Published online May 21, 2014. doi: 10.3748/wjg.v20.i19.5773
Revised: January 14, 2014
Accepted: March 4, 2014
Published online: May 21, 2014
Processing time: 222 Days and 1.9 Hours
Autophagy is a lysosome-associated, degradative process that catabolizes cytosolic components to recycle nutrients for further use and maintain cell homeostasis. Hepatitis C virus (HCV) is a major cause of chronic hepatitis, which often leads to end-stage liver-associated diseases and is a significant burden on worldwide public health. Emerging lines of evidence indicate that autophagy plays an important role in promoting the HCV life cycle in host cells. Moreover, the diverse impacts of autophagy on a variety of signaling pathways in HCV-infected cells suggest that the autophagic process is required for balancing HCV-host cell interactions and involved in the pathogenesis of HCV-related liver diseases. However, the detailed molecular mechanism underlying how HCV activates autophagy to benefit viral growth is still enigmatic. Additionally, how the autophagic response contributes to disease progression in HCV-infected cells remains largely unknown. Hence, in this review, we overview the interplay between autophagy and the HCV life cycle and propose possible mechanisms by which autophagy may promote the pathogenesis of HCV-associated chronic liver diseases. Moreover, we outline the related studies on how autophagy interplays with HCV replication and discuss the possible implications of autophagy and viral replication in the progression of HCV-induced liver diseases, e.g., steatosis and hepatocellular carcinoma. Finally, we explore the potential therapeutics that target autophagy to cure HCV infection and its related liver diseases.
Core tip: Hepatitis C virus (HCV) is a major cause of chronic liver disease and is associated with over 170 million infected individuals worldwide. However, a successful strategy for completely eradicating HCV infection is still limited. Autophagy is a catabolic process that delivers cytosolic components to lysosomes for breakdown. HCV has been shown to activate autophagy to promote viral growth in vitro. In this review, we outline the recent findings on the physiological significance of autophagy in the HCV life cycle and propose a potential role of autophagy in the development of HCV-related liver diseases as well as a perspective on therapeutics targeting autophagy to cure HCV infection.
- Citation: Ke PY, Chen SSL. Autophagy in hepatitis C virus-host interactions: Potential roles and therapeutic targets for liver-associated diseases. World J Gastroenterol 2014; 20(19): 5773-5793
- URL: https://www.wjgnet.com/1007-9327/full/v20/i19/5773.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i19.5773
Hepatitis C virus (HCV) infection is a global public health burden. Approximately 170 million people are infected with HCV worldwide, and most of these patients become persistently infected. Furthermore, HCV infection in some patients may progress into chronic liver diseases, such as steatosis, cirrhosis, and hepatocellular carcinoma[1,2]. The successful rate of curing HCV infection using the current therapy that combines interferon (IFN) and ribavirin is still limited due to its low efficacy, drug resistance, and severe side effects in a particular population of infected individuals[3]. Additionally, preventive vaccines against HCV are not yet available[4,5]. Hence, a new antiviral drug with high potency and/or a protective vaccination against HCV infection are urgently needed. Autophagy is an evolutionarily conserved, catabolic pathway by which eukaryotic cells degrade unnecessary cytoplasmic compartments to recycle nutrients and maintain cellular homeostasis[6,7]. Recent studies collectively indicate that HCV activates autophagy to promote viral growth through regulating different steps of the viral life cycle by affecting different host cellular signaling processes[8-11]. Because autophagy has widely been shown to contribute to the progression of human diseases[12,13], HCV-activated autophagy could be physiologically significant in the pathogenesis of HCV-associated liver diseases. Most importantly, interference with the autophagic process can suppress HCV replication[8-11], suggesting that inhibition of autophagy can serve as a novel therapeutic strategy against HCV infection. Therefore, in this review, we outline the current findings on the functional roles of autophagy at each stage of the HCV life cycle and the molecular mechanism by which HCV activates the autophagic response. Lastly, we also discuss the possible impacts of the autophagic response on the development of HCV-related liver disorders as well as provide a perspective on the implications of modulating autophagy to control HCV infection.
An unknown infectious agent that caused non-A, non-B post-transfusion hepatitis was first discovered in the mid-1970s[14]. In 1989, the nucleic acid sequence of this unidentified virus was cloned, reported, and formally named HCV[1]. It is estimated that over 3% of the human population is infected by HCV. Most of the infected individuals become chronically infected, and HCV infection often progresses to severe, liver-associated diseases, such as cirrhosis, steatosis, and hepatocellular carcinoma[2]. To date, due to the low efficacy of the combined therapy of pegylated IFN-α and ribavirin[3], options for complete eradication of HCV infection and a preventive strategy are still absent[4,5]; therefore, HCV infection is a global public health problem.
HCV is a membrane-enveloped, positive-sense, single-stranded RNA virus belonging to the Hepacivirus genus and the Flaviviridae family[2]. Until now, the known isolates of HCV were classified into seven genotypes, i.e., genotypes 1 through 7, with 20%-30% sequence divergence, and an array of subtypes could be grouped within each genotype[15]. The genetic heterogeneity of these HCV genotypes could result in the variable degree of risk for progressive liver diseases and the different treatment outcomes of IFN-based therapy[16]. For instance, a higher prevalence of progression into hepatosteatosis and cirrhosis occurs in cases infected with HCV genotype 3[17,18]. Regarding the efficacy of standard IFN/ribavirin treatment, the successful rates of patients infected with genotypes 2 and 3 are higher than those of patients infected with genotypes 1 and 4[19-22].
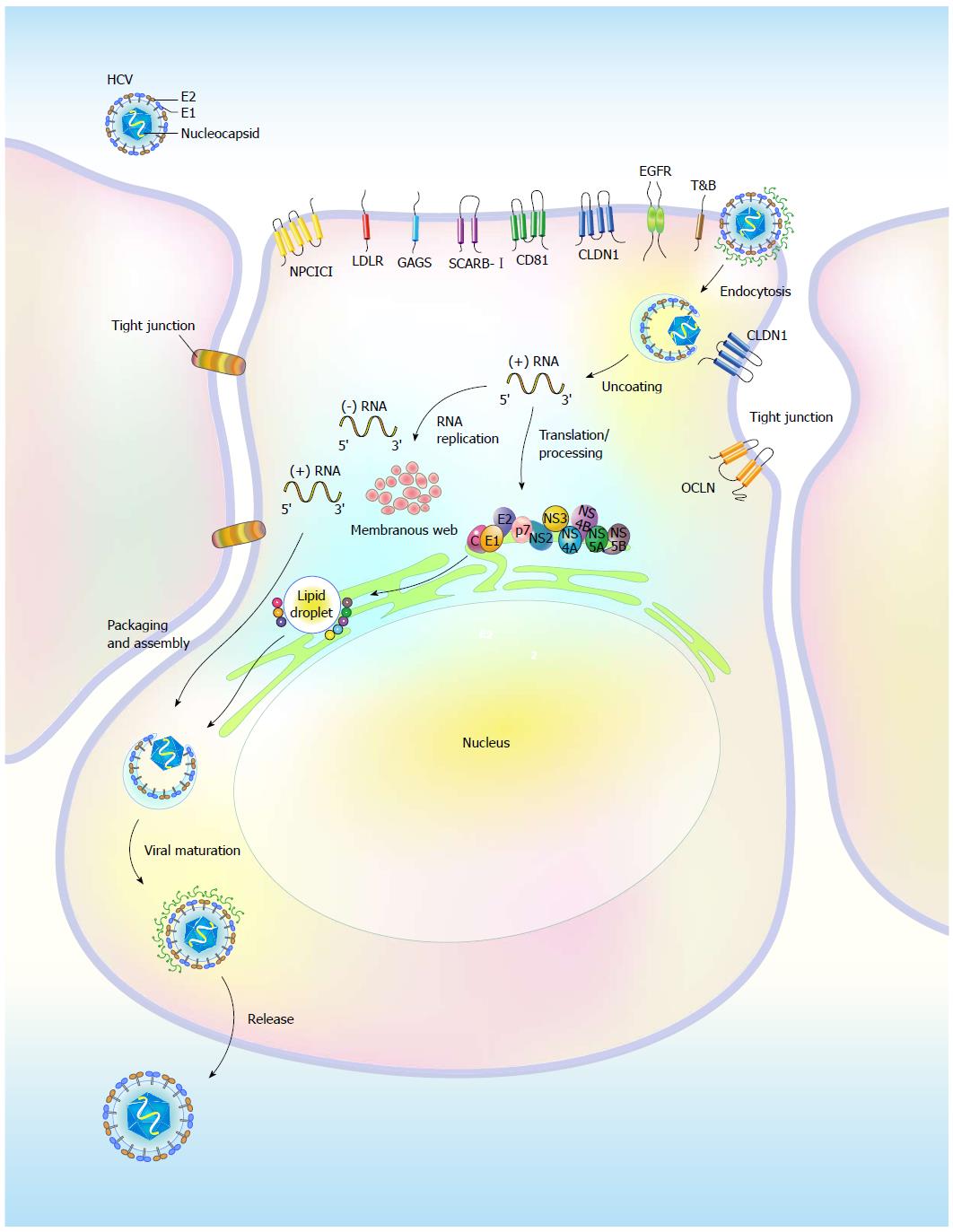
Hepatocytes in the liver are the predominant targets of HCV infection, and the entry of HCV into hepatocytes is a stringently coordinated process that relies on successive and concerted interactions between the envelope glycoproteins E1 and E2 and host cellular factors that are present on the cell surface, i.e., the so called “entry (co)receptors” (Figure 1). The known (co)receptors, including the tetraspanin CD81[23-25], scavenger receptor class B member I (SCRAB-I)[26,27], Claudin 1 (CLDN1)[28], and Occludin (OCLN)[29], have been shown to mediate HCV entry into hepatocytes (Figure 1). In addition, the low-density lipoprotein receptor (LDLR)[30], highly sulfated heparin[31], and the dendritic cell-specific intercellular adhesion molecule three grabbing non antigen[32] were reported to be involved in attachment to and concentration of lipoprotein-associated viral particles on the cell surface of infected cells (Figure 1). After attachment to the cell surface, the virions bind to CD81 and SCRAB-I on the plasma membrane through the interaction between the E2 protein and these two entry (co)factors[33-35]. Subsequently, the association of CD81 or SCARB-I with CLDN1 on the basolateral surfaces of hepatocytes facilitates the formation of entry complexes[36-38] (Figure 1), thus promoting the internalization process of viral particles via the clathrin-mediated and pH-dependent endocytosis pathway[39,40]. Following internalization into cells, the envelopes of the virions fuse with the endosomal membrane, allowing uncoating and release of the viral genomes into the cytoplasm, where the translation of viral proteins and replication of viral RNA occur[39,40] (Figure 1). The exact physiological role of OCLN in the entry of the HCV virion is still unclear, although the second extracellular loop of this protein, along with CD81, has been shown to determine the host tropism of HCV infection[29]. In addition to CLDN1 and OCLN, other tight junction proteins such as CLDN6 and CLDN9 have been reported to participate in the entry of HCV into peripheral blood mononuclear cells, which lack CLDN1 expression[41,42]. This represents an alternative route for HCV infection in extrahepatic compartments[42].
In addition to these entry (co)receptors, epidermal growth factor receptor (EGFR) and ephrin receptor A2 (EphA2) have recently been identified as additional (co)factors for HCV entry by facilitating the CD81-CLDN1 interaction[43] (Figure 1). Additionally, the receptor tyrosine kinase activities of these two molecules were shown to enhance the membrane fusogenic activity of HCV envelope glycoproteins[43]. In addition, the entry of HCV virions into host cells can be mediated by an association with cholesterol via the Niemann-Pick C1-like L1 (NPC1L1) cholesterol uptake receptor[44]. Likewise, transferrin receptor (TfR), which is an iron absorption receptor, was recently demonstrated to be involved in the internalization of HCV virions into hepatocytes[45] (Figure 1). These studies collectively indicate that infection of target cells by HCV is a highly regulated process.
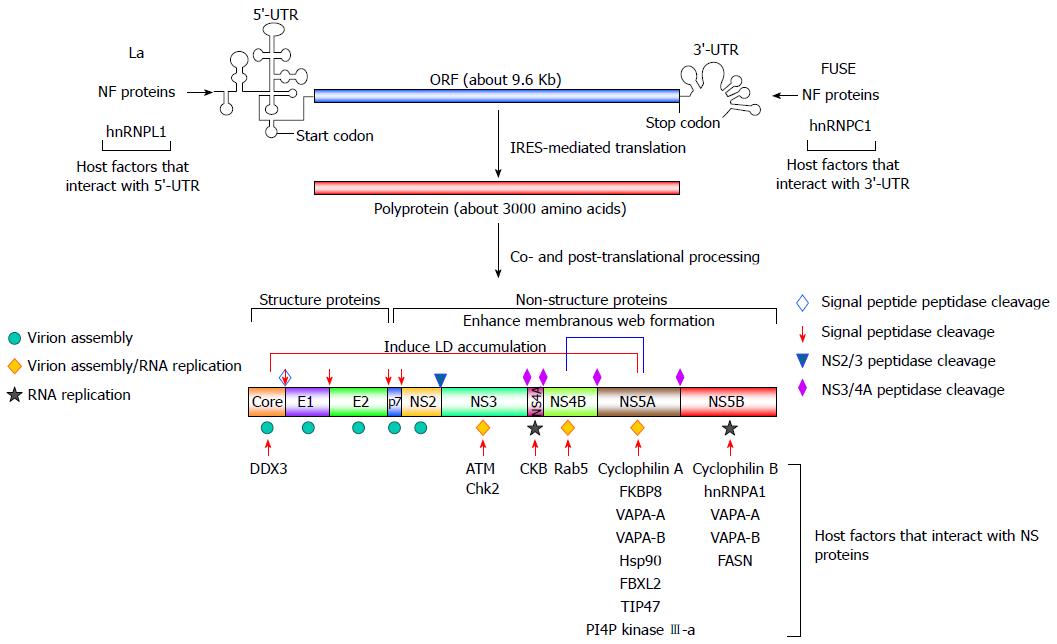
The viral genome of HCV is a positive-stranded RNA of approximately 9.6 Kb in length[46] that contains two untranslated regions (UTRs) located on the 5’ and 3’ termini, which flank a major open reading frame (ORF) region[46] (Figure 2). The ORF of the HCV genome can be translated into a polypeptide of approximately 3300 amino acids using an internal ribosome entry site (IRES) that is located within the 5’-UTR. The large polyprotein is then co-translationally processed into structural (core and the envelope glycoproteins E1 and E2) and nonstructural (NS) proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) via a combination of viral and cellular proteases[46] (Figure 2). The structural proteins constitute the viral particle[2,46], whereas the NS proteins, in conjunction with cellular proteins, participate in viral RNA replication by organizing the replication complex within a multi-vesiculated, membranous web[47,48] (Figure 2). The NS proteins are also concertedly localized on the surface of lipid droplets (LDs), which is a process that is required for virion assembly[49,50] (Figure 2).
In addition to the involvement of viral proteins that function as cis-acting elements for HCV replication, many host cellular proteins have been known to regulate viral RNA replication through their interaction with viral proteins and RNAs. Playing a pivotal role in the IRES-mediated translation of a precursor polypeptide that is subsequently processed into individual viral proteins, the eukaryotic translational initiation factors and RNA-binding proteins, such as the La autoantigen[51,52], nuclear factors (NF) NF45, NF90, and NF110[53], the far upstream element-binding proteins[54], and heterogeneous nuclear ribonucleoproteins[55-57], have been implicated in viral RNA translation and replication via interactions with the 5’- and 3’-UTRs (Figure 2), thus promoting HCV replication. In addition to these host factors that modulate viral translation, other host factors, such as cyclophilin B and hnRNP A1[57,58], may directly modulate viral RNA replication by forming a protein complex with the RNA-dependent RNA polymerase (RdRp), NS5B, and regulating replicase activity (Figure 2). The other cyclophilin family proteins, including cyclophilin A and FK506-binding protein 8 (FKBP8), have been reported to interact with NS5A, and this association recruits heat shock protein 90 to form a protein complex that promotes the efficiency of viral RNA replication[58-62] (Figure 2). On the other hand, a variety of vesicle-associated membrane proteins (VAPA), such as VAPA-A and VAPA-B, have been reported to positively regulate HCV replication by interacting with NS5A and NS5B[63,64] (Figure 2). Additionally, the geranylgeranylated protein F-box/LRR-repeat protein 2 is a host protein that interacts with NS5A to promote HCV replication[65,66] (Figure 2). In addition, other host factors, such as DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked[67], ataxia telangiectasia mutated (ATM), checkpoint Chk2 kinases[68], creatine kinase B (CKB)[69], and the small Ras GTPase-binding protein 5 (Rab5)[70], may regulate HCV replication via binding to core, NS3, NS4A, and NS4B, respectively (Figure 2).
In addition to cellular proteins that interact with viral proteins, several host cellular proteins exert their trans-acting function in the HCV life cycle by altering lipid metabolism. The findings that support this notion originate from observations suggesting that the expression of sterol regulatory element-binding proteins can be enhanced by HCV infection and the ectopic expression of individual viral proteins, e.g., core, NS2, and NS4B[71-73] (Figure 3). The SREBP-mediated transactivation of lipogenic genes enhances cholesterol biogenesis and biosynthesis of fatty acids, which, in turn, promote the storage of neutral lipids within LDs[74]. Therefore, these results indicate that HCV infection may activate the gene expression of lipogenes, thus modulating the metabolic pathways of lipids to support the HCV life cycle. In line with this, the gene expression of fatty acid synthase, which is involved in the synthesis and transport of fatty acids, was reported to be upregulated and required for HCV viral RNA replication[75]. Recently, FASN was demonstrated to interact with NS5B to enhance RdRp replicase activity, thereby promoting HCV replication[76] (Figure 2).
Apart from these proteins that directly function in the modulation of lipid biosynthesis, a new subset of host cellular factors has emerged based on their roles in HCV replication via altering the expression and subcellular distribution of phosphatidylinositol-4-phosphate (PI4P). HCV infection was reported to increase the intracellular level of PI4P via PI4P kinase (PI4PK) IIIα and PI4PKIIIβ[77-83]. Interference with the gene expression of these two PI4P kinases dramatically inhibits HCV replication[77-83]. Furthermore, recent studies indicated that the NS5A protein can recruit (PI4PK) IIIα to the membranous web, which is a multi-vesiculated structure that supports efficient replication of HCV viral RNA, thus upregulating the level of PI4P to maintain the membranous web architecture[77,78,81,84,85]. Reciprocally, (PI4PK) IIIα can modulate the phosphorylation status of NS5A, thus regulating the morphogenesis of viral replication compartments[86]. On the other hand, annexin A2 and proline-serine-threonine phosphatase-interacting protein 2 (PSTPIP2), two host membrane-associated proteins, were also shown to regulate HCV replication via facilitating the formation of the membranous web[87,88]. In addition, the tail interacting protein of 47 kDa (TIP47), which is an LD-associated protein, has recently been shown to positively modulate HCV RNA replication by interacting with NS5A[89,90] (Figure 2). Collectively, these studies indicate that host cellular factors may regulate HCV replication via directly facilitating the reconstitution of the membranous web or modulating LDs by interacting with viral NS proteins.
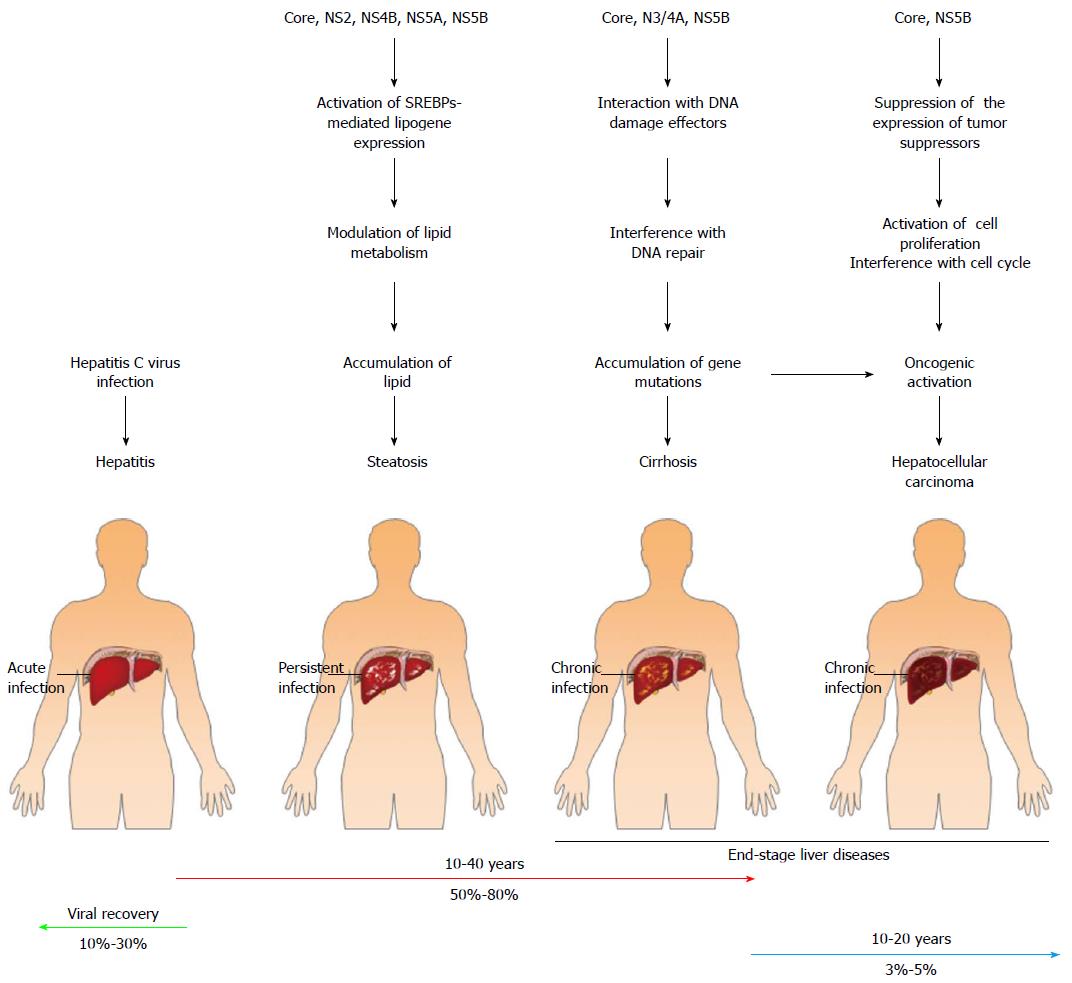
The disease progression in HCV-infected patients reveals a nonlinear and long-term period mode. At the beginning of infection, most infected patients are non-symptomatic, although hepatitis, jaundice, and fulminant hepatic failure can be detected in some cases of acute infection[16,91] (Figure 3). However, more than 50% of HCV infection cases develop viral persistence, which often leads to liver steatosis, fibrosis, cirrhosis, and ultimately to hepatocellular carcinoma (HCC)[16,91-95] (Figure 3). In the majority of infected individuals, the disease progression to end-stage HCV-related diseases, such as liver cirrhosis and HCC, often occurs 20-30 years after acute infection, and approximately 20%-30% of the infected patients will progress to end-stage liver diseases[16,91-95] (Figure 3). In addition to the end-stage liver diseases, chronic HCV infection in some patients is also highly associated with extrahepatic diseases such as mixed cryoglobulinemia vasculitis, which is an inflammatory symptom of small blood vessels, due to the precipitation of cryoglobulin-containing immune complexes in blood vessels of the skin and other tissues[96,97]. Additionally, chronic HCV infection is associated with metabolic syndromes such as diabetes and insulin resistance[98,99]. The development of these diseases often requires long periods of time, thus affecting the health quality of patients and imposing a heavy burden on medical care for the treatment of HCV-related diseases. Therefore, it is urgent to design a new, efficacious therapeutic strategy and/or to develop prophylactic vaccines.
Many HCV proteins are known to participate in the pathogenesis of HCV-related diseases. For instance, the HCV core protein was reported to coat the surface of LDs and induce the emergence of LDs from the endoplasmic reticulum (ER) and the clustering of LDs in a cultured cell model[100-102]. On the other hand, accumulating lines of evidence have shown that the HCV core protein can induce hepatic steatosis in an in vivo transgenic model[103-106]. Most importantly, the genetic variation within the core sequences of different HCV genotypes has been reported to critically determine the status of hepatic steatosis. The prevalence and severity of hepatic steatosis are higher in patients infected with HCV genotype 3 than in those infected with other genotypes[107-109]. This greater extent of steatosis in genotype 3-infected individuals may be due to the substitution of phenylalanine for tyrosine at amino acid residue 164 of the core protein in the genome of HCV genotype 3[110]. Despite this amino acid variation, several specific polymorphisms in the core protein of different HCV genotypes have been shown to increase the intracellular lipid levels and, thus, contribute to hepatic steatosis[111,112]. Cumulatively, these studies suggest that HCV infection may lead to hepatic steatosis through core-induced LD accumulation. Apart from the impacts of altering intracellular lipids, HCV core was shown to suppress the expression of the tumor suppressors p53 and cyclin-dependent kinase (CDK) inhibitor p21, thereby enhancing CDK2 activity and increasing the phosphorylation status of retinoblastoma, RB, in cells[113] (Figure 3). In turn, phosphorylated RB stimulates the DNA binding ability of E2F transcriptional factor 1 and activates the expression of downstream genes, such as S phase kinase-interacting protein 2, which is an initiating signal for cell proliferation[113] (Figure 3). Moreover, HCV NS5B has also been demonstrated to interact with RB and target it for proteolysis, thus activating downstream E2F-responsive promoters and cell proliferation[114] (Figure 3). Due to their suppression of the expression of tumor suppressors and promotion of cell proliferation, the HCV core and NS proteins may contribute to the progression of uncontrolled hepatocyte growth, thereby increasing the occurrence of hepatocellular carcinoma.
The interaction between HCV NS3/4A with ATM kinase has been shown to lead to the cytoplasmic retention and dephosphorylation of ATM, thus interfering with the activation of the DNA repair mechanism and desensitizing Huh7 cells to ionization[115] (Figure 3). In addition, the ATM and Chk2 kinases bind to the HCV NS5B protein to promote HCV viral RNA replication[68] (Figure 3). On the other hand, recent studies indicate that HCV infection may interfere with multiple signaling pathways of DNA repair via interactions between HCV core and the Nijmegen breakage syndrome protein 1 (NBS1), which is a downstream effector of the ATM-associated DNA damage response[116] (Figure 3). Taken together, these findings imply that HCV infection may interfere with the host DNA damage/repair response, thus benefiting viral growth. Interference with the integrity of the DNA repair mechanism may introduce error-prone effects on DNA replication, which leads to the accumulation of gene mutations, gene instability, and oncogenic activation in infected cells and, eventually, promotion of the progression of infected cells into hepatocellular carcinoma.
Innovation of new therapeutic strategies against HCV infection relies on a comprehensive understanding of the entire viral life cycle and HCV-host interactions. Although HCV was identified more than two decades ago, our knowledge of how HCV infection leads to a homeostatic balance with host cells is still limited due to the lack of an in vitro cell culture model that can support the complete HCV cycle. The replication assay utilizing a subgenomic replicon that harbors only the HCV nonstructural genome in human hepatoma Huh7 cells was established in the late 1990s. This model allows one to study HCV RNA replication and the biological functions of each of the viral NS proteins[117-120]. The advent of a replicon system also facilitates the identification of adaptive mutations in the HCV viral genome and promotes the discovery of potent anti-HCV agents[118,121]. On the other hand, the establishment of an HCV pseudoparticle system (HCVpp), in which the HCV E1 and E2 glycoproteins are incorporated onto retro- or lentiviral particles, provides an efficient system to study HCV entry, identify entry (co)receptors, and screen for neutralizing antibodies[122-125]. In 2005, the robust production of infectious HCV in a cell culture system (HCVcc) based on the entire genome of the JFH1 strain, which is an HCV genotype 2a virus that was isolated from a fulminant hepatitis patient in Japan, was developed[126-128], thus marking a great achievement in the HCV research field. The generation of infectious HCVcc allows one to investigate each step of the viral life cycle and HCV-host cell interactions in vitro and will be useful for the screening and testing of new antiviral drugs. Nevertheless, the availability of an in vivo model for HCV research was limited to chimpanzee, which has been used as a model for studying viral replication kinetics, the immune response, and vaccine development[129-135]. The immune-deficient mouse system transplanted with human hepatocytes serves as an additional research tool to analyze the HCV life cycle in a humanized, small animal model[135,136]; however, the study using these two animal models is circumscribed by their high cost and the inconvenience of the experimental manipulations. Recently, a model that allows investigation of the complete HCV life cycle in an immune-competent mouse system was successfully developed by genetically engineering human CD81, SCARB-I, CLDN1 and/or OCLN into mice[137,138]. This system provides a new research platform for studying HCV infection in vivo and screening anti-HCV drug and vaccine candidates. Nevertheless, the low level of viral replication and virion production in this HCV-rodent model hampers the use of this in vivo model. Thus, further efforts are needed to develop an improved version of this rodent system.
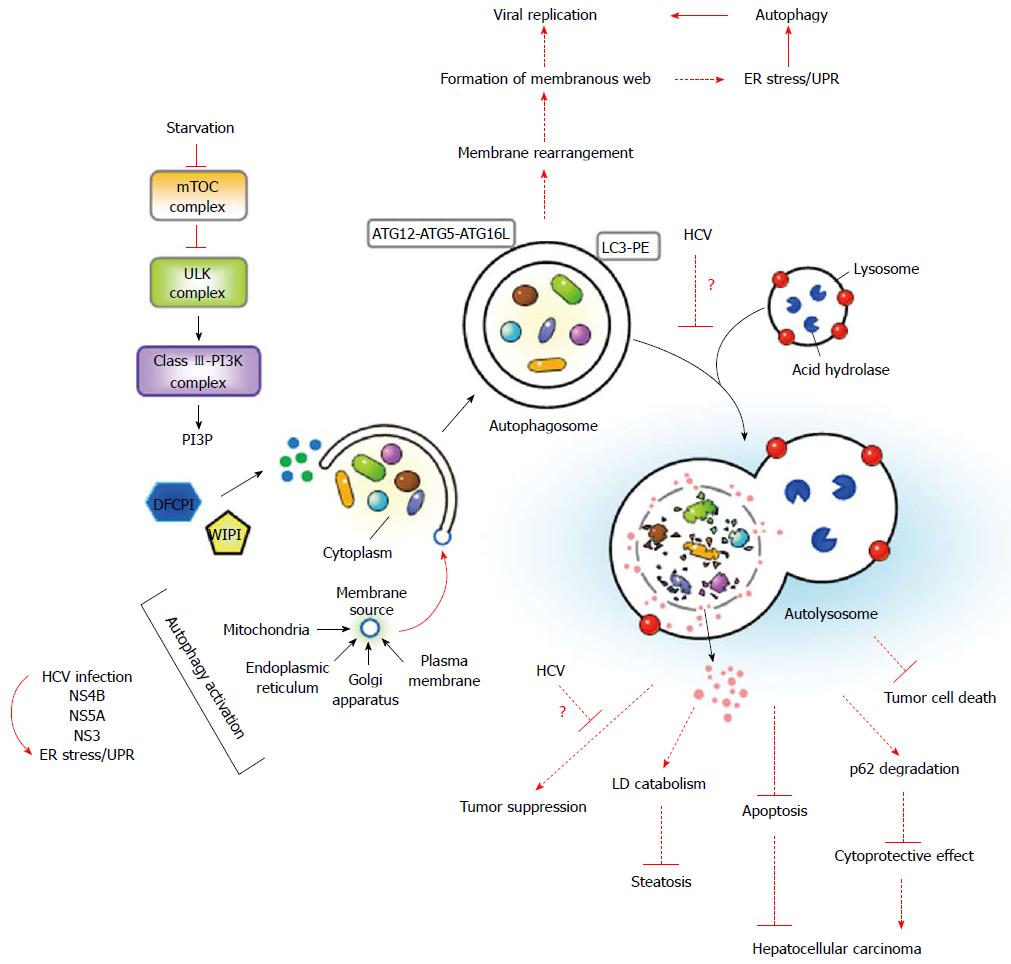
Autophagy is considered a “self-eating” process in eukaryotic cells that engulfs unwanted cytoplasmic components within double-membranous vacuoles and delivers these cargos to lysosomes for breakdown. The autophagic process promotes the turnover of damaged organelles and aggregated proteins through lysosomal degradation to ensure the recycling of cellular constituents, thus maintaining cellular homeostasis[6,7]. The concept of “self-eating” was originally described in the mid-1950s in Christine de Duve’s work on biochemical characterization of the lysosome in liver tissue[139,140]. Soon after this study, she and other researchers independently utilized transmission electron microscopy to show that dense bodies similar in size to mitochondria are present in the cytosol of renal and hepatic cells[141-146]. These observed dense bodies formed unique single- and double-membranous vesicle structures that were associated with lysosomes and contained mitochondria and endoplasmic reticulum (ER)[141,144,145]. Based on these observations, de Duve proposed a new term, “autophagy”, to illustrate this de novo process of sequestrating cytoplasmic organelles within a double membrane-enclosed vesicle termed an “autophagosome”. Per Seglen’s group then investigated the process prior to autophagosome formation in autophagy and identified the expansion of the “phagophore”, which is an initial, membrane-rearranged structure, into the autophagosome[147-149]. Additionally, Mortimore and Schworer showed that amino acid deprivation activates autophagy and proteolysis in rat liver, and they were the first to suggest that energy imbalance and/or an insufficient nutrient supply can stimulate the initiation of the autophagic process[150-153]. In the early 1990s, the detailed molecular basis of autophagy began to be uncovered through Yoshinori Ohsumi’s study using the yeast Saccharomyces cerevisiae[154,155]. Using the advantage of well-established genetic manipulation techniques and the well-known genomic background in the yeast model system, Ohsumi’s and Klionsky’s groups began to identify the genes involved in the autophagic process (ATGs)[156-168]. Most homologues of the yeast ATG genes also exist in humans and other eukaryotes, and the human orthologs of the yeast ATG genes can carry out similar functions[161,169]. Finally, a unified nomenclature for all ATGs in the different model systems was denoted[170-172]. These significant breakthroughs enhanced the understanding of the mechanism underlying how autophagy initiates and terminates and provided a crucial foundation for further investigation of autophagy-related processes.
Autophagy is the concerted action of the core molecular machineries that are involved in the initiation stage of the isolation membrane (IM)/phagophore, the middle step of elongation and enclosure of the autophagosome, and the late stage of autophagosome fusion with a lysosome and the subsequent degradation of the sequestrated components within the autolysosome by acidic proteases[6,7] (Figure 4). The completion of autophagy requires a dynamic membrane rearrangement process and multiple signaling pathways[171-174] (Figure 4). Upon nutrient deprivation, autophagy begins with the suppression of mammalian target of rapamycin (mTOR), which is a serine/threonine protein kinase that is required for cellular metabolism. Inhibition of mTOR leads to translocation of the downstream unc-51 like-kinase (ULK) complex, i.e., ULK1-ATG13-FIP200-ATG101, from the cytoplasm to the unique ER membrane-associated compartments[175,176] (Figure 4). The class III phosphatidylinositol-3-OH kinase (class III-PI3K) complex, i.e., PI3K-Vps15-Beclin-ATG14, is then recruited to the ER-derived nucleation site, where the P3K complex catalyzes the formation of phosphatidylinositol-3-phosphate (PI3P)[177] (Figure 4). The newly synthesized PI3P, in turn, recruits downstream effectors, including the double-FYVE-containing protein 1 (DFCP1) and the WD-repeat domain PI3P-interacting (WIPI) family proteins, resulting in the formation of an ER-associated Ω-like structure called the “omegasome”[178]. After the nucleation step, elongation and enclosure of the IM/phagophore to form an autophagosome require two ubiquitin-like conjugation complexes, i.e., ATG12-ATG5-ATG16L and ATG4-ATG3-LC3II (Figure 4). The ATG12-ATG5 conjugate is first produced through the concerted action of the ATG7 (E1-like) and ATG10 (E2-like) enzymes, and it then binds to ATG16L, which forms the ATG12-ATG5-ATG16L trimeric complex. The conjugation of microtubule-associated protein 1 light chain 3 (LC3), i.e., the ATG8 homologue in mammals, to phosphatidylethanolamine (PE) begins with the proteolytic cleavage of its C-terminus by ATG4. Subsequently, the cleaved form of LC3 is conjugated to PE through the catalytic cascade of ATG7 and ATG3, thus generating the lipidated form of LC3, i.e., LC3-II[179]. Finally, the autophagosome fuses with the endosome and lysosome to form a mature autophagolysosome, named an “autolysosome”, in which the engulfed materials are broken down and recycled for further use by cells[7]. Despite the emergence of an IM/phagophore from the ER[180,181], a variety of organelles, such as the plasma membrane (PM)[182], mitochondria[183], and Golgi apparatus[184], can also serve as membrane sources for the initiation of autophagy in mammalian and yeast cells (Figure 4). Nonetheless, what and how signaling pathways regulate each step of the membrane rearrangement processes must be further investigated.
Apart from nutrient starvation, multiple stresses, including ER stress, accumulation of aggregated proteins and damaged organelles, and pathogen infections, can activate the autophagic response[7,185,186]. Upon infection by bacteria and viruses, the autophagic process is triggered in cells to directly engulf incoming pathogens and deliver them to the lysosome for degradation, i.e., xenophagy[187-189]. In addition, autophagy may induce the innate immune defense to repress microbial infection, such as enhancing Toll-like receptor-mediated innate immune signaling and promoting the presentation of antigens derived from viruses such as vesicular stomatitis virus and Epstein-Barr virus onto a major histocompatibility complex class II molecule[6,190-193]. Thus, autophagy can function as a restrictive route to eliminate pathogens. In addition, autophagy was shown to be exploited by many RNA viruses, such as mouse hepatitis virus, poliovirus, and rhinovirus, to promote their life cycle by serving as a membranous compartment for RNA replication[194]. On the other hand, autophagy also plays critical physiological roles in the pathogenesis of various diseases, including neurodegenerative disorders[12,13], inflammatory diseases[195,196], liver-associated diseases[197], and cancers[198]. Because of its impacts on a wide array of physiological and pathological conditions, autophagy has become an attractive field of biomedical research. Nevertheless, further studies are needed to better understand the regulation of autophagy and its exact physiological significance in the pathogenicities of various diseases.
Infections of flaviviruses such as HCV, Dengue virus, and Japanese encephalitis virus were shown to activate autophagy in vitro[8-11,199-202]. Many studies have reported that HCV induces the autophagic response in HCV viral RNA-expressing and HCVcc-infected cells[8-11]. By analyzing the viral expression of HCV genotype 1a H77 strain in immortalized human hepatocytes (IHH), Ait-Goughoulte’s group demonstrated that autophagic vesicles and GFP-LC3-labeled punctate structures accumulate in viral RNA-transfected cells[8] (Table 1). These authors also found that the H77 HCV-induced autophagic response is accompanied by increases in Beclin expression and the ATG12-ATG5 conjugate[8]. Shortly after their study, Sir et al[11] reported that HCV triggers autophagosome formation in full length, JFH1 (genotype 2a) viral RNA genome-transfected Huh7 cells (Table 1). They further showed that HCV triggers incomplete autophagy, i.e., autophagolysosome maturation is interrupted. This conclusion was based on the observation that the degradation of long-lived proteins and p62, which is a substrate of autophagic degradation, was inhibited despite increased numbers of LC3-II and GFP-LC3-labeled dot-like vesicles in JFH1 RNA-transfected cells[11] (Table 1). Their study also demonstrated, for the first time, that autophagy positively regulates HCV growth via promoting viral replication because knockdown of ATG7 and LC3 dramatically suppressed viral RNA expression in HCV RNA-transfected cells[11] (Table 1). Whether autophagy is analogously activated by HCV in the HCV infection system had not been determined until Druex and her colleagues investigated autophagic activation in Huh7.5 cells infected with the JFH1 HCVcc virus in 2009[9] (Table 1). These authors provided first-line evidence demonstrating that HCV infection can induce the autophagic response to enhance the translation of the incoming viral RNA, rather than regulate viral growth, when virus RNA replication is established[9].
| HCV genotype | Expression | Model | Analysis of autophagy activation | Physiological significance | Ref. |
| HCV-H77 (1a) | Transfection of viral RNA | IHH cells | 1 Detection of GFP-LC3 punctate structure formation | Promotion of viral RNA replication | Ait-Goughoulte et al[8] |
| 2 Accumulated autophagosome in TEM analysis | |||||
| 3 Upregulation of Beclin expression and ATG5-ATG12 conjugate | |||||
| HCV-JFH1 (2a) | Transfection of viral RNA | Huh7.5 cells | 1 Upregulated expression of LC3-II | Promotion of viral RNA replication | Sir et al[11] |
| 2 No overlapping signal of GFP-LC3 punctate with lysosome | |||||
| 3 Autophagic activation by UPR | |||||
| 4 An incomplete autophagic process lacking enhanced autophagic degradation of long-lived proteins and p62 | |||||
| HCV-JFH1 (2a) | HCVcc infection | Huh7 cells | 1 Upregulation of LC3-II | Enhanced translation of the incoming viral RNA | Dreux et al[9] |
| 2 Accumulation of GFP-LC3 dot-like vesicles | |||||
| 3 No colocalization of autophagic vacuoles with viral proteins | |||||
| HCV-JFH1 (2a) | HCVcc infection | Huh7.5-1 cells | 1 Increase of GFP-LC3 dot-like structures | Promotion of virion assembly | Tanida et al[207] |
| 2 No colocalization of autophagic vacuoles with viral proteins | |||||
| HCV-JFH1 (2a) | HCVcc infection | Huh7 cells | 1 Transient interaction of ATG5 with NS5B and NS4B | Promotion of viral RNA replication by organizing membranous web | Guévin et al[205] |
| 2 Association of ATG5 with membranous web | |||||
| HCV-JFH1 (2a) infection | HCVcc infection | Huh7 cells | 1 Detection of early- and late-stage autophagic vacuoles by TEM analysis | Promotion of viral RNA replication by suppressing antiviral immunity | Ke et al[10] |
| 2 Colocalization of autophagic vacuoles with lysosome | |||||
| 3 Accumulated LC3B-II expression by interference with autolysosome maturation | |||||
| 4 Complete autophagic process by HCV infection | |||||
| HCV-H77 (1a); HCV-JFH1 (2a) | HCVcc infection | IHH | 1 Activated IFN response in the HCV-infected cells by silencing of Beclin and ATG7 | Promotion of viral RNA replication by suppressing antiviral immunity | Shrivastava et al[203] |
| 2 Increased caspase-dependent apoptosis by knockdown of Beclin and ATG7 in the HCV-infection cells | |||||
| HCV-Con1 (1b) and JFH1 (2a) | Replicon viral RNA transfection | Huh7 cells; Huh7.5-1 cells; Liver biopsy | 1 An inverse correlation between hepatic steatosis and activation of autophagy in liver biopsy samples of infected patients | Promotion of catabolism of LDs | Vescovo et al[208] |
| 2 Colocalization of autophagic vacuoles with LDs | |||||
| HCV-JFH1 (2a) | Replicon viral RNA transfection | Huh7 cells; HCV-transgenic mice | 1 Enhanced ROS level in mitochondria in HCV viral RNA-transfected cells | Regulation of oxidative response in mitochondria | Chu et al[210] |
| 2 Activated autophagy by expression of HCV NS proteins | |||||
| 3 Alteration of antioxidant response by upregulation of antioxidant enzymes in HCV NS protein-expressing cells | |||||
| HCV-JFH1 (2a) | HCVcc infection | Huh7 cells; Huh7.5-1 cells | 1 Accumulation of mito autolysosome in HCV-infected cells | Elimination of damaged mitochondria and promotion of viral RNA replication | Kim et al[209] |
| 2 Stimulation of Parkin and Pink 1 expression in HCV-infected cells | |||||
| 3 Activation of mitophagy via a Parkin-dependent pathway | |||||
| HCV-JFH1 (2a) HCV-N (1b) | HCVcc infection; Replicon viral RNA transfection | Huh7 cells | 1 Activation of autophagy through AKT1-TSC-mTORC1 signaling | Promotion of viral RNA replication | Huang et al[214] |
| 2 Activation of autophagy via UPR | |||||
| HCV-JC1 (2a) infection; HCV NS4B | HCVcc infection; Ectopic overexpression | Huh7.5 cells | 1 Activation of autophagy by HCV NS4B amino acid 1-190 | Organization of virus replication site | Su et al[216] |
| 2 Requirement of Rab5 and PI3K for autophagic activation | |||||
| HCV-JFH1 (2a) | HCVcc infection | IHH cells | 1 Transcriptional activation of Beclin gene expression | Promotion of viral RNA replication | Shrivastava et al[215] |
| 2 Autophagy activation in a Bcl2-Beclin dissociation- and mTOR inhibition-independent manner | |||||
| HCV-JFH1 (2a) | HCVcc infection | Huh7.5 cells | 1 Activation of autophagy through IRGM | Promotion of viral RNA replication; modulation of innate immunity (?) | Grégoire et al[237] |
| 2 Interaction of HCV NS3 with IRGM |
Utilizing the JFH1 infection system, we demonstrated that HCV infection of Huh7 cells enhances the autophagic flux and triggers the complete autophagic process throughout the formation of the mature autophagolysosome[9,10] (Table 1). Several lines of evidence supported this conclusion, including (1) the detection of early and late-stage autophagic vacuoles in the TEM analysis of HCV-infected cells; (2) the accumulation of LC3-II expression by blocking the fusion of the autophagosome with a lysosome; (3) the predominant expression of RFP, but not GFP, fluorescence of a mRFP-GFP-LC3 reporter in the infected cells; and (4) the high degree of colocalization of GFP-LC3 puncta with lysosomes in infected cells[9,10] (Table 1). Moreover, we showed that silencing of the ATG genes and treatment with pharmacological inhibitors of autophagolysosome maturation repress HCV viral RNA replication[9,10]. However, interfering with autophagy had no detectable effect on virus entry or the translation of viral RNA[9,10]. Most importantly, inhibition of the HCV-activated complete autophagy drastically upregulated the IFN response that was mediated by the HCV pathogen-associated molecular pattern (PAMP), which is located within the poly-U/UC region of HCV 3′-UTR[9,10]. Consistent with our study, Shrivastava et al[203] reported that gene silencing of Beclin or ATG7 inhibits HCV growth and activates IFN and interferon-stimulated gene expression in HCV-infected human IHH cells (Table 1). Together, these two studies imply that autophagy may represent a repressive mode to protect HCV-infected cells against an excessive IFN antiviral response, thereby promoting viral RNA replication.
In addition to its suppressive effect on antiviral immunity, autophagy was reported to promote viral RNA replication through other mechanisms[204,205]. Guevin and colleagues showed the transient interaction of ATG5 with NS4B and NS5B as well as the detection of ATG5 in the HCV-induced membranous web, which suggested a proviral role of the autophagic machinery in the formation of the HCV replication complex[205] (Table 1 and Figure 4). Sir et al[204] further showed that NS5A, NS5B, and nascent viral RNA were colocalized with the autophagosome and argued that the HCV-induced autophagic membrane can be used as a membrane-associated compartment for the replication of viral RNA (Table 1). In addition to its pivotal role in viral RNA replication, autophagy was shown to regulate the assembly of infectious virions and protection of infected cells from death[206,207] (Table 1). Tanida and colleagues first reported that knockdown of ATG7 and Beclin gene expression moderately downregulates the extracellular titer of HCV virions without showing an apparent effect on the intracellular level of viral proteins and RNAs[207] (Table 1). This study suggests that HCV-activated autophagy may modulate the egress of HCV virions into infected cells. Additionally, HCV was shown to activate autophagy to protect infected cells from cell death[206] (Table 1). In this study, severe cytoplasmic vacuolation and cell death accumulated in Con1-HCV (genotype 1b)-transfected cells through interference with autophagy via ectopic expression of a protease-inactive mutant ATG4BC47A[206], which implied that HCV may exploit autophagy as a cellular surveillance machinery to counteract the overloaded stress that is triggered by viral replication.
The HCV-induced autophagic process was also shown to regulate host cellular metabolism, including eliminating excess lipids and degrading damaged mitochondria[208,209] (Table 1 and Figure 4). Vescovo et al[208] studied the correlation of autophagy markers with the clinical parameters of lipid metabolism in liver biopsies of patients chronically infected with HCV and found an inverse relationship between autophagy activation and the extent of steatosis in those patients (Table 1). The authors further showed that autophagy participates in the catabolism of LDs in cells transfected with the HCV subgenomic RNA replicon[208], implying that HCV may utilize autophagic degradation to promote LD breakdown and circumvent virus-triggered lipid accumulation in host cells (Figure 4). In addition to its degradation role in lipid metabolism, a unique form of autophagy, termed “mitophagy”, was recently shown to eliminate damaged mitochondria in HCV-infected cells in a Parkin-dependent manner[209,210] (Table 1). Knockdown of Parkin and Pink gene expression suppresses HCV viral RNA replication[209], suggesting a critical role of mitophagy in HCV replication.
The molecular mechanism for how HCV initiates autophagy is not fully understood, although several studies have shown that ER stress and the unfolded protein response (UPR) can stimulate autophagy activation[211-213] (Table 1 and Figure 4). Remarkably, two independent reports showed that the UPR is required for activation of autophagy by HCV[10,11]. Recently, Huang et al[214] showed that HCV can inhibit the protein kinase B (PKB)-tuberous sclerosis (TSC)-mTOR complex 1 (mTORC1) signaling pathway via virus-induced ER stress, thus activating autophagy. Nevertheless, Shrivastava et al[215] demonstrated that HCV induces autophagy by transcriptionally activating the expression of the Beclin mRNA and triggering mTOR signaling. In addition to virus-triggered ER stress and the UPR, viral protein expression seems to be another signal for HCV-activated autophagy (Figure 4). Su et al[216] showed that HCV NS4B can trigger incomplete autophagy via an interaction with Rab5 and Vps34 (Table 1). Moreover, HCV NS5A was reported to be sufficient to trigger the autophagic response[215] (Figure 4). On the other hand, Gregoire and colleagues demonstrated that several RNA viruses, including HCV, could modulate autophagy via the interaction of the immunity-associated GTPase family M (IRGM) with ATG5 and LC3[217] (Table 1). They also showed that HCV NS3 is sufficient to activate IRGM-mediated autophagy[217]. Collectively, these studies reveal that multiple signaling pathways may be involved in the HCV-activated autophagic response. However, further investigations are necessary to determine how HCV RNA or proteins cooperate with those cellular signaling pathways to modulate autophagy.
Although HCV infection is shown to positively induce the autophagic process in the cultured human hepatocyte system[8-11], the evidence for autophagy activation in an in vivo animal model and liver specimens from infected patients is still limited. Autophagy activated by HCV infection has been demonstrated to promote HCV growth in host cells via regulating RNA replication, the translation of incoming viral RNA, and the assembly of infectious viral particles[9-11,207]. In addition to its proviral role in the HCV life cycle, upregulation of autophagy functions in suppressing innate immunity[10,203,218], altering the apoptosis pathway[215] (Figure 4), and maintaining the surveillance of infected cells[206]. In addition, recent studies provide a new horizon for autophagy and its role in protection of host cells from excess LDs due to HCV infection and the elimination of damaged mitochondria via the degradative process[208-210]. These studies also suggest that the autophagic response is utilized to maintain cell homeostasis via promoting the breakdown of excess lipids and damaged organelles that are induced by HCV[208-210] (Figure 4). However, how these cell-signaling pathways, in turn, affect cellular metabolism or alter cell homeostasis, which lead to the development of HCV-associated liver diseases, still remains to be investigated (Figure 4).
The potential role of autophagy in the progression of HCV-induced steatosis and fibrosis emerges from the recent findings of Singh et al[219]. This group showed that autophagy regulates lipid metabolism in hepatocytes via a selective degradation process, i.e., lipophagy[219] (Figure 4). Lipophagy represents a new mode of autophagy in lipid metabolism that catabolizes LDs in the liver[219]. Moreover, Singh et al[220] proposed another function of autophagy in the control of body lipids through regulating the differentiation of adipose tissues. Collectively, their studies imply that modulation of autophagy in the liver may affect the metabolic cycle of lipids. In line with their findings, Vescovo’s group reported that HCV might subvert the degradative process of autophagy to promote the catabolism of LDs[208]. Based on the in vitro HCV replicon study and in vivo investigation of liver biopsies from patients chronically infected with HCV[208], Vescovo et al[208] concluded an inverse interrelationship between the extent of autophagy activation and the level of steatosis in HCV patients (Figure 4). This notion was based on their observations that the autophagic process facilitates LD breakdown in HCV replicon cells and that interference with autophagy leads to an elevated cholesterol level in HCV JFH1-infected cells. Although HCV-activated autophagy acts as a counteracting mechanism to prevent excessive accumulation of lipids that are induced by virus infection, it remains to be determined whether virus-induced autophagy affects the cell metabolism balance during the enhanced catabolism of LDs in liver cells, such as by altering the homeostatic levels of related lipids or interfering with the balance between the lipogenesis and lipolysis pathways. Moreover, whether activation of lipophagy by HCV infection affects the regular cellular functions and leads to pathological changes in the infected hepatocytes warrants further investigation. Nevertheless, the autophagy-mediated regulation of lipid metabolism may represent a mechanism of deregulation that interferes with metabolic homeostasis, thus promoting the progression of HCV-associated metabolic syndrome.
A recent study has shown that ectopic expression of HCV NS4B is sufficient to activate incomplete autophagy by interacting with Rab5 and Vps34 in human hepatoma cells[216], which suggests that activation of autophagy by the NS4B protein may be related to membranous web formation (Figure 4). Reciprocally, the HCV NS4B-induced membranous web accumulation could trigger a stress response, such as ER stress, which was indicated to be an inducer of HCV-triggered autophagy[10,11] (Figure 4). It is still unknown whether HCV NS4B can utilize the autophagy-mediated membrane rearrangement process to generate double-membrane vesicles (DMV) within the membranous web, which is required for HCV replication. Notably, several studies indicated that HCV activates incomplete autophagy[11,216], which may serve as a means of inducing the accumulation of DMV. Moreover, the autophagosomal membrane has recently been demonstrated to be a site for HCV viral RNA replication[204]. In addition to NS4B, HCV NS5A was shown to activate autophagy via enhancing phospho-mTOR expression and its downstream target 4EBP1 in IHH cells[215]. HCV NS5A being a critical regulator for modulating the local concentration of PI4P, which is a critical component of the membranous web[77,78,81,84,85], implies again that HCV may exploit autophagy to regulate the formation of the membranous web (Figure 4). These studies collectively imply that the extent of host cellular autophagy may affect the pathogenesis of HCV-associated liver diseases through modulating the status of HCV replication and membranous web formation (Figure 4).
In addition to the possibility of participating in the development of liver-associated diseases by altering lipid metabolism, HCV-activated autophagy may contribute to the development of hepatocellular carcinoma (Figure 4). The role of autophagy in inhibiting tumor development originated from the investigation of the functional impact of knocking out the ATG genes in mice[221-225]. The heterozygous loss of the Beclin gene with a repressed autophagic process in mice promoted tumorigenesis and increased the occurrence of spontaneous malignancies, which indicated, for the first time, that Beclin may serve as a tumor suppressor in tumor progression[221]. Likewise, mosaic knockout of ATG5 and the conditional depletion of ATG7 in mice also resulted in spontaneous formation of benign liver cancer[223]. Additionally, inhibition of tumor suppressor genes, such as phosphatase and tensin homolog (PTEN) and p53, suppresses the basal autophagic response[223,226,227], suggesting an association of autophagy with tumor formation. On the other hand, recent studies from Komatsu’s group showed that the accumulation of p62, which is a substrate of autophagy, by interference with autophagy promotes the formation of hepatocellular carcinoma[224,228] (Figure 4). This process occurs through direct interaction of p62 with Kelch-like ECH-associated protein 1 (Keap1), which is a component of Cullin3-associated ubiquitin E3 ligase, and ablation of the Keap1-mediated degradation of activating nuclear factor (erythroid-derived 2)-like factor 2 (Nrf2) and, therefore, leads to persistent activation of the expression of Nrf2 downstream cytoprotective genes[224,228]. These studies collectively unveil a novel role of autophagy in tumor suppression (Figure 4). Along with these findings, it would be interesting to investigate whether HCV-activated autophagy interferes with the suppressive effect of basal autophagy in tumor progression, thus promoting the development of liver cancer. Currently, how HCV-induced autophagy interacts with and/or affects basal autophagy in vivo to promote tumor formation is still unclear and needs to be studied.
On the other hand, accumulated evidence has indicated that cancer cells may activate autophagy to alter the tumor microenvironment and promote cell surveillance, which would, therefore, protect tumor cells from cell death[198,229-235] (Figure 4). The tumor microenvironment often faces stringent stress conditions of hypoxia and restricted nutrients; thus, tumor cells activate autophagy to counteract these stress responses[198,232,235]. Additionally, activation of autophagy is exploited by cancer cells to trigger resistance against anti-cancer therapy[229-231]. Hence, suppression of autophagy has been shown to synergistically enhance the efficacy of anti-cancer drugs to kill cancer cells[229,230]. The potential role of the HCV-activated autophagic response in the establishment of the tumor microenvironment and chemo-resistance of hepatocellular carcinoma has not yet been determined, but such a hypothesis is conceivably reasonable. For instance, the autophagy that is triggered by the virus may protect chronically HCV-infected cells from stress-induced cell death, such as through apoptosis, which would promote cell survival and possibly result in the development of tumors (Figure 4). Nevertheless, further investigations on the relationship between autophagy and the pathogenesis of HCV-related liver diseases and tumor progression are urgently needed. Without a convenient small animal model that supports the entire HCV life cycle and allows the monitoring of the HCV-associated disease progression, a large gap must be crossed before investigation on the in vivo relevance of autophagy in the development of end-stage HCV-associated liver diseases becomes feasible.
Suppression of autophagy has emerged as a means to inhibit HCV replication[9-11,207]; therefore, the implications of repressed autophagic activity in anti-HCV therapy can be envisioned. Our recent studies indicated that pharmacological inhibitors of autophagy, such as chloroquine (CQ) and bafilomycin A1 (BAF-A1), can specifically inhibit HCV infection through activation of type I IFN antiviral immunity in the in vitro HCVcc model[10]. CQ and BAF-A1 were also shown to inhibit HCV entry via inhibiting the endocytosis pathway[39], and CQ has been demonstrated to inhibit the development of pancreatic tumor formation in a rodent model[233,234]. Recently, in vivo gene transfer of transcriptional factor EB, which is a master gene that regulates autophagy in the livers of mice, can promote clearance of mutant, hepatotoxic alpha-1-anti-trypsin, which is a protein aggregate that commonly causes liver injury[236]. This finding implicates that modulation of cellular autophagy may provide an innovative and feasible therapeutic strategy for curing liver-associated diseases. Therefore, it is anticipated that these autophagic inhibitors, along with small molecule of inhibiting autophagy, could be therapeutically applied in the treatment of HCV infection and possibly HCV-associated liver diseases. Again, an in vivo small animal model for studying HCV infection and the progression of liver-related diseases is required for screening and testing the efficacy and safety of a potential therapeutic strategy.
Autophagy has emerged as an important topic in HCV research. However, the detailed mechanistic action of how HCV activates the autophagic process and comprehensive knowledge of the physiological significance of autophagy at each step of the HCV life cycle still remain to be investigated. Moreover, autophagy may contribute to the pathogenesis of HCV-associated liver diseases. In the future, studies on the exploration of the clinical relevance of autophagy in HCV-infected patients and in vivo investigations using small animal models that can support the complete HCV replication cycle shall provide mechanistic insights into the functional impacts of autophagy-HCV interactions in the pathogenesis of HCV-derived liver diseases. The results of these studies will benefit the development of new therapeutic strategies that are capable of curing HCV infection and elucidate the pathogenesis of HCV-associated liver diseases. These results will also facilitate the design of an efficacious vaccine that can protect the human population against HCV infection.
P- Reviewers: Bonino F, Ikuo S S- Editor: Wen LL L- Editor: A E- Editor: Zhang DN
| 1. | Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362. [PubMed] |
| 3. | Gao B, Hong F, Radaeva S. Host factors and failure of interferon-alpha treatment in hepatitis C virus. Hepatology. 2004;39:880-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 131] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 4. | Liang TJ. Current progress in development of hepatitis C virus vaccines. Nat Med. 2013;19:869-878. [PubMed] |
| 5. | Liang TJ, Ghany MG. Current and future therapies for hepatitis C virus infection. N Engl J Med. 2013;368:1907-1917. [PubMed] |
| 6. | Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527-549. [PubMed] |
| 7. | Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069-1075. [PubMed] |
| 8. | Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82:2241-2249. [PubMed] |
| 9. | Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:14046-14051. [PubMed] |
| 10. | Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37-56. [PubMed] |
| 11. | Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054-1061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 297] [Cited by in RCA: 292] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 12. | Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983-997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1310] [Cited by in RCA: 1561] [Article Influence: 130.1] [Reference Citation Analysis (0)] |
| 13. | Harris H, Rubinsztein DC. Control of autophagy as a therapy for neurodegenerative disease. Nat Rev Neurol. 2012;8:108-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 360] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 14. | Feinstone SM, Kapikian AZ, Purcell RH, Alter HJ, Holland PV. Transfusion-associated hepatitis not due to viral hepatitis type A or B. 1975. Rev Med Virol. 2001;11:3-8; discussion 8-9. [PubMed] |
| 15. | Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, Halfon P, Inchauspé G, Kuiken C, Maertens G. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962-973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1070] [Cited by in RCA: 1083] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 16. | Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21-S29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 354] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 17. | Chayama K, Hayes CN. Hepatitis C virus: How genetic variability affects pathobiology of disease. J Gastroenterol Hepatol. 2011;26 Suppl 1:83-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Roingeard P. Hepatitis C virus diversity and hepatic steatosis. J Viral Hepat. 2013;20:77-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 19. | Trepo C. Genotype and viral load as prognostic indicators in the treatment of hepatitis C. J Viral Hepat. 2000;7:250-257. [PubMed] |
| 20. | Nelson DR, Davis GL. Treatment of chronic hepatitis C. Compr Ther. 1997;23:269-276. [PubMed] |
| 21. | Manns MP, Wedemeyer H, Cornberg M. Treating viral hepatitis C: efficacy, side effects, and complications. Gut. 2006;55:1350-1359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 470] [Cited by in RCA: 491] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 22. | Davis GL, Lau JY. Factors predictive of a beneficial response to therapy of hepatitis C. Hepatology. 1997;26:122S-127S. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 230] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 23. | Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G. Binding of hepatitis C virus to CD81. Science. 1998;282:938-941. [PubMed] |
| 24. | McKeating JA, Zhang LQ, Logvinoff C, Flint M, Zhang J, Yu J, Butera D, Ho DD, Dustin LB, Rice CM. Diverse hepatitis C virus glycoproteins mediate viral infection in a CD81-dependent manner. J Virol. 2004;78:8496-8505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 133] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 25. | Zhang J, Randall G, Higginbottom A, Monk P, Rice CM, McKeating JA. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J Virol. 2004;78:1448-1455. [PubMed] |
| 26. | Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017-5025. [PubMed] |
| 27. | Voisset C, Callens N, Blanchard E, Op De Beeck A, Dubuisson J, Vu-Dac N. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J Biol Chem. 2005;280:7793-7799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 183] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 28. | Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wölk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 991] [Cited by in RCA: 942] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 29. | Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882-886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 774] [Cited by in RCA: 730] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 30. | Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA. 1999;96:12766-12771. [PubMed] |
| 31. | Barth H, Schafer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem. 2003;278:41003-41012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 350] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 32. | Pöhlmann S, Zhang J, Baribaud F, Chen Z, Leslie GJ, Lin G, Granelli-Piperno A, Doms RW, Rice CM, McKeating JA. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J Virol. 2003;77:4070-4080. [PubMed] |
| 33. | Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol. 2006;80:5308-5320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 332] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 34. | Zeisel MB, Koutsoudakis G, Schnober EK, Haberstroh A, Blum HE, Cosset FL, Wakita T, Jaeck D, Doffoel M, Royer C. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology. 2007;46:1722-1731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 190] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 35. | Sharma NR, Mateu G, Dreux M, Grakoui A, Cosset FL, Melikyan GB. Hepatitis C virus is primed by CD81 protein for low pH-dependent fusion. J Biol Chem. 2011;286:30361-30376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 36. | Harris HJ, Davis C, Mullins JG, Hu K, Goodall M, Farquhar MJ, Mee CJ, McCaffrey K, Young S, Drummer H. Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem. 2010;285:21092-21102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 161] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 37. | Harris HJ, Farquhar MJ, Mee CJ, Davis C, Reynolds GM, Jennings A, Hu K, Yuan F, Deng H, Hubscher SG. CD81 and claudin 1 coreceptor association: role in hepatitis C virus entry. J Virol. 2008;82:5007-5020. [PubMed] |
| 38. | Schwarz AK, Grove J, Hu K, Mee CJ, Balfe P, McKeating JA. Hepatoma cell density promotes claudin-1 and scavenger receptor BI expression and hepatitis C virus internalization. J Virol. 2009;83:12407-12414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 39. | Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. 2006;80:1734-1741. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 308] [Cited by in RCA: 308] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 40. | Helle F, Dubuisson J. Hepatitis C virus entry into host cells. Cell Mol Life Sci. 2008;65:100-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 97] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 41. | Meertens L, Bertaux C, Cukierman L, Cormier E, Lavillette D, Cosset FL, Dragic T. The tight junction proteins claudin-1, -6, and -9 are entry cofactors for hepatitis C virus. J Virol. 2008;82:3555-3560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 156] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 42. | Zheng A, Yuan F, Li Y, Zhu F, Hou P, Li J, Song X, Ding M, Deng H. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J Virol. 2007;81:12465-12471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 176] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 43. | Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 603] [Cited by in RCA: 569] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 44. | Sainz B, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med. 2012;18:281-285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 349] [Cited by in RCA: 353] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 45. | Martin DN, Uprichard SL. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc Natl Acad Sci USA. 2013;110:10777-10782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 166] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 46. | Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453-463. [PubMed] |
| 47. | Egger D, Wölk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974-5984. [PubMed] |
| 48. | Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487-5492. [PubMed] |
| 49. | Lindenbach BD. Virion assembly and release. Curr Top Microbiol Immunol. 2013;369:199-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 50. | Suzuki T. Assembly of hepatitis C virus particles. Microbiol Immunol. 2011;55:12-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 51. | Ali N, Pruijn GJ, Kenan DJ, Keene JD, Siddiqui A. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J Biol Chem. 2000;275:27531-27540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 126] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 52. | Ali N, Siddiqui A. The La antigen binds 5’ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249-2254. [PubMed] |
| 53. | Isken O, Baroth M, Grassmann CW, Weinlich S, Ostareck DH, Ostareck-Lederer A, Behrens SE. Nuclear factors are involved in hepatitis C virus RNA replication. RNA. 2007;13:1675-1692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 133] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 54. | Zhang Z, Harris D, Pandey VN. The FUSE binding protein is a cellular factor required for efficient replication of hepatitis C virus. J Virol. 2008;82:5761-5773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 38] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 55. | Gontarek RR, Gutshall LL, Herold KM, Tsai J, Sathe GM, Mao J, Prescott C, Del Vecchio AM. hnRNP C and polypyrimidine tract-binding protein specifically interact with the pyrimidine-rich region within the 3’NTR of the HCV RNA genome. Nucleic Acids Res. 1999;27:1457-1463. [PubMed] |
| 56. | Hahm B, Kim YK, Kim JH, Kim TY, Jang SK. Heterogeneous nuclear ribonucleoprotein L interacts with the 3’ border of the internal ribosomal entry site of hepatitis C virus. J Virol. 1998;72:8782-8788. [PubMed] |
| 57. | Kim CS, Seol SK, Song OK, Park JH, Jang SK. An RNA-binding protein, hnRNP A1, and a scaffold protein, septin 6, facilitate hepatitis C virus replication. J Virol. 2007;81:3852-3865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 85] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 58. | Kaul A, Stauffer S, Berger C, Pertel T, Schmitt J, Kallis S, Zayas M, Lohmann V, Luban J, Bartenschlager R. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009;5:e1000546. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 199] [Cited by in RCA: 205] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 59. | Liu X, Sun L, Yu M, Wang Z, Xu C, Xue Q, Zhang K, Ye X, Kitamura Y, Liu W. Cyclophilin A interacts with influenza A virus M1 protein and impairs the early stage of the viral replication. Cell Microbiol. 2009;11:730-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 101] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 60. | Liu Z, Yang F, Robotham JM, Tang H. Critical role of cyclophilin A and its prolyl-peptidyl isomerase activity in the structure and function of the hepatitis C virus replication complex. J Virol. 2009;83:6554-6565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 119] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 61. | Okamoto T, Nishimura Y, Ichimura T, Suzuki K, Miyamura T, Suzuki T, Moriishi K, Matsuura Y. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 2006;25:5015-5025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 207] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 62. | Okamoto T, Omori H, Kaname Y, Abe T, Nishimura Y, Suzuki T, Miyamura T, Yoshimori T, Moriishi K, Matsuura Y. A single-amino-acid mutation in hepatitis C virus NS5A disrupting FKBP8 interaction impairs viral replication. J Virol. 2008;82:3480-3489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 63. | Gao L, Aizaki H, He JW, Lai MM. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J Virol. 2004;78:3480-3488. [PubMed] |
| 64. | Hamamoto I, Nishimura Y, Okamoto T, Aizaki H, Liu M, Mori Y, Abe T, Suzuki T, Lai MM, Miyamura T. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J Virol. 2005;79:13473-13482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 165] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 65. | Wang C, Gale M, Keller BC, Huang H, Brown MS, Goldstein JL, Ye J. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol Cell. 2005;18:425-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 233] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 66. | Ye J, Wang C, Sumpter R, Brown MS, Goldstein JL, Gale M. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc Natl Acad Sci USA. 2003;100:15865-15870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 292] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 67. | Ariumi Y, Kuroki M, Abe K, Dansako H, Ikeda M, Wakita T, Kato N. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J Virol. 2007;81:13922-13926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 212] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 68. | Ariumi Y, Kuroki M, Dansako H, Abe K, Ikeda M, Wakita T, Kato N. The DNA damage sensors ataxia-telangiectasia mutated kinase and checkpoint kinase 2 are required for hepatitis C virus RNA replication. J Virol. 2008;82:9639-9646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 55] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 69. | Hara H, Aizaki H, Matsuda M, Shinkai-Ouchi F, Inoue Y, Murakami K, Shoji I, Kawakami H, Matsuura Y, Lai MM. Involvement of creatine kinase B in hepatitis C virus genome replication through interaction with the viral NS4A protein. J Virol. 2009;83:5137-5147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 70. | Stone M, Jia S, Heo WD, Meyer T, Konan KV. Participation of rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J Virol. 2007;81:4551-4563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 100] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 71. | Oem JK, Jackel-Cram C, Li YP, Zhou Y, Zhong J, Shimano H, Babiuk LA, Liu Q. Activation of sterol regulatory element-binding protein 1c and fatty acid synthase transcription by hepatitis C virus non-structural protein 2. J Gen Virol. 2008;89:1225-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 91] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 72. | Park CY, Jun HJ, Wakita T, Cheong JH, Hwang SB. Hepatitis C virus nonstructural 4B protein modulates sterol regulatory element-binding protein signaling via the AKT pathway. J Biol Chem. 2009;284:9237-9246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 115] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 73. | Waris G, Felmlee DJ, Negro F, Siddiqui A. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J Virol. 2007;81:8122-8130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 212] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 74. | Jeon TI, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol Metab. 2012;23:65-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 402] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 75. | Yang W, Hood BL, Chadwick SL, Liu S, Watkins SC, Luo G, Conrads TP, Wang T. Fatty acid synthase is up-regulated during hepatitis C virus infection and regulates hepatitis C virus entry and production. Hepatology. 2008;48:1396-1403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 159] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 76. | Huang JT, Tseng CP, Liao MH, Lu SC, Yeh WZ, Sakamoto N, Chen CM, Cheng JC. Hepatitis C virus replication is modulated by the interaction of nonstructural protein NS5B and fatty acid synthase. J Virol. 2013;87:4994-5004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 77. | Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:7577-7582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 287] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 78. | Berger KL, Randall G. Potential roles for cellular cofactors in hepatitis C virus replication complex formation. Commun Integr Biol. 2009;2:471-473. [PubMed] |
| 79. | Borawski J, Troke P, Puyang X, Gibaja V, Zhao S, Mickanin C, Leighton-Davies J, Wilson CJ, Myer V, Cornellataracido I. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J Virol. 2009;83:10058-10074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 163] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 80. | Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci USA. 2009;106:16410-16415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 298] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 81. | Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298-307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 361] [Cited by in RCA: 354] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 82. | Trotard M, Lepère-Douard C, Régeard M, Piquet-Pellorce C, Lavillette D, Cosset FL, Gripon P, Le Seyec J. Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. FASEB J. 2009;23:3780-3789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 114] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 83. | Vaillancourt FH, Pilote L, Cartier M, Lippens J, Liuzzi M, Bethell RC, Cordingley MG, Kukolj G. Identification of a lipid kinase as a host factor involved in hepatitis C virus RNA replication. Virology. 2009;387:5-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 128] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 84. | Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32-45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 426] [Cited by in RCA: 406] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 85. | Lim YS, Hwang SB. Hepatitis C virus NS5A protein interacts with phosphatidylinositol 4-kinase type IIIalpha and regulates viral propagation. J Biol Chem. 2011;286:11290-11298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 82] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 86. | Reiss S, Harak C, Romero-Brey I, Radujkovic D, Klein R, Ruggieri A, Rebhan I, Bartenschlager R, Lohmann V. The lipid kinase phosphatidylinositol-4 kinase III alpha regulates the phosphorylation status of hepatitis C virus NS5A. PLoS Pathog. 2013;9:e1003359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 105] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 87. | Chao TC, Su WC, Huang JY, Chen YC, Jeng KS, Wang HD, Lai MM. Proline-serine-threonine phosphatase-interacting protein 2 (PSTPIP2), a host membrane-deforming protein, is critical for membranous web formation in hepatitis C virus replication. J Virol. 2012;86:1739-1749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 88. | Saxena V, Lai CK, Chao TC, Jeng KS, Lai MM. Annexin A2 is involved in the formation of hepatitis C virus replication complex on the lipid raft. J Virol. 2012;86:4139-4150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 89. | Ploen D, Hafirassou ML, Himmelsbach K, Sauter D, Biniossek ML, Weiss TS, Baumert TF, Schuster C, Hildt E. TIP47 plays a crucial role in the life cycle of hepatitis C virus. J Hepatol. 2013;58:1081-1088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 90. | Vogt DA, Camus G, Herker E, Webster BR, Tsou CL, Greene WC, Yen TS, Ott M. Lipid droplet-binding protein TIP47 regulates hepatitis C Virus RNA replication through interaction with the viral NS5A protein. PLoS Pathog. 2013;9:e1003302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 93] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 91. | Hoofnagle JH. Hepatitis C: the clinical spectrum of disease. Hepatology. 1997;26:15S-20S. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 481] [Cited by in RCA: 481] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 92. | Di Bisceglie AM, Goodman ZD, Ishak KG, Hoofnagle JH, Melpolder JJ, Alter HJ. Long-term clinical and histopathological follow-up of chronic posttransfusion hepatitis. Hepatology. 1991;14:969-974. [PubMed] |
| 93. | Di Bisceglie AM, Order SE, Klein JL, Waggoner JG, Sjogren MH, Kuo G, Houghton M, Choo QL, Hoofnagle JH. The role of chronic viral hepatitis in hepatocellular carcinoma in the United States. Am J Gastroenterol. 1991;86:335-338. [PubMed] |
| 94. | Poynard T, Bedossa P, Opolon P. Natural history of liver fibrosis progression in patients with chronic hepatitis C. The OBSVIRC, METAVIR, CLINIVIR, and DOSVIRC groups. Lancet. 1997;349:825-832. [PubMed] |
| 95. | Villano SA, Vlahov D, Nelson KE, Cohn S, Thomas DL. Persistence of viremia and the importance of long-term follow-up after acute hepatitis C infection. Hepatology. 1999;29:908-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 327] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 96. | Agnello V, Chung RT, Kaplan LM. A role for hepatitis C virus infection in type II cryoglobulinemia. N Engl J Med. 1992;327:1490-1495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 925] [Cited by in RCA: 861] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 97. | Charles ED, Dustin LB. Hepatitis C virus-induced cryoglobulinemia. Kidney Int. 2009;76:818-824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 135] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 98. | Clément S, Pascarella S, Negro F. Hepatitis C virus infection: molecular pathways to steatosis, insulin resistance and oxidative stress. Viruses. 2009;1:126-143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 99. | Negro F, Alaei M. Hepatitis C virus and type 2 diabetes. World J Gastroenterol. 2009;15:1537-1547. [PubMed] |
| 100. | Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, Eder G, Schaff Z, Chapman MJ, Miyamura T. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci USA. 1997;94:1200-1205. [PubMed] |
| 101. | Depla M, Uzbekov R, Hourioux C, Blanchard E, Le Gouge A, Gillet L, Roingeard P. Ultrastructural and quantitative analysis of the lipid droplet clustering induced by hepatitis C virus core protein. Cell Mol Life Sci. 2010;67:3151-3161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 102. | Moradpour D, Englert C, Wakita T, Wands JR. Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology. 1996;222:51-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 172] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 103. | Lerat H, Honda M, Beard MR, Loesch K, Sun J, Yang Y, Okuda M, Gosert R, Xiao SY, Weinman SA. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology. 2002;122:352-365. [PubMed] |
| 104. | Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T, Koike K. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065-1067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 931] [Cited by in RCA: 912] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 105. | Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, Ishibashi K, Matsuura Y, Miyamura T, Koike K. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J Gen Virol. 1997;78:1527-1531. [PubMed] |
| 106. | Perlemuter G, Lettéron P, Carnot F, Zavala F, Pessayre D, Nalpas B, Bréchot C. Alcohol and hepatitis C virus core protein additively increase lipid peroxidation and synergistically trigger hepatic cytokine expression in a transgenic mouse model. J Hepatol. 2003;39:1020-1027. [PubMed] |
| 107. | Adinolfi LE, Gambardella M, Andreana A, Tripodi MF, Utili R, Ruggiero G. Steatosis accelerates the progression of liver damage of chronic hepatitis C patients and correlates with specific HCV genotype and visceral obesity. Hepatology. 2001;33:1358-1364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 778] [Cited by in RCA: 774] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 108. | Rubbia-Brandt L, Fabris P, Paganin S, Leandro G, Male PJ, Giostra E, Carlotto A, Bozzola L, Smedile A, Negro F. Steatosis affects chronic hepatitis C progression in a genotype specific way. Gut. 2004;53:406-412. [PubMed] |
| 109. | Rubbia-Brandt L, Quadri R, Abid K, Giostra E, Malé PJ, Mentha G, Spahr L, Zarski JP, Borisch B, Hadengue A. Hepatocyte steatosis is a cytopathic effect of hepatitis C virus genotype 3. J Hepatol. 2000;33:106-115. [PubMed] |
| 110. | Hourioux C, Patient R, Morin A, Blanchard E, Moreau A, Trassard S, Giraudeau B, Roingeard P. The genotype 3-specific hepatitis C virus core protein residue phenylalanine 164 increases steatosis in an in vitro cellular model. Gut. 2007;56:1302-1308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 77] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 111. | Jhaveri R, McHutchison J, Patel K, Qiang G, Diehl AM. Specific polymorphisms in hepatitis C virus genotype 3 core protein associated with intracellular lipid accumulation. J Infect Dis. 2008;197:283-291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 51] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 112. | Tachi Y, Katano Y, Honda T, Hayashi K, Ishigami M, Itoh A, Hirooka Y, Nakano I, Samejima Y, Goto H. Impact of amino acid substitutions in the hepatitis C virus genotype 1b core region on liver steatosis and hepatic oxidative stress in patients with chronic hepatitis C. Liver Int. 2010;30:554-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 113. | Hassan M, Ghozlan H, Abdel-Kader O. Activation of RB/E2F signaling pathway is required for the modulation of hepatitis C virus core protein-induced cell growth in liver and non-liver cells. Cell Signal. 2004;16:1375-1385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 114. | Munakata T, Nakamura M, Liang Y, Li K, Lemon SM. Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 2005;102:18159-18164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 110] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 115. | Lai CK, Jeng KS, Machida K, Cheng YS, Lai MM. Hepatitis C virus NS3/4A protein interacts with ATM, impairs DNA repair and enhances sensitivity to ionizing radiation. Virology. 2008;370:295-309. [PubMed] |
| 116. | Machida K, McNamara G, Cheng KT, Huang J, Wang CH, Comai L, Ou JH, Lai MM. Hepatitis C virus inhibits DNA damage repair through reactive oxygen and nitrogen species and by interfering with the ATM-NBS1/Mre11/Rad50 DNA repair pathway in monocytes and hepatocytes. J Immunol. 2010;185:6985-6998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 83] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 117. | Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110-113. [PubMed] |
| 118. | Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972-1974. [PubMed] |
| 119. | Evans MJ, Rice CM, Goff SP. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc Natl Acad Sci USA. 2004;101:13038-13043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 255] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 120. | Evans MJ, Rice CM, Goff SP. Genetic interactions between hepatitis C virus replicons. J Virol. 2004;78:12085-12089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 121. | Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J Virol. 2003;77:3007-3019. [PubMed] |
| 122. | Hsu M, Zhang J, Flint M, Logvinoff C, Cheng-Mayer C, Rice CM, McKeating JA. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci USA. 2003;100:7271-7276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 655] [Cited by in RCA: 644] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 123. | Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197:633-642. [PubMed] |
| 124. | Bartosch B, Cosset FL. Studying HCV cell entry with HCV pseudoparticles (HCVpp). Methods Mol Biol. 2009;510:279-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 125. | Castet V, Moradpour D. A model for the study of hepatitis C virus entry. Hepatology. 2003;38:771-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 126. | Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294-9299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1475] [Cited by in RCA: 1466] [Article Influence: 73.3] [Reference Citation Analysis (0)] |
| 127. | Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791-796. [PubMed] |
| 128. | Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933-938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 602] [Cited by in RCA: 601] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 129. | Weiner A, Erickson AL, Kansopon J, Crawford K, Muchmore E, Hughes AL, Houghton M, Walker CM. Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc Natl Acad Sci USA. 1995;92:2755-2759. [PubMed] |
| 130. | Lanford RE, Bigger C, Bassett S, Klimpel G. The chimpanzee model of hepatitis C virus infections. ILAR J. 2001;42:117-126. [PubMed] |
| 131. | Forns X, Ampurdanès S, Sanchez-Tapias JM, Guilera M, Sans M, Sánchez-Fueyo A, Quintó L, Joya P, Bruguera M, Rodés J. Long-term follow-up of chronic hepatitis C in patients diagnosed at a tertiary-care center. J Hepatol. 2001;35:265-271. [PubMed] |
| 132. | Bukh J, Forns X, Emerson SU, Purcell RH. Studies of hepatitis C virus in chimpanzees and their importance for vaccine development. Intervirology. 2001;44:132-142. [PubMed] |
| 133. | Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570-574. [PubMed] |
| 134. | Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94:8738-8743. [PubMed] |
| 135. | Bukh J. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology. 2012;142:1279-1287.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 136. | Mercer DF. Animal models for studying hepatitis C and alcohol effects on liver. World J Gastroenterol. 2011;17:2515-2519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 7] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 137. | Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, Jones CT, Schoggins JW, Catanese MT, Burton DR. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208-211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 306] [Cited by in RCA: 287] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 138. | Dorner M, Horwitz JA, Donovan BM, Labitt RN, Budell WC, Friling T, Vogt A, Catanese MT, Satoh T, Kawai T. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature. 2013;501:237-241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 179] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 139. | De Duve C, Pressman BC, Gianetto R, Wattiaux R, Appelmans F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J. 1955;60:604-617. [PubMed] |
| 140. | Gianetto R, De Duve C. Tissue fractionation studies. 4. Comparative study of the binding of acid phosphatase, beta-glucuronidase and cathepsin by rat-liver particles. Biochem J. 1955;59:433-438. [PubMed] |
| 141. | Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198-202. [PubMed] |
| 142. | Clark SL. Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. J Biophys Biochem Cytol. 1957;3:349-362. [PubMed] |
| 143. | De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2158] [Cited by in RCA: 2099] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 144. | Novikoff AB. The proximal tubule cell in experimental hydronephrosis. J Biophys Biochem Cytol. 1959;6:136-138. [PubMed] |
| 145. | Novikoff AB, Essner E. Pathological changes in cytoplasmic organelles. Fed Proc. 1962;21:1130-1142. [PubMed] |
| 146. | Novikoff AB, ESSNER E. Cytolysomes and mitochondrial degeneration. J Cell Biol. 1962;15:140-146. [PubMed] |
| 147. | Gordon PB, Kovacs AL, Seglen PO. Temperature dependence of protein degradation, autophagic sequestration and mitochondrial sugar uptake in rat hepatocytes. Biochim Biophys Acta. 1987;929:128-133. [PubMed] |
| 148. | Gordon PB, Seglen PO. Prelysosomal convergence of autophagic and endocytic pathways. Biochem Biophys Res Commun. 1988;151:40-47. [PubMed] |
| 149. | Plomp PJ, Wolvetang EJ, Groen AK, Meijer AJ, Gordon PB, Seglen PO. Energy dependence of autophagic protein degradation in isolated rat hepatocytes. Eur J Biochem. 1987;164:197-203. [PubMed] |
| 150. | Mortimore GE, Schworer CM. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature. 1977;270:174-176. [PubMed] |
| 151. | Schworer CM, Cox JR, Mortimore GE. Alteration of lysosomal density by sequestered glycogen during deprivation-induced autophagy in rat liver. Biochem Biophys Res Commun. 1979;87:163-170. [PubMed] |
| 152. | Schworer CM, Mortimore GE. Glucagon-induced autophagy and proteolysis in rat liver: mediation by selective deprivation of intracellular amino acids. Proc Natl Acad Sci USA. 1979;76:3169-3173. [PubMed] |
| 153. | Ward WF, Chua BL, Li JB, Morgan HE, Mortimore GE. Inhibition of basal and deprivation-induced proteolysis by leupeptin and pepstatin in perfused rat liver and heart. Biochem Biophys Res Commun. 1979;87:92-98. [PubMed] |
| 154. | Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301-311. [PubMed] |
| 155. | Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124:903-913. [PubMed] |
| 156. | Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687-1695. [PubMed] |
| 157. | Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192:207-213. [PubMed] |
| 158. | Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem. 1998;273:22284-22291. [PubMed] |
| 159. | Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245-250. [PubMed] |
| 160. | Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963-3966. [PubMed] |
| 161. | Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1205] [Cited by in RCA: 1292] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 162. | Guan J, Stromhaug PE, George MD, Habibzadegah-Tari P, Bevan A, Dunn WA, Klionsky DJ. Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol Biol Cell. 2001;12:3821-3838. [PubMed] |
| 163. | Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol Biol Cell. 1999;10:1337-1351. [PubMed] |
| 164. | Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381-396. [PubMed] |
| 165. | Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148:465-480. [PubMed] |
| 166. | Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001;7:1131-1141. [PubMed] |
| 167. | Scott SV, Nice DC, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000;275:25840-25849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 190] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 168. | Wang CW, Kim J, Huang WP, Abeliovich H, Stromhaug PE, Dunn WA, Klionsky DJ. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J Biol Chem. 2001;276:30442-30451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 125] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 169. | Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273:33889-33892. [PubMed] |
| 170. | Klionsky DJ, Cregg JM, Dunn WA, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539-545. [PubMed] |
| 171. | Klionsky DJ, Baehrecke EH, Brumell JH, Chu CT, Codogno P, Cuervo AM, Debnath J, Deretic V, Elazar Z, Eskelinen EL. A comprehensive glossary of autophagy-related molecules and processes (2nd edition). Autophagy. 2011;7:1273-1294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 227] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 172. | Klionsky DJ, Codogno P, Cuervo AM, Deretic V, Elazar Z, Fueyo-Margareto J, Gewirtz DA, Kroemer G, Levine B, Mizushima N. A comprehensive glossary of autophagy-related molecules and processes. Autophagy. 2010;6:438-448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 131] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 173. | Levine B, Ranganathan R. Autophagy: Snapshot of the network. Nature. 2010;466:38-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 174. | Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2082] [Cited by in RCA: 2348] [Article Influence: 167.7] [Reference Citation Analysis (0)] |
| 175. | Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol. 2013;14:133-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 597] [Cited by in RCA: 689] [Article Influence: 57.4] [Reference Citation Analysis (0)] |
| 176. | Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 743] [Cited by in RCA: 835] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 177. | Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511-521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 385] [Cited by in RCA: 369] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 178. | Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685-701. [PubMed] |
| 179. | Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092-2100. [PubMed] |
| 180. | Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433-1437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 769] [Cited by in RCA: 819] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 181. | Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180-1185. [PubMed] |
| 182. | Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747-757. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 723] [Cited by in RCA: 694] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 183. | Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005;1:101-109. [PubMed] |
| 184. | Yen WL, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, Hughson FM, Baba M, Klionsky DJ. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188:101-114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 160] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 185. | Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280-293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2886] [Cited by in RCA: 2757] [Article Influence: 183.8] [Reference Citation Analysis (0)] |
| 186. | Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5765] [Cited by in RCA: 5746] [Article Influence: 338.0] [Reference Citation Analysis (1)] |
| 187. | Orvedahl A, MacPherson S, Sumpter R, Tallóczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115-127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 462] [Cited by in RCA: 444] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 188. | Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567-577. [PubMed] |
| 189. | Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586-8596. [PubMed] |
| 190. | Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398-1401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 661] [Cited by in RCA: 698] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 191. | Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Münz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 644] [Cited by in RCA: 663] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 192. | Münz C. Enhancing immunity through autophagy. Annu Rev Immunol. 2009;27:423-449. [PubMed] |
| 193. | Orvedahl A, Levine B. Eating the enemy within: autophagy in infectious diseases. Cell Death Differ. 2009;16:57-69. [PubMed] |
| 194. | Wileman T. Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu Rev Microbiol. 2007;61:149-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 140] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 195. | Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323-335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2701] [Cited by in RCA: 2577] [Article Influence: 184.1] [Reference Citation Analysis (1)] |
| 196. | Nys K, Agostinis P, Vermeire S. Autophagy: a new target or an old strategy for the treatment of Crohn’s disease? Nat Rev Gastroenterol Hepatol. 2013;10:395-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 197. | Czaja MJ, Ding WX, Donohue TM, Friedman SL, Kim JS, Komatsu M, Lemasters JJ, Lemoine A, Lin JD, Ou JH. Functions of autophagy in normal and diseased liver. Autophagy. 2013;9:1131-1158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 368] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 198. | Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999-2010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 475] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 199. | Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422-432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 575] [Cited by in RCA: 549] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
| 200. | Lee YR, Lei HY, Liu MT, Wang JR, Chen SH, Jiang-Shieh YF, Lin YS, Yeh TM, Liu CC, Liu HS. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008;374:240-248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 287] [Cited by in RCA: 304] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 201. | Li JK, Liang JJ, Liao CL, Lin YL. Autophagy is involved in the early step of Japanese encephalitis virus infection. Microbes Infect. 2012;14:159-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 202. | Panyasrivanit M, Greenwood MP, Murphy D, Isidoro C, Auewarakul P, Smith DR. Induced autophagy reduces virus output in dengue infected monocytic cells. Virology. 2011;418:74-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 203. | Shrivastava S, Raychoudhuri A, Steele R, Ray R, Ray RB. Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology. 2011;53:406-414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 172] [Cited by in RCA: 170] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 204. | Sir D, Kuo CF, Tian Y, Liu HM, Huang EJ, Jung JU, Machida K, Ou JH. Replication of hepatitis C virus RNA on autophagosomal membranes. J Biol Chem. 2012;287:18036-18043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 148] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 205. | Guévin C, Manna D, Bélanger C, Konan KV, Mak P, Labonté P. Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection. Virology. 2010;405:1-7. [PubMed] |
| 206. | Taguwa S, Kambara H, Fujita N, Noda T, Yoshimori T, Koike K, Moriishi K, Matsuura Y. Dysfunction of autophagy participates in vacuole formation and cell death in cells replicating hepatitis C virus. J Virol. 2011;85:13185-13194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 207. | Tanida I, Fukasawa M, Ueno T, Kominami E, Wakita T, Hanada K. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy. 2009;5:937-945. [PubMed] |
| 208. | Vescovo T, Romagnoli A, Perdomo AB, Corazzari M, Ciccosanti F, Alonzi T, Nardacci R, Ippolito G, Tripodi M, Garcia-Monzon C. Autophagy protects cells from HCV-induced defects in lipid metabolism. Gastroenterology. 2012;142:644-653.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 209. | Kim SJ, Syed GH, Siddiqui A. Hepatitis C virus induces the mitochondrial translocation of Parkin and subsequent mitophagy. PLoS Pathog. 2013;9:e1003285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 162] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 210. | Chu VC, Bhattacharya S, Nomoto A, Lin J, Zaidi SK, Oberley TD, Weinman SA, Azhar S, Huang TT. Persistent expression of hepatitis C virus non-structural proteins leads to increased autophagy and mitochondrial injury in human hepatoma cells. PLoS One. 2011;6:e28551. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 211. | Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7:1013-1030. [PubMed] |
| 212. | Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220-9231. [PubMed] |
| 213. | Schröder M. Endoplasmic reticulum stress responses. Cell Mol Life Sci. 2008;65:862-894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 504] [Cited by in RCA: 496] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 214. | Huang H, Kang R, Wang J, Luo G, Yang W, Zhao Z. Hepatitis C virus inhibits AKT-tuberous sclerosis complex (TSC), the mechanistic target of rapamycin (MTOR) pathway, through endoplasmic reticulum stress to induce autophagy. Autophagy. 2013;9:175-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 215. | Shrivastava S, Bhanja Chowdhury J, Steele R, Ray R, Ray RB. Hepatitis C virus upregulates Beclin1 for induction of autophagy and activates mTOR signaling. J Virol. 2012;86:8705-8712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 135] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 216. | Su WC, Chao TC, Huang YL, Weng SC, Jeng KS, Lai MM. Rab5 and class III phosphoinositide 3-kinase Vps34 are involved in hepatitis C virus NS4B-induced autophagy. J Virol. 2011;85:10561-10571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 136] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 217. | Grégoire IP, Richetta C, Meyniel-Schicklin L, Borel S, Pradezynski F, Diaz O, Deloire A, Azocar O, Baguet J, Le Breton M. IRGM is a common target of RNA viruses that subvert the autophagy network. PLoS Pathog. 2011;7:e1002422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 157] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 218. | Ke PY, Chen SS. Active RNA replication of hepatitis C virus downregulates CD81 expression. PLoS One. 2013;8:e54866. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 219. | Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131-1135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3239] [Cited by in RCA: 3105] [Article Influence: 194.1] [Reference Citation Analysis (0)] |
| 220. | Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329-3339. [PubMed] |
| 221. | Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809-1820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1582] [Cited by in RCA: 1835] [Article Influence: 83.4] [Reference Citation Analysis (0)] |
| 222. | Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077-15082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 223. | Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 923] [Cited by in RCA: 1059] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 224. | Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol. 2011;193:275-284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 430] [Cited by in RCA: 500] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 226. | Balaburski GM, Hontz RD, Murphy ME. p53 and ARF: unexpected players in autophagy. Trends Cell Biol. 2010;20:363-369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 84] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 227. | Degtyarev M, De Mazière A, Orr C, Lin J, Lee BB, Tien JY, Prior WW, van Dijk S, Wu H, Gray DC. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008;183:101-116. [PubMed] |
| 228. | Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1546] [Cited by in RCA: 1912] [Article Influence: 127.5] [Reference Citation Analysis (0)] |
| 229. | Amaravadi RK, Thompson CB. The roles of therapy-induced autophagy and necrosis in cancer treatment. Clin Cancer Res. 2007;13:7271-7279. [PubMed] |
| 230. | Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 832] [Cited by in RCA: 917] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 231. | Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51-64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1605] [Cited by in RCA: 1586] [Article Influence: 83.5] [Reference Citation Analysis (0)] |
| 232. | Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961-967. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1483] [Cited by in RCA: 1456] [Article Influence: 80.9] [Reference Citation Analysis (0)] |
| 233. | Yang S, Kimmelman AC. A critical role for autophagy in pancreatic cancer. Autophagy. 2011;7:912-913. [PubMed] |
| 234. | Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1005] [Cited by in RCA: 1189] [Article Influence: 84.9] [Reference Citation Analysis (0)] |
| 235. | Maes H, Rubio N, Garg AD, Agostinis P. Autophagy: shaping the tumor microenvironment and therapeutic response. Trends Mol Med. 2013;19:428-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 223] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 236. | Pastore N, Blomenkamp K, Annunziata F, Piccolo P, Mithbaokar P, Maria Sepe R, Vetrini F, Palmer D, Ng P, Polishchuk E. Gene transfer of master autophagy regulator TFEB results in clearance of toxic protein and correction of hepatic disease in alpha-1-anti-trypsin deficiency. EMBO Mol Med. 2013;5:397-412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 114] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 237. | Grégoire IP, Rabourdin-Combe C, Faure M. Autophagy and RNA virus interactomes reveal IRGM as a common target. Autophagy. 2012;8:1136-1137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |