H, Deng R, Feng GK, Zeng YX, Zhu XF. Silencing of LMP1 induces cell cycle arrest and enhances chemosensitivity through inhibition of AKT signaling pathway in EBV-positive nasopharyngeal carcinoma cells. Cell Cycle. 2007;6:1379-1385. [PubMed] |
| 50. | Qiu H, Yashiro M, Shinto O, Matsuzaki T, Hirakawa K. DNA methyltransferase inhibitor 5-aza-CdR enhances the radiosensitivity of gastric cancer cells. Cancer Sci. 2009;100:181-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 49] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 51. | Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769-784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2201] [Cited by in RCA: 2301] [Article Influence: 121.1] [Reference Citation Analysis (0)] |
| 52. | Fiskus W, Pranpat M, Bali P, Balasis M, Kumaraswamy S, Boyapalle S, Rocha K, Wu J, Giles F, Manley PW. Combined effects of novel tyrosine kinase inhibitor AMN107 and histone deacetylase inhibitor LBH589 against Bcr-Abl-expressing human leukemia cells. Blood. 2006;108:645-652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 112] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 53. | Jung EJ, Lee YM, Lee BL, Chang MS, Kim WH. Lytic induction and apoptosis of Epstein-Barr virus-associated gastric cancer cell line with epigenetic modifiers and ganciclovir. Cancer Lett. 2007;247:77-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 54. | Zhao J, Jin H, Cheung KF, Tong JH, Zhang S, Go MY, Tian L, Kang W, Leung PP, Zeng Z. Zinc finger E-box binding factor 1 plays a central role in regulating Epstein-Barr virus (EBV) latent-lytic switch and acts as a therapeutic target in EBV-associated gastric cancer. Cancer. 2012;118:924-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 55. | Yoo CB, Cheng JC, Jones PA. Zebularine: a new drug for epigenetic therapy. Biochem Soc Trans. 2004;32:910-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 131] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 56. | Lübbert M, Wijermans P, Kunzmann R, Verhoef G, Bosly A, Ravoet C, Andre M, Ferrant A. Cytogenetic responses in high-risk myelodysplastic syndrome following low-dose treatment with the DNA methylation inhibitor 5-aza-2’-deoxycytidine. Br J Haematol. 2001;114:349-357. [PubMed] |
| 57. | Yang AS, Estecio MR, Garcia-Manero G, Kantarjian HM, Issa JP. Comment on “Chromosomal instability and tumors promoted by DNA hypomethylation” and “Induction of tumors in nice by genomic hypomethylation”. Science. 2003;302:1153; author reply 1153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 53] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 58. | Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse HJ, Luftig MA. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012;8:e1002484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 292] [Cited by in RCA: 287] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 59. | Riley KJ, Rabinowitz GS, Yario TA, Luna JM, Darnell RB, Steitz JA. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012;31:2207-2221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 232] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 60. | Xia T, O’Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436-1442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 256] [Cited by in RCA: 258] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 61. | Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jäker C, Höck J, Meister G, Grässer FA. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666-675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 270] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 62. | Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 379] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 63. | Dölken L, Malterer G, Erhard F, Kothe S, Friedel CC, Suffert G, Marcinowski L, Motsch N, Barth S, Beitzinger M. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe. 2010;7:324-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 169] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 64. | Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, Kwong DL, Tsao SW, Jin DY. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205:2551-2560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 340] [Cited by in RCA: 367] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 65. | Iizasa H, Wulff BE, Alla NR, Maragkakis M, Megraw M, Hatzigeorgiou A, Iwakiri D, Takada K, Wiedmer A, Showe L. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J Biol Chem. 2010;285:33358-33370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 196] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 66. | Marquitz AR, Mathur A, Nam CS, Raab-Traub N. The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology. 2011;412:392-400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 159] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 67. | Hino R, Uozaki H, Murakami N, Ushiku T, Shinozaki A, Ishikawa S, Morikawa T, Nakaya T, Sakatani T, Takada K. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009;69:2766-2774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 288] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 68. | Sudo M, Chong JM, Sakuma K, Ushiku T, Uozaki H, Nagai H, Funata N, Matsumoto Y, Fukayama M. Promoter hypermethylation of E-cadherin and its abnormal expression in Epstein-Barr virus-associated gastric carcinoma. Int J Cancer. 2004;109:194-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 57] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 69. | Zazula M, Ferreira AM, Czopek JP, Kolodziejczyk P, Sinczak-Kuta A, Klimkowska A, Wojcik P, Okon K, Bialas M, Kulig J. CDH1 gene promoter hypermethylation in gastric cancer: relationship to Goseki grading, microsatellite instability status, and EBV invasion. Diagn Mol Pathol. 2006;15:24-29. [PubMed] |
| 70. | Kang GH, Lee S, Cho NY, Gandamihardja T, Long TI, Weisenberger DJ, Campan M, Laird PW. DNA methylation profiles of gastric carcinoma characterized by quantitative DNA methylation analysis. Lab Invest. 2008;88:161-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 136] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 71. | Liu X, Wang Y, Wang X, Sun Z, Li L, Tao Q, Luo B. Epigenetic silencing of WNT5A in Epstein-Barr virus-associated gastric carcinoma. Arch Virol. 2013;158:123-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 72. | Kuo HK, Griffith JD, Kreuzer KN. 5-Azacytidine induced methyltransferase-DNA adducts block DNA replication in vivo. Cancer Res. 2007;67:8248-8254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 89] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 73. | Singh N, Dueñas-González A, Lyko F, Medina-Franco JL. Molecular modeling and molecular dynamics studies of hydralazine with human DNA methyltransferase 1. ChemMedChem. 2009;4:792-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 78] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 74. | Shanafelt TD, Call TG, Zent CS, Leis JF, LaPlant B, Bowen DA, Roos M, Laumann K, Ghosh AK, Lesnick C. Phase 2 trial of daily, oral Polyphenon E in patients with asymptomatic, Rai stage 0 to II chronic lymphocytic leukemia. Cancer. 2013;119:363-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 140] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 75. | McLarty J, Bigelow RL, Smith M, Elmajian D, Ankem M, Cardelli JA. Tea polyphenols decrease serum levels of prostate-specific antigen, hepatocyte growth factor, and vascular endothelial growth factor in prostate cancer patients and inhibit production of hepatocyte growth factor and vascular endothelial growth factor in vitro. Cancer Prev Res (Phila). 2009;2:673-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 179] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 76. | Wang JS, Luo H, Wang P, Tang L, Yu J, Huang T, Cox S, Gao W. Validation of green tea polyphenol biomarkers in a phase II human intervention trial. Food Chem Toxicol. 2008;46:232-240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 40] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 77. | Beumer JH, Parise RA, Newman EM, Doroshow JH, Synold TW, Lenz HJ, Egorin MJ. Concentrations of the DNA methyltransferase inhibitor 5-fluoro-2’-deoxycytidine (FdCyd) and its cytotoxic metabolites in plasma of patients treated with FdCyd and tetrahydrouridine (THU). Cancer Chemother Pharmacol. 2008;62:363-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 56] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 78. | Zhao Q, Fan J, Hong W, Li L, Wu M. Inhibition of cancer cell proliferation by 5-fluoro-2’-deoxycytidine, a DNA methylation inhibitor, through activation of DNA damage response pathway. Springerplus. 2012;1:65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 79. | Zhou L, Cheng X, Connolly BA, Dickman MJ, Hurd PJ, Hornby DP. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J Mol Biol. 2002;321:591-599. [PubMed] |
| 80. | Villar-Garea A, Fraga MF, Espada J, Esteller M. Procaine is a DNA-demethylating agent with growth-inhibitory effects in human cancer cells. Cancer Res. 2003;63:4984-4989. [PubMed] |
| 81. | Hiusen TJ, Kamble-Shripat T. Delayed toxicity of two chitinolytic enzyme inhibitors (psammaplin a and pentoxifylline) against eastern subterranean termites (Isoptera: Rhinotermitidae). J Econ Entomol. 2013;106:1788-1793. [PubMed] |
| 82. | Baud MG, Leiser T, Petrucci V, Gunaratnam M, Neidle S, Meyer-Almes FJ, Fuchter MJ. Thioester derivatives of the natural product psammaplin A as potent histone deacetylase inhibitors. Beilstein J Org Chem. 2013;9:81-88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 83. | Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305-6311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 388] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 84. | Suzuki T, Tanaka R, Hamada S, Nakagawa H, Miyata N. Design, synthesis, inhibitory activity, and binding mode study of novel DNA methyltransferase 1 inhibitors. Bioorg Med Chem Lett. 2010;20:1124-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 85. | Savickiene J, Treigyte G, Jazdauskaite A, Borutinskaite VV, Navakauskiene R. DNA methyltransferase inhibitor RG108 and histone deacetylase inhibitors cooperate to enhance NB4 cell differentiation and E-cadherin re-expression by chromatin remodelling. Cell Biol Int. 2012;36:1067-1078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 86. | Savickiene J, Treigyte G, Borutinskaite VV, Navakauskiene R. Antileukemic activity of combined epigenetic agents, DNMT inhibitors zebularine and RG108 with HDAC inhibitors, against promyelocytic leukemia HL-60 cells. Cell Mol Biol Lett. 2012;17:501-525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 87. | Cheng JC, Matsen CB, Gonzales FA, Ye W, Greer S, Marquez VE, Jones PA, Selker EU. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J Natl Cancer Inst. 2003;95:399-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 392] [Cited by in RCA: 373] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 88. | Billam M, Sobolewski MD, Davidson NE. Effects of a novel DNA methyltransferase inhibitor zebularine on human breast cancer cells. Breast Cancer Res Treat. 2010;120:581-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 103] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 89. | Flotho C, Claus R, Batz C, Schneider M, Sandrock I, Ihde S, Plass C, Niemeyer CM, Lübbert M. The DNA methyltransferase inhibitors azacitidine, decitabine and zebularine exert differential effects on cancer gene expression in acute myeloid leukemia cells. Leukemia. 2009;23:1019-1028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 241] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 90. | Fatkins DG, Zheng W. Substituting N(epsilon)-thioacetyl-lysine for N(epsilon)-acetyl-lysine in peptide substrates as a general approach to inhibiting human NAD(+)-dependent protein deacetylases. Int J Mol Sci. 2008;9:1-11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 91. | Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci USA. 2004;101:1241-1246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 453] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 92. | Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26:28-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 389] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 93. | Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, Yang H, Rosner G, Verstovsek S, Rytting M, Wierda WG, Ravandi F, Koller C. Phase 1/2 study of the combination of 5-aza-2’-deoxycytidine with valproic acid in patients with leukemia. Blood. 2006;108:3271-3279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 376] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 94. | Nemunaitis JJ, Orr D, Eager R, Cunningham CC, Williams A, Mennel R, Grove W, Olson S. Phase I study of oral CI-994 in combination with gemcitabine in treatment of patients with advanced cancer. Cancer J. 2003;9:58-66. [PubMed] |
| 95. | Undevia SD, Kindler HL, Janisch L, Olson SC, Schilsky RL, Vogelzang NJ, Kimmel KA, Macek TA, Ratain MJ. A phase I study of the oral combination of CI-994, a putative histone deacetylase inhibitor, and capecitabine. Ann Oncol. 2004;15:1705-1711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 96. | Medda F, Russell RJ, Higgins M, McCarthy AR, Campbell J, Slawin AM, Lane DP, Lain S, Westwood NJ. Novel cambinol analogs as sirtuin inhibitors: synthesis, biological evaluation, and rationalization of activity. J Med Chem. 2009;52:2673-2682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 103] [Article Influence: 6.4] [Reference Citation Analysis (0)] |