Published online Apr 14, 2014. doi: 10.3748/wjg.v20.i14.3916
Revised: January 8, 2014
Accepted: February 16, 2014
Published online: April 14, 2014
Processing time: 167 Days and 21.6 Hours
Gastric cancer is a leading cause of cancer death worldwide, and significant effort has been focused on clarifying the pathology of gastric cancer. In particular, the development of genome-wide analysis tools has enabled the detection of genetic and epigenetic alterations in gastric cancer; for example, aberrant DNA methylation in gene promoter regions is thought to play a crucial role in gastric carcinogenesis. The etiological viewpoint is also essential for the study of gastric cancers, and two distinct pathogens, Helicobacter pylori (H. pylori) and Epstein-Barr virus (EBV), are known to participate in gastric carcinogenesis. Chronic inflammation of the gastric epithelium due to H. pylori infection induces aberrant polyclonal methylation that may lead to an increased risk of gastric cancer. In addition, EBV infection is known to cause extensive methylation, and EBV-positive gastric cancers display a high methylation epigenotype, in which aberrant methylation extends to not only Polycomb repressive complex (PRC)-target genes in embryonic stem cells but also non-PRC-target genes. Here, we review aberrant DNA methylation in gastric cancer and the association between methylation and infection with H. pylori and EBV.
Core tip: Recent technological advances in genome-wide analysis tools have revealed various molecular aberrations in cancer. Although gastric cancer involves multiple genetic and epigenetic alterations, aberrant DNA methylation in gene promoter regions is thought to play a critical role in gastric carcinogenesis. From the etiological viewpoint, two pathogens, Helicobacter pylori (H. pylori) and Epstein-Barr virus (EBV), are known to participate in gastric carcinogenesis. Chronic inflammation in the gastric mucosa due to H. pylori and EBV infection of gastric epithelial cells has been reported to cause aberrant promoter methylation, which may contribute to the tumorigenic mechanisms of these pathogens.
-
Citation: Matsusaka K, Funata S, Fukayama M, Kaneda A. DNA methylation in gastric cancer, related to
Helicobacter pylori and Epstein-Barr virus. World J Gastroenterol 2014; 20(14): 3916-3926 - URL: https://www.wjgnet.com/1007-9327/full/v20/i14/3916.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i14.3916
Gastric cancer is a leading cause of cancer death worldwide[1]. Malignant tumors, including gastric cancer, are known to arise through multiple genetic and epigenetic alterations[2], and these molecular changes eventually impact the expression of cancer-associated genes, such as oncogenes and tumor-suppressor genes. Historically, one of the most common genetic alterations in cancer is mutation of the TP53 gene[3,4]. TP53 is a core tumor-suppressor gene, and more than half of all gastric cancers demonstrate loss of TP53 function due to genetic alterations[5]. Another example is CDH1, the gene encoding a calcium-dependent cell-to-cell adhesion glycoprotein that is responsible for familial diffuse type gastric cancers due to germline mutations[6]. However, sporadic gastric cancers also display CDH1 somatic mutations at a constant rate[7]. Moreover, recent whole-genome exome analyses in gastric cancer have identified mutations in several genes, including ARID1A, PIK3CA, and FAT4[8,9].
Although gastric cancer involves various molecular alterations, aberrant promoter methylation plays a major role in gastric carcinogenesis[10-15]. p16INK4A is the most well-known tumor-suppressor gene that is silenced by promoter methylation; the promoter region of p16INK4A is aberrantly methylated in 25%-42% of gastric cancers[10,11,16,17], while mutations or deletions are very rare[16]. RUNX3 is also a significant tumor-suppressor gene in gastric cancer[18], and approximately half of all gastric cancer cases lose RUNX3 expression due to hemizygous deletion and promoter hypermethylation, while point mutations are rarely reported. Although mutations in DNA mismatch-repair genes such as MLH1 and MSH2 are quite rare in gastric cancers[19,20], promoter methylation of MLH1 represents a major cause of microsatellite instability (MSI)[21,22], which is observed in 31%-67% of gastric cancers[19,23].
Several scanning methods have been developed to identify novel tumor-suppressor genes silenced by promoter methylation[24-30], and genome-wide analysis has demonstrated unusual clustering of aberrant methylation in a subset of cancer cases. The phenotype presenting atypical methylation of cytosine-phosphate-guanine (CpG) islands, termed the CpG island methylator phenotype (CIMP), was first described in colorectal cancers[31]. Gastric cancer was also evaluated using methylation markers for colorectal cancer CIMP, and CIMP was also found to be present in gastric cancer[10]. Genome-wide analysis of aberrant DNA methylation in gastric cancer was first performed using the methylation-sensitive-representational difference analysis (MS-RDA) method to identify methylation-associated silenced genes, including novel tumor-suppressor genes[32-34]. Using silenced genes as markers, a subset of gastric cancers was demonstrated to harbor unusual accumulation of aberrant methylation in promoter CpG islands[32].
Environmental factors are also significantly related to the induction of aberrant DNA methylation, and etiological studies have provided evidence that two distinct infectious agents, Helicobacter pylori (H. pylori) and Epstein-Barr virus (EBV), are closely associated with gastric carcinogenesis[35-37]. Here, we review aberrant DNA methylation in gastric cancer and the association between methylation and infection with these two unique pathogens.
“Epigenetics”, as compared to “genetics”, is defined as the study of genomic DNA modifications that are heritable during cell division but do not involve a change in the DNA sequence itself, such as DNA methylation and histone modification[12,13,38]. DNA methylation is the covalent modification of a methyl group on the 5-position of cytosine at CpG dinucleotides[38,39]. CpG islands are genomic regions that contain dense CpG dinucleotides, and they are located in the promoter regions of approximately half of all genes. CpG islands are generally free from DNA methylation, allowing for the expression of downstream genes whose transcription is regulated by histone modification[13].
In normal cellular processes, DNA methylation is used for robust gene silencing, such as genome imprinting[40] and X-chromosome inactivation[41]. Moreover, tissue-specific patterns of methylation or changes in methylation during cellular differentiation have been discovered at CpG-poor promoters[42] and inter- and intragenic CpG islands[43]. In addition, when cells encounter foreign nucleic acid, such as viral DNA, host cells take advantage of DNA methylation as a defensive system to inactivate foreign nucleic acid[44].
Broadly speaking, aberrant DNA methylation in cancer is divided into two categories: “genome-overall hypomethylation” and “regional hypermethylation”. The former, global hypomethylation, was discovered in the 1980s[45] and can be defined as a decrease in 5-methylcytosine content throughout the genome. CpG dinucleotides show heterogeneous distribution, especially in repetitive sequences, which are typically methylated in normal tissue[46,47]. In cancers, these repetitive sequences demonstrate aberrant hypomethylation[48], promoting genomic instability and cancer progression[49-51]. Loss of imprinting is another example of an epigenetic alteration related to aberrant hypomethylation[52], and loss of imprinting in IGF2 was shown to be involved in the early events of carcinogenesis and was associated with increased colorectal cancer risk[53,54]. A subset of male germ line-specific genes, specifically the MAGE gene families, was discovered to be a cancer antigen in malignant melanoma[55]. These genes are repressed by promoter methylation in normal somatic tissues but are activated through promoter hypomethylation in several types of cancers[56,57].
The latter type of DNA methylation, regional hypermethylation, arises in CpG islands[58-60]. Aberrant methylation of promoter CpG islands leads to inappropriate transcriptional silencing, and this phenomenon is regarded as one of the major mechanisms for inactivating tumor-suppressor genes[2,12-14]. Promoter methylation in tumor-suppressor genes has been discovered in various cancers, including RB in sporadic retinoblastoma[61], VHL in renal cell carcinoma[62], CDH1 in hepatocellular carcinoma[63], and p16INK4A in various cancers[64].
In embryonic stem (ES) cells, the Polycomb repressive complex (PRC) plays a significant role in reversibly repressing gene expression. In ES cells, these PRC-target genes are frequently methylated compared to non-PRC-target genes in various cancers[65]. Our comprehensive methylation analysis of gastric cancer revealed significant enrichment of aberrant methylation in PRC-target genes in a subset of gastric cancers with a high-methylation epigenotype[37]. However, another subset of gastric cancer demonstrated an extensively high methylation epigenotype that displayed extended methylation in both PRC-target genes and non-PRC-target genes. This phenotype was detected in EBV-positive gastric cancer, and it will be discussed in detail later in this review.
Among the factors known to cause aberrant DNA methylation in non-cancerous tissues, aging is known to promote the accumulation of DNA methylation[66,67]. Indeed, age-dependent promoter methylation could explain the association between cancer and aging[68]. Recent whole-genome bisulfite sequencing comparing newborn and centenarian genomes demonstrated that centenarian DNA had a lower DNA methylation content throughout the genome and showed the more hypomethylated CpGs in promoters, exonic, intronic, and intergenic regions, whereas a greater level of DNA methylation was observed in CpG island promoters[69]. Another report showed that replicative senescent human cells exhibited features similar to the cancer epigenome, such as widespread DNA hypomethylation and focal hypermethylation[70]. Epidemiological studies have also revealed that the epigenetic status is influenced by various environmental factors[67] and can be associated with cancer incidence or prognosis[71,72].
Among environmental factors, chronic inflammation is a significant inducer of aberrant DNA methylation, as demonstrated by the analysis of non-cancerous tissues, such as colonic mucosae with ulcerative colitis[73], liver tissue with chronic hepatitis[74], esophageal mucosae with inflammatory reflux esophagitis[75], and gastric mucosae with chronic gastritis[76]. In a mouse colitis model induced by dextran sodium sulfate, aberrant CpG island methylation in colonic epithelial cells was shown to accumulate gradually on a monthly basis[77]. Interestingly, even in severe combined immunodeficiency (SCID) mice lacking functional T and B lymphocytes, DNA methylation was induced at the same level as in the background strain of mice, suggesting that functional T and B lymphocytes are not essential for methylation accumulation.
Two distinct pathogens, H. pylori and EBV, are known to be involved in gastric carcinogenesis. First, we will discuss the association between chronic inflammation due to H. pylori and DNA methylation.
H. pylori, discovered in 1983 by Marshall BJ and Warren JR[78], is a helix-shaped Gram-negative bacterium present in the stomach of approximately half of the world’s population[79,80]. Recent prospective cohort studies indicate that H. pylori infection plays an essential role in various disorders, including gastric cancer[35,36], chronic gastritis[78], intestinal metaplasia[81,82], and gastric lymphoma[83]. In 1994, the World Health Organization concluded that “H. pylori is a definite carcinogen” based on epidemiological evidence[79,80].
Two pathways have been proposed to play a role in gastric carcinogenesis resulting from H. pylori infection: the direct interaction of H. pylori with the gastric epithelium and its indirect involvement through chronic inflammation. Ultimately, however, both of these pathways cooperate to promote gastric carcinogenesis.
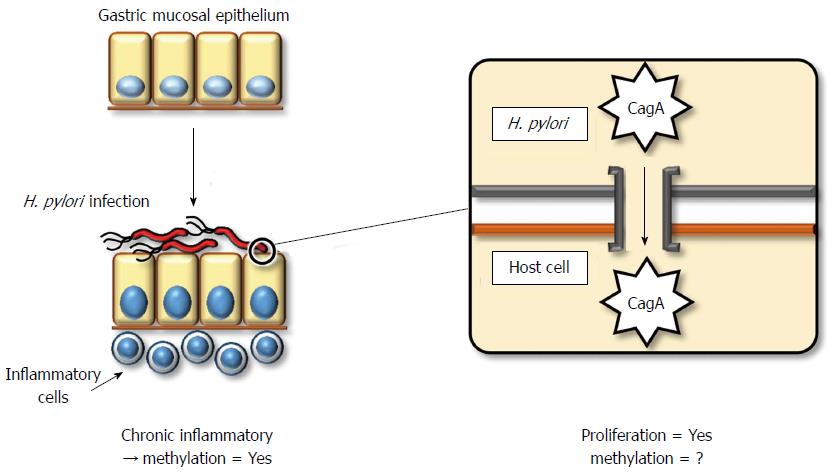
The direct mechanism by which H. pylori contributes to gastric carcinogenesis is attributed to its pathogenicity. Most Gram-negative bacteria exert pathogenicity through the acquisition of an exogenous gene cluster, termed a pathogenicity island (PAI). H. pylori also contains a cag PAI, which consists of an approximate 40-kbp stretch of DNA encoding approximately 30 genes, including those of the type IV secretion system[84]. The pathogenicity of H. pylori depends on whether it contains cytotoxin-associated gene A (CagA) protein or not. Almost all strains of H. pylori in East Asia contain the CagA protein, whereas this frequency in Western strains is limited to 60%. The pathogenicity of the CagA protein is exerted via injection into gastric epithelial cells through type IV secretion systems (Figure 1). The CagA protein contains a conserved motif in the C-terminus, the EPIYA motif, which dictates the severity of its pathogenicity. East Asian strains of CagA exert more aggressive cytotoxicity compared to Western strains[85].
Host cellular responses against injected CagA protein display several patterns. These include (1) enhanced cell motility that induces a growth-factor-like phenotype, termed hummingbird, in host gastric cells[86]; (2) disruption of the epithelial apical-junctional complex[87]; and (3) epithelial proliferative and proinflammatory responses associated with the development of chronic gastritis and gastric cancer[88]. Therefore, CagA plays a key role in gastric carcinogenesis, although the direct involvement of CagA or other components of H. pylori in the induction of DNA methylation has not been clarified.
H. pylori indirectly promotes pathogenicity by inducing chronic inflammation. Chronic inflammation generally involves the accumulation of molecular damage through a variety of mechanisms, such as DNA damage by free radicals[89] or aberrant expression of activation-induced cytidine deaminase (AID)[90]. Moreover, chronic inflammation induces aberrant DNA methylation[74]. Rather than H. pylori infection itself, inflammatory cell infiltration by H. pylori might be a more significant factor for the induction of aberrant DNA methylation[91]. In a Mongolian gerbil model, suppression of aberrant DNA methylation by 5-aza-dC treatment reduced, but did not entirely prevent, the incidence of H. pylori-induced gastric cancers[92]. This result demonstrates that aberrant DNA methylation contributes to H. pylori-related gastric carcinogenesis, although some direct influences of H. pylori, without aberrant DNA methylation, may also be significant.
H. pylori infection induces aberrant promoter methylation in tumor-suppressor genes, including p16INK4A, LOX, and CDH1[34,93]. Although eradication of H. pylori can reduce the level of promoter methylation, a certain amount of methylation remains[91,94]. This observation suggests that not only fully differentiated gastric epithelial cells but also stem/progenitor cells might acquire aberrant methylation. In human ulcerative colitis and hepatitis, increased expression of IL-1β, IL-8, NOS2, and TNF was observed[95-98], and these genes may represent a common factor associated with the induction of aberrant DNA methylation during chronic inflammation. In particular, IL-1β is thought to be significant, as a specific single-nucleotide polymorphism of IL-1β is associated with increased gastric cancer risk and increased incidence of CDH1 promoter methylation in gastric cancers[99,100]. Furthermore, the role of IL-1β in H. pylori-induced gastric inflammation and DNA methylation was confirmed using IL-1 receptor type 1 knockout mice[101].
EBV is another pathogen known to be involved in gastric carcinogenesis.
EBV is a gamma-herpes virus consisting of a double-strand DNA genome approximately 170 kbp in length. EBV may cause infectious mononucleosis during initial infection, and more than 90% of adult individuals become EBV carriers[102], as this virus can be maintained asymptomatically in a latent form in memory B lymphocytes. However, EBV displays the characteristics of an oncogenic virus; indeed, it was initially discovered in human neoplastic cells, specifically a Burkitt’s lymphoma cell line, in 1964[103]. Subsequently, EBV was associated with several types of malignant tumors, such as nasopharyngeal carcinoma[104], Hodgkin lymphoma[105], and opportunistic lymphoma in immunocompromised individuals[106,107]. Moreover, a subgroup of gastric cancer patients infected with EBV was discovered in 1990[108], and this unique subgroup is distributed throughout the world, without regional or racial deviation, at a rate of 7%-15%[109,110].
EBV-positive (EBV+) gastric cancers show distinct clinicopathological features. First, EBV+ gastric cancers demonstrate EBV infection in almost all neoplastic cells of the tumor, which has been confirmed by in situ hybridization for a non-coding small RNA, EBER, which is abundantly expressed in the nuclei of infected neoplastic cells. In addition, the clinical features of EBV+ gastric cancers differ from EBV-negative (EBV-) gastric cancers due to their male predominance, proximal location, and relatively favorable prognosis[111]. Histopathologically EBV+ gastric cancers demonstrate characteristic features of a poorly differentiated adenocarcinoma with marked infiltration of lymphocytes into the stromal tissue, which has been reported as “gastric cancer with lymphoid stroma”[112].
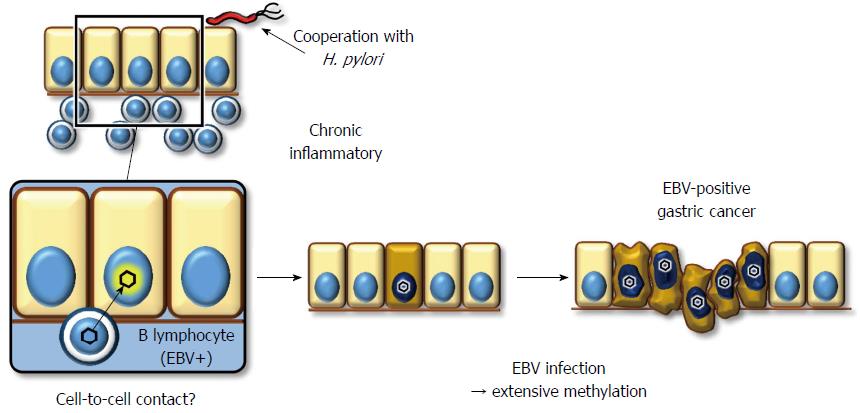
EBV has a double-stranded DNA genome that exists in a linear form in viral particles. After EBV enters the host cell, the viral DNA circularizes via the fusion of terminal repeats at both ends, and it maintains its circular form in the nuclei of latently infected cells without integration into the host genome[113]. Southern blot analysis for terminal repeats has demonstrated that EBV present in neoplastic cells is mono- or oligo-clonal, even in advanced stages[114-116]. Moreover, all of the cancerous cells are positive for EBER-in situ hybridization in all cases of EBV+ gastric cancer. This fact indicates that EBV infection occurs at the initial, or a very early stage, of carcinoma development, and it implies a profound association of EBV with gastric carcinogenesis.
The mechanism underlying EBV infection in the gastric mucosal epithelium remains unclear, while the viral receptor molecule for CD21 in B lymphocytes is not expressed on epithelial cells[117]. Because co-cultivation of virus-producing lymphocytes demonstrates a much greater efficiency of infection (up to 800-fold) compared to cell-free infection, direct cell-to-cell contact between B lymphocytes and gastric epithelial cells is the most likely model to explain how EBV infects epithelial cells in vivo (Figure 2)[118]. This hypothesis supports histopathological data showing that the background mucosa of EBV+ gastric cancer presents atrophic gastritis with lymphocyte infiltration due to H. pylori infection[119]. However, it remains unclear whether chronic inflammation with H. pylori is a prerequisite for EBV to infect gastric epithelial cells.
EBV+ gastric cancer forms a distinct subgroup of gastric cancer. Previous reports have indicated that promoter methylation is observed more frequently in EBV+ gastric cancers than in EBV gastric cancers, despite analyzing a limited number of cancer-associated genes[120-122]. We performed a comprehensive analysis of promoter methylation in clinical gastric cancers and found that gastric cancers clustered into three distinct subgroups. Interestingly, EBV+ gastric cancers displayed an extremely high methylation phenotype, termed the EBV+ epigenotype[37]. Moreover, genes specifically methylated in EBV+ gastric cancers were shown to expand not only within PRC-target genes in ES cells but also to non-PRC-target genes. This result implies that EBV+ gastric cancer is methylated via a unique mechanism(s). Subsequently, to clarify the causal role of EBV infection, we performed in vitro EBV infection experiments in low-methylation MKN7 gastric cancer cells to determine whether these cells would acquire extensive methylation and, as a result, the EBV+-specific methylation epigenotype. The induced methylation repressed multiple genes, including multiple tumor-suppressor genes, suggesting a role for EBV in tumorigenesis[37].
The inducer of aberrant DNA methylation remains elusive. EBV exists in three latent forms defined by the expression pattern of latent genes. Lymphoblastoid cell line (LCL) and transformed primary B lymphocytes infected with EBV express all latent genes, LMPs (1, 2A, 2B), EBNAs (1, 2, 3A, 3B, 3C, LP), EBERs (1, 2), and BARTs, and this expression program is referred to as type III latency. In contrast, Burkitt’s lymphoma shows type I latency, with the minimum expression of EBNA1, EBERs, and BARTs only. Type II latency, in which LMP1 and LMP2 are expressed in addition to latency I genes, is observed in EBV-associated Hodgkin lymphoma, peripheral natural killer/T-cell lymphoma, and nasopharyngeal carcinoma. EBV+ gastric cancer shows type I (or II) latency and expresses EBNA1, EBERs, BARTs, and LMP2A[105,111,123].
Several studies have elucidated the function of latent genes for promoter methylation. LMP1 was reported to down-regulate CDH1 gene expression and induce cell migration using cellular DNA methylation machinery[124,125]. LMP2A plays an essential role in epigenetic abnormalities by inducing promoter methylation of PTEN[126]. EBER1 and EBER2 are small non-coding RNAs of approximately 170 bases in length that are abundantly expressed in the nuclei of latently infected cells, up to 107 copies per cell. Although some oncogenic properties of EBERs have been reported, such as the contribution of efficient growth transformation of B lymphocytes[127,128] or the induction of insulin-like growth factor 1 (IGF-I) acting as an autocrine growth factor in gastric cancer or nasopharyngeal carcinoma cells[129,130], the distinct influence of epigenetic modification remains unclear. Moreover, while these viral genes may play a role in aberrant methylation, methylation induction at the genome-wide scale, which can result from EBV infection, has not been demonstrated through the forced expression of any viral gene[37].
Rather than viral factors, host cellular mechanisms may play more important roles in the induction of aberrant methylation. In type I latency, while host cells induce dense methylation in the viral genome to silence most viral genes, the host genome itself is also extensively hypermethylated[131,132]. In type III latency, such as LCL, neither the viral genome nor the host cellular genome is significantly hypermethylated[132], and this observation implies that a host-driven mechanism that induces DNA methylation in the viral genome may affect methylation of the host genome. Recent exome sequencing analysis demonstrated that ARID1A was frequently mutated in EBV+ gastric cancer[8,133], and other chromatin remodelers were also mutated[9]. While it is not known whether mutation of these genes is causally associated with aberrant methylation in EBV+ gastric cancer, these chromatin remodelers may play a role in protecting the epigenomic status of the host genome from the pressure of methylation induction. Further investigation is necessary to clarify the roles of host cellular factors in methylation induction.
In gastric carcinogenesis, aberrant promoter methylation plays a major role by inactivating tumor-suppressor genes. Two pathogens, H. pylori and EBV, may contribute to carcinogenesis through the induction of aberrant methylation in gastric epithelial cells, although further study is necessary to elucidate the detailed molecular mechanisms underlying the induction of aberrant promoter methylation in response to infection with these two pathogens. Understanding these mechanisms could clarify the process of gastric carcinogenesis, and application of this knowledge for clinical use could aid in diagnosis, risk management, and prevention. Epigenetic aberrations can accumulate at early stages of carcinogenesis, preceding genomic mutations in polyclonal tissues; aberrant DNA methylation is therefore a powerful biomarker for the early detection of cancers and/or cancer risk. Moreover, from prophylactic or therapeutic viewpoints, aberrant DNA methylation could represent an attractive target due to its reversible nature. For example, a patient with persistent DNA methylation after H. pylori eradication might be a candidate for demethylation therapy to prevent gastric cancer. Moreover, in EBV+ gastric cancers, aberrations in chromatin remodeling factors in background fields may promote EBV infection or carcinogenesis and could represent a target for the prevention of EBV+ gastric cancer.
P- Reviewers: Leal MF, Lee HC, Zou CL S- Editor: Qi Y L- Editor: A E- Editor: Ma S
| 1. | Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. [PubMed] |
| 2. | Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5906] [Cited by in RCA: 5601] [Article Influence: 466.8] [Reference Citation Analysis (0)] |
| 3. | Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217-221. [PubMed] |
| 4. | Malkin D, Li FP, Strong LC, Fraumeni JF, Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233-1238. [PubMed] |
| 5. | Tamura G, Kihana T, Nomura K, Terada M, Sugimura T, Hirohashi S. Detection of frequent p53 gene mutations in primary gastric cancer by cell sorting and polymerase chain reaction single-strand conformation polymorphism analysis. Cancer Res. 1991;51:3056-3058. [PubMed] |
| 6. | Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, Taite H, Scoular R, Miller A, Reeve AE. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1135] [Cited by in RCA: 1140] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 7. | Becker KF, Atkinson MJ, Reich U, Becker I, Nekarda H, Siewert JR, Höfler H. E-cadherin gene mutations provide clues to diffuse type gastric carcinomas. Cancer Res. 1994;54:3845-3852. [PubMed] |
| 8. | Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, Chan TL, Kan Z, Chan AS, Tsui WY. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219-1223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 550] [Cited by in RCA: 631] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 9. | Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 458] [Cited by in RCA: 512] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 10. | Toyota M, Ahuja N, Suzuki H, Itoh F, Ohe-Toyota M, Imai K, Baylin SB, Issa JP. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999;59:5438-5442. [PubMed] |
| 11. | Kaneda A, Kaminishi M, Yanagihara K, Sugimura T, Ushijima T. Identification of silencing of nine genes in human gastric cancers. Cancer Res. 2002;62:6645-6650. [PubMed] |
| 12. | Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168-174. [PubMed] |
| 13. | Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3397] [Cited by in RCA: 3722] [Article Influence: 161.8] [Reference Citation Analysis (0)] |
| 14. | Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1674] [Cited by in RCA: 1533] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 15. | Ushijima T, Sasako M. Focus on gastric cancer. Cancer Cell. 2004;5:121-125. [PubMed] |
| 16. | Lee YY, Kang SH, Seo JY, Jung CW, Lee KU, Choe KJ, Kim BK, Kim NK, Koeffler HP, Bang YJ. Alterations of p16INK4A and p15INK4B genes in gastric carcinomas. Cancer. 1997;80:1889-1896. [PubMed] |
| 17. | Shim YH, Kang GH, Ro JY. Correlation of p16 hypermethylation with p16 protein loss in sporadic gastric carcinomas. Lab Invest. 2000;80:689-695. [PubMed] |
| 18. | Li QL, Ito K, Sakakura C, Fukamachi H, Inoue Ki, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113-124. [PubMed] |
| 19. | Akiyama Y, Nakasaki H, Nihei Z, Iwama T, Nomizu T, Utsunomiya J, Yuasa Y. Frequent microsatellite instabilities and analyses of the related genes in familial gastric cancers. Jpn J Cancer Res. 1996;87:595-601. [PubMed] |
| 20. | Keller G, Grimm V, Vogelsang H, Bischoff P, Mueller J, Siewert JR, Höfler H. Analysis for microsatellite instability and mutations of the DNA mismatch repair gene hMLH1 in familial gastric cancer. Int J Cancer. 1996;68:571-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 21. | Leung SY, Yuen ST, Chung LP, Chu KM, Chan AS, Ho JC. hMLH1 promoter methylation and lack of hMLH1 expression in sporadic gastric carcinomas with high-frequency microsatellite instability. Cancer Res. 1999;59:159-164. [PubMed] |
| 22. | Fleisher AS, Esteller M, Wang S, Tamura G, Suzuki H, Yin J, Zou TT, Abraham JM, Kong D, Smolinski KN. Hypermethylation of the hMLH1 gene promoter in human gastric cancers with microsatellite instability. Cancer Res. 1999;59:1090-1095. [PubMed] |
| 23. | Rhyu MG, Park WS, Meltzer SJ. Microsatellite instability occurs frequently in human gastric carcinoma. Oncogene. 1994;9:29-32. [PubMed] |
| 24. | Hayashizaki Y, Hirotsune S, Okazaki Y, Hatada I, Shibata H, Kawai J, Hirose K, Watanabe S, Fushiki S, Wada S. Restriction landmark genomic scanning method and its various applications. Electrophoresis. 1993;14:251-258. [PubMed] |
| 25. | Ushijima T, Morimura K, Hosoya Y, Okonogi H, Tatematsu M, Sugimura T, Nagao M. Establishment of methylation-sensitive-representational difference analysis and isolation of hypo- and hypermethylated genomic fragments in mouse liver tumors. Proc Natl Acad Sci USA. 1997;94:2284-2289. [PubMed] |
| 26. | Gonzalgo ML, Liang G, Spruck CH, Zingg JM, Rideout WM, Jones PA. Identification and characterization of differentially methylated regions of genomic DNA by methylation-sensitive arbitrarily primed PCR. Cancer Res. 1997;57:594-599. [PubMed] |
| 27. | Huang TH, Laux DE, Hamlin BC, Tran P, Tran H, Lubahn DB. Identification of DNA methylation markers for human breast carcinomas using the methylation-sensitive restriction fingerprinting technique. Cancer Res. 1997;57:1030-1034. [PubMed] |
| 28. | Toyota M, Ho C, Ahuja N, Jair KW, Li Q, Ohe-Toyota M, Baylin SB, Issa JP. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307-2312. [PubMed] |
| 29. | Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet. 1999;8:459-470. [PubMed] |
| 30. | Shiraishi M, Chuu YH, Sekiya T. Isolation of DNA fragments associated with methylated CpG islands in human adenocarcinomas of the lung using a methylated DNA binding column and denaturing gradient gel electrophoresis. Proc Natl Acad Sci USA. 1999;96:2913-2918. [PubMed] |
| 31. | Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681-8686. [PubMed] |
| 32. | Kaneda A, Kaminishi M, Nakanishi Y, Sugimura T, Ushijima T. Reduced expression of the insulin-induced protein 1 and p41 Arp2/3 complex genes in human gastric cancers. Int J Cancer. 2002;100:57-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 33. | Kaneda A, Takai D, Kaminishi M, Okochi E, Ushijima T. Methylation-sensitive representational difference analysis and its application to cancer research. Ann N Y Acad Sci. 2003;983:131-141. [PubMed] |
| 34. | Kaneda A, Wakazono K, Tsukamoto T, Watanabe N, Yagi Y, Tatematsu M, Kaminishi M, Sugimura T, Ushijima T. Lysyl oxidase is a tumor suppressor gene inactivated by methylation and loss of heterozygosity in human gastric cancers. Cancer Res. 2004;64:6410-6415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 139] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 35. | Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3126] [Cited by in RCA: 3187] [Article Influence: 132.8] [Reference Citation Analysis (0)] |
| 36. | Ohnishi N, Yuasa H, Tanaka S, Sawa H, Miura M, Matsui A, Higashi H, Musashi M, Iwabuchi K, Suzuki M. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci USA. 2008;105:1003-1008. [PubMed] |
| 37. | Matsusaka K, Kaneda A, Nagae G, Ushiku T, Kikuchi Y, Hino R, Uozaki H, Seto Y, Takada K, Aburatani H. Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res. 2011;71:7187-7197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 191] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 38. | Feinberg AP. Cancer epigenetics takes center stage. Proc Natl Acad Sci USA. 2001;98:392-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 109] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 39. | Laird PW, Jaenisch R. The role of DNA methylation in cancer genetic and epigenetics. Annu Rev Genet. 1996;30:441-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 358] [Cited by in RCA: 346] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 40. | Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1588] [Cited by in RCA: 1577] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 41. | Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature. 1961;190:372-373. [PubMed] |
| 42. | Nagae G, Isagawa T, Shiraki N, Fujita T, Yamamoto S, Tsutsumi S, Nonaka A, Yoshiba S, Matsusaka K, Midorikawa Y. Tissue-specific demethylation in CpG-poor promoters during cellular differentiation. Hum Mol Genet. 2011;20:2710-2721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 60] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 43. | Illingworth R, Kerr A, Desousa D, Jørgensen H, Ellis P, Stalker J, Jackson D, Clee C, Plumb R, Rogers J. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 468] [Cited by in RCA: 464] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 44. | Taniguchi Y, Nosaka K, Yasunaga J, Maeda M, Mueller N, Okayama A, Matsuoka M. Silencing of human T-cell leukemia virus type I gene transcription by epigenetic mechanisms. Retrovirology. 2005;2:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 150] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 45. | Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883-6894. [PubMed] |
| 46. | Dunn BK. Hypomethylation: one side of a larger picture. Ann N Y Acad Sci. 2003;983:28-42. [PubMed] |
| 47. | Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol. 2002;196:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 497] [Cited by in RCA: 495] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 48. | Jürgens B, Schmitz-Dräger BJ, Schulz WA. Hypomethylation of L1 LINE sequences prevailing in human urothelial carcinoma. Cancer Res. 1996;56:5698-5703. [PubMed] |
| 49. | Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 694] [Cited by in RCA: 664] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 50. | Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1012] [Cited by in RCA: 944] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 51. | Yamada Y, Jackson-Grusby L, Linhart H, Meissner A, Eden A, Lin H, Jaenisch R. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci USA. 2005;102:13580-13585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 188] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 52. | Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR, Feinberg AP. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753-1755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 555] [Cited by in RCA: 514] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 53. | Sakatani T, Kaneda A, Iacobuzio-Donahue CA, Carter MG, de Boom Witzel S, Okano H, Ko MS, Ohlsson R, Longo DL, Feinberg AP. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307:1976-1978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 232] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 54. | Kaneda A, Feinberg AP. Loss of imprinting of IGF2: a common epigenetic modifier of intestinal tumor risk. Cancer Res. 2005;65:11236-11240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 103] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 55. | van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643-1647. [PubMed] |
| 56. | De Smet C, Loriot A, Boon T. Promoter-dependent mechanism leading to selective hypomethylation within the 5’ region of gene MAGE-A1 in tumor cells. Mol Cell Biol. 2004;24:4781-4790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 148] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 57. | Loriot A, De Plaen E, Boon T, De Smet C. Transient down-regulation of DNMT1 methyltransferase leads to activation and stable hypomethylation of MAGE-A1 in melanoma cells. J Biol Chem. 2006;281:10118-10126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 64] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 58. | Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2674] [Cited by in RCA: 2761] [Article Influence: 70.8] [Reference Citation Analysis (0)] |
| 59. | Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261-282. [PubMed] |
| 60. | Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99:3740-3745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1070] [Cited by in RCA: 1024] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 61. | Ohtani-Fujita N, Fujita T, Aoike A, Osifchin NE, Robbins PD, Sakai T. CpG methylation inactivates the promoter activity of the human retinoblastoma tumor-suppressor gene. Oncogene. 1993;8:1063-1067. [PubMed] |
| 62. | Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, Samid D, Duan DS, Gnarra JR, Linehan WM. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci USA. 1994;91:9700-9704. [PubMed] |
| 63. | Yoshiura K, Kanai Y, Ochiai A, Shimoyama Y, Sugimura T, Hirohashi S. Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc Natl Acad Sci USA. 1995;92:7416-7419. [PubMed] |
| 64. | Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5’ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686-692. [PubMed] |
| 65. | Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 830] [Cited by in RCA: 794] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 66. | Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 864] [Cited by in RCA: 831] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 67. | Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 799] [Cited by in RCA: 779] [Article Influence: 48.7] [Reference Citation Analysis (0)] |
| 69. | Heyn H, Li N, Ferreira HJ, Moran S, Pisano DG, Gomez A, Diez J, Sanchez-Mut JV, Setien F, Carmona FJ. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci USA. 2012;109:10522-10527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 569] [Cited by in RCA: 581] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 70. | Cruickshanks HA, McBryan T, Nelson DM, Vanderkraats ND, Shah PP, van Tuyn J, Singh Rai T, Brock C, Donahue G, Dunican DS. Senescent cells harbour features of the cancer epigenome. Nat Cell Biol. 2013;15:1495-1506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 264] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 71. | Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 596] [Cited by in RCA: 564] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 72. | Marsit CJ, Houseman EA, Schned AR, Karagas MR, Kelsey KT. Promoter hypermethylation is associated with current smoking, age, gender and survival in bladder cancer. Carcinogenesis. 2007;28:1745-1751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 59] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 73. | Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61:3573-3577. [PubMed] |
| 74. | Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis--A comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2000;32:970-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 196] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 75. | Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, Peters JH, DeMeester TR, Danenberg KD, Danenberg PV. Fields of aberrant CpG island hypermethylation in Barrett’s esophagus and associated adenocarcinoma. Cancer Res. 2000;60:5021-5026. [PubMed] |
| 76. | Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12:989-995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 474] [Cited by in RCA: 480] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 77. | Katsurano M, Niwa T, Yasui Y, Shigematsu Y, Yamashita S, Takeshima H, Lee MS, Kim YJ, Tanaka T, Ushijima T. Early-stage formation of an epigenetic field defect in a mouse colitis model, and non-essential roles of T- and B-cells in DNA methylation induction. Oncogene. 2012;31:342-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 78. | Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311-1315. [PubMed] |
| 79. | Nomura A, Stemmermann GN, Chyou PH, Kato I, Perez-Perez GI, Blaser MJ. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991;325:1132-1136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1302] [Cited by in RCA: 1234] [Article Influence: 36.3] [Reference Citation Analysis (1)] |
| 80. | Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127-1131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2805] [Cited by in RCA: 2740] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 81. | Craanen ME, Dekker W, Blok P, Ferwerda J, Tytgat GN. Intestinal metaplasia and Helicobacter pylori: an endoscopic bioptic study of the gastric antrum. Gut. 1992;33:16-20. [PubMed] |
| 82. | Rugge M, Cassaro M, Leandro G, Baffa R, Avellini C, Bufo P, Stracca V, Battaglia G, Fabiano A, Guerini A. Helicobacter pylori in promotion of gastric carcinogenesis. Dig Dis Sci. 1996;41:950-955. [PubMed] |
| 83. | Weber DM, Dimopoulos MA, Anandu DP, Pugh WC, Steinbach G. Regression of gastric lymphoma of mucosa-associated lymphoid tissue with antibiotic therapy for Helicobacter pylori. Gastroenterology. 1994;107:1835-1838. [PubMed] |
| 84. | Bourzac KM, Guillemin K. Helicobacter pylori-host cell interactions mediated by type IV secretion. Cell Microbiol. 2005;7:911-919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 82] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 85. | Hatakeyama M. Anthropological and clinical implications for the structural diversity of the Helicobacter pylori CagA oncoprotein. Cancer Sci. 2011;102:36-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 86. | Segal ED, Cha J, Lo J, Falkow S, Tompkins LS. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc Natl Acad Sci USA. 1999;96:14559-14564. [PubMed] |
| 87. | Amieva MR, Vogelmann R, Covacci A, Tompkins LS, Nelson WJ, Falkow S. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science. 2003;300:1430-1434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 598] [Cited by in RCA: 584] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 88. | Suzuki M, Mimuro H, Kiga K, Fukumatsu M, Ishijima N, Morikawa H, Nagai S, Koyasu S, Gilman RH, Kersulyte D. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe. 2009;5:23-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 266] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 89. | Tretyakova NY, Burney S, Pamir B, Wishnok JS, Dedon PC, Wogan GN, Tannenbaum SR. Peroxynitrite-induced DNA damage in the supF gene: correlation with the mutational spectrum. Mutat Res. 2000;447:287-303. [PubMed] |
| 90. | Matsumoto Y, Marusawa H, Kinoshita K, Endo Y, Kou T, Morisawa T, Azuma T, Okazaki IM, Honjo T, Chiba T. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat Med. 2007;13:470-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 382] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 91. | Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, Ichinose M, Tatematsu M, Ushijima T. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70:1430-1440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 311] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 92. | Niwa T, Toyoda T, Tsukamoto T, Mori A, Tatematsu M, Ushijima T. Prevention of Helicobacter pylori-induced gastric cancers in gerbils by a DNA demethylating agent. Cancer Prev Res (Phila). 2013;6:263-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 93. | Ushijima T, Nakajima T, Maekita T. DNA methylation as a marker for the past and future. J Gastroenterol. 2006;41:401-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 114] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 94. | Nakajima T, Enomoto S, Yamashita S, Ando T, Nakanishi Y, Nakazawa K, Oda I, Gotoda T, Ushijima T. Persistence of a component of DNA methylation in gastric mucosae after Helicobacter pylori eradication. J Gastroenterol. 2010;45:37-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 102] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 95. | Cappello M, Keshav S, Prince C, Jewell DP, Gordon S. Detection of mRNAs for macrophage products in inflammatory bowel disease by in situ hybridisation. Gut. 1992;33:1214-1219. [PubMed] |
| 96. | Llorent L, Richaud-Patin Y, Alcocer-Castillejos N, Ruiz-Soto R, Mercado MA, Orozco H, Gamboa-Domínguez A, Alcocer-Varela J. Cytokine gene expression in cirrhotic and non-cirrhotic human liver. J Hepatol. 1996;24:555-563. [PubMed] |
| 97. | McLaughlan JM, Seth R, Vautier G, Robins RA, Scott BB, Hawkey CJ, Jenkins D. Interleukin-8 and inducible nitric oxide synthase mRNA levels in inflammatory bowel disease at first presentation. J Pathol. 1997;181:87-92. [PubMed] |
| 98. | Mihm S, Fayyazi A, Ramadori G. Hepatic expression of inducible nitric oxide synthase transcripts in chronic hepatitis C virus infection: relation to hepatic viral load and liver injury. Hepatology. 1997;26:451-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 53] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 99. | El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, Herrera J, Lissowska J, Yuan CC, Rothman N. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1690] [Cited by in RCA: 1676] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 100. | Chan AO, Chu KM, Huang C, Lam KF, Leung SY, Sun YW, Ko S, Xia HH, Cho CH, Hui WM. Association between Helicobacter pylori infection and interleukin 1beta polymorphism predispose to CpG island methylation in gastric cancer. Gut. 2007;56:595-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 29] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 101. | Huang FY, Chan AO, Lo RC, Rashid A, Wong DK, Cho CH, Lai CL, Yuen MF. Characterization of interleukin-1β in Helicobacter pylori-induced gastric inflammation and DNA methylation in interleukin-1 receptor type 1 knockout (IL-1R1(-/-)) mice. Eur J Cancer. 2013;49:2760-2770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 102. | Niedobitek G, Agathanggelou A, Steven N, Young LS. Epstein-Barr virus (EBV) in infectious mononucleosis: detection of the virus in tonsillar B lymphocytes but not in desquamated oropharyngeal epithelial cells. Mol Pathol. 2000;53:37-42. [PubMed] |
| 103. | Epstein MA, Barr YM. Cultivation in vitro of human lymphoblasts from Burkitt’s malignant lymphoma. Lancet. 1964;1:252-253. [PubMed] |
| 104. | zur Hausen H, Schulte-Holthausen H, Klein G, Henle W, Henle G, Clifford P, Santesson L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature. 1970;228:1056-1058. [PubMed] |
| 105. | Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1539] [Cited by in RCA: 1598] [Article Influence: 76.1] [Reference Citation Analysis (0)] |
| 106. | Nalesnik MA, Jaffe R, Starzl TE, Demetris AJ, Porter K, Burnham JA, Makowka L, Ho M, Locker J. The pathology of posttransplant lymphoproliferative disorders occurring in the setting of cyclosporine A-prednisone immunosuppression. Am J Pathol. 1988;133:173-192. [PubMed] |
| 107. | Duval A, Raphael M, Brennetot C, Poirel H, Buhard O, Aubry A, Martin A, Krimi A, Leblond V, Gabarre J. The mutator pathway is a feature of immunodeficiency-related lymphomas. Proc Natl Acad Sci USA. 2004;101:5002-5007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 63] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 108. | Burke AP, Yen TS, Shekitka KM, Sobin LH. Lymphoepithelial carcinoma of the stomach with Epstein-Barr virus demonstrated by polymerase chain reaction. Mod Pathol. 1990;3:377-380. [PubMed] |
| 109. | Shibata D, Weiss LM, Hernandez AM, Nathwani BN, Bernstein L, Levine AM. Epstein-Barr virus-associated non-Hodgkin’s lymphoma in patients infected with the human immunodeficiency virus. Blood. 1993;81:2102-2109. [PubMed] |
| 110. | Tokunaga M, Land CE, Uemura Y, Tokudome T, Tanaka S, Sato E. Epstein-Barr virus in gastric carcinoma. Am J Pathol. 1993;143:1250-1254. [PubMed] |
| 111. | Fukayama M, Ushiku T. Epstein-Barr virus-associated gastric carcinoma. Pathol Res Pract. 2011;207:529-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 105] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 112. | Watanabe H, Enjoji M, Imai T. Gastric carcinoma with lymphoid stroma. Its morphologic characteristics and prognostic correlations. Cancer. 1976;38:232-243. [PubMed] |
| 113. | Kutok JL, Wang F. Spectrum of Epstein-Barr virus-associated diseases. Annu Rev Pathol. 2006;1:375-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 369] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 114. | Imai S, Koizumi S, Sugiura M, Tokunaga M, Uemura Y, Yamamoto N, Tanaka S, Sato E, Osato T. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc Natl Acad Sci USA. 1994;91:9131-9135. [PubMed] |
| 115. | Fukayama M, Hayashi Y, Iwasaki Y, Chong J, Ooba T, Takizawa T, Koike M, Mizutani S, Miyaki M, Hirai K. Epstein-Barr virus-associated gastric carcinoma and Epstein-Barr virus infection of the stomach. Lab Invest. 1994;71:73-81. [PubMed] |
| 116. | Uozaki H, Fukayama M. Epstein-Barr virus and gastric carcinoma--viral carcinogenesis through epigenetic mechanisms. Int J Clin Exp Pathol. 2008;1:198-216. [PubMed] |
| 117. | Yoshiyama H, Imai S, Shimizu N, Takada K. Epstein-Barr virus infection of human gastric carcinoma cells: implication of the existence of a new virus receptor different from CD21. J Virol. 1997;71:5688-5691. [PubMed] |
| 118. | Imai S, Nishikawa J, Takada K. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J Virol. 1998;72:4371-4378. [PubMed] |
| 119. | Kaizaki Y, Sakurai S, Chong JM, Fukayama M. Atrophic gastritis, Epstein-Barr virus infection, and Epstein-Barr virus-associated gastric carcinoma. Gastric Cancer. 1999;2:101-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 120. | Kang GH, Lee S, Kim WH, Lee HW, Kim JC, Rhyu MG, Ro JY. Epstein-barr virus-positive gastric carcinoma demonstrates frequent aberrant methylation of multiple genes and constitutes CpG island methylator phenotype-positive gastric carcinoma. Am J Pathol. 2002;160:787-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 260] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 121. | Chang MS, Uozaki H, Chong JM, Ushiku T, Sakuma K, Ishikawa S, Hino R, Barua RR, Iwasaki Y, Arai K. CpG island methylation status in gastric carcinoma with and without infection of Epstein-Barr virus. Clin Cancer Res. 2006;12:2995-3002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 153] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 122. | Ushiku T, Chong JM, Uozaki H, Hino R, Chang MS, Sudo M, Rani BR, Sakuma K, Nagai H, Fukayama M. p73 gene promoter methylation in Epstein-Barr virus-associated gastric carcinoma. Int J Cancer. 2007;120:60-66. [PubMed] |
| 123. | Fukayama M, Hino R, Uozaki H. Epstein-Barr virus and gastric carcinoma: virus-host interactions leading to carcinoma. Cancer Sci. 2008;99:1726-1733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 126] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 124. | Tsai CN, Tsai CL, Tse KP, Chang HY, Chang YS. The Epstein-Barr virus oncogene product, latent membrane protein 1, induces the downregulation of E-cadherin gene expression via activation of DNA methyltransferases. Proc Natl Acad Sci USA. 2002;99:10084-10089. [PubMed] |
| 125. | Tsai CL, Li HP, Lu YJ, Hsueh C, Liang Y, Chen CL, Tsao SW, Tse KP, Yu JS, Chang YS. Activation of DNA methyltransferase 1 by EBV LMP1 Involves c-Jun NH(2)-terminal kinase signaling. Cancer Res. 2006;66:11668-11676. [PubMed] |
| 126. | Hino R, Uozaki H, Murakami N, Ushiku T, Shinozaki A, Ishikawa S, Morikawa T, Nakaya T, Sakatani T, Takada K. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009;69:2766-2774. [PubMed] |
| 127. | Yajima M, Kanda T, Takada K. Critical role of Epstein-Barr Virus (EBV)-encoded RNA in efficient EBV-induced B-lymphocyte growth transformation. J Virol. 2005;79:4298-4307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 89] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 128. | Wu Y, Maruo S, Yajima M, Kanda T, Takada K. Epstein-Barr virus (EBV)-encoded RNA 2 (EBER2) but not EBER1 plays a critical role in EBV-induced B-cell growth transformation. J Virol. 2007;81:11236-11245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 69] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 129. | Iwakiri D, Eizuru Y, Tokunaga M, Takada K. Autocrine growth of Epstein-Barr virus-positive gastric carcinoma cells mediated by an Epstein-Barr virus-encoded small RNA. Cancer Res. 2003;63:7062-7067. [PubMed] |
| 130. | Iwakiri D, Sheen TS, Chen JY, Huang DP, Takada K. Epstein-Barr virus-encoded small RNA induces insulin-like growth factor 1 and supports growth of nasopharyngeal carcinoma-derived cell lines. Oncogene. 2005;24:1767-1773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 97] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 131. | Fernandez AF, Rosales C, Lopez-Nieva P, Graña O, Ballestar E, Ropero S, Espada J, Melo SA, Lujambio A, Fraga MF. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res. 2009;19:438-451. [PubMed] |
| 132. | Kaneda A, Matsusaka K, Aburatani H, Fukayama M. Epstein-Barr virus infection as an epigenetic driver of tumorigenesis. Cancer Res. 2012;72:3445-3450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 150] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 133. | Abe H, Maeda D, Hino R, Otake Y, Isogai M, Ushiku AS, Matsusaka K, Kunita A, Ushiku T, Uozaki H. ARID1A expression loss in gastric cancer: pathway-dependent roles with and without Epstein-Barr virus infection and microsatellite instability. Virchows Arch. 2012;461:367-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |