Published online Apr 14, 2014. doi: 10.3748/wjg.v20.i14.3804
Revised: January 24, 2014
Accepted: March 8, 2014
Published online: April 14, 2014
Processing time: 170 Days and 18 Hours
Colorectal cancer (CRC) is the second most common cause of cancer-related deaths in Europe and other Western countries, mainly due to the lack of well-validated clinically useful biomarkers with enough sensitivity and specificity to detect this disease at early stages. Although it is well known that the pathogenesis of CRC is a progressive accumulation of mutations in multiple genes, much less is known at the proteome level. Therefore, in the last years many proteomic studies have been conducted to find new candidate protein biomarkers for diagnosis, prognosis and as therapeutic targets for this malignancy, as well as to elucidate the molecular mechanisms of colorectal carcinogenesis. An important advantage of the proteomic approaches is the capacity to look for multiple differentially expressed proteins in a single study. This review provides an overview of the recent reports describing the different proteomic tools used for the discovery of new protein markers for CRC such as two-dimensional electrophoresis methods, quantitative mass spectrometry-based techniques or protein microarrays. Additionally, we will also focus on the diverse biological samples used for CRC biomarker discovery such as tissue, serum and faeces, besides cell lines and murine models, discussing their advantages and disadvantages, and summarize the most frequently identified candidate CRC markers.
Core tip: Proteomics is an important tool for the identification of candidate cancer biomarkers since it allows the simultaneous analysis of multiple differentially expressed proteins in a single study. This review provides an overview of recent reports focused on the different proteomic tools used for the discovery of candidate protein markers for colorectal cancer (CRC), such as two-dimensional electrophoresis methods, quantitative mass spectrometry-based techniques or protein microarrays. We also emphasize the use of different samples including cell lines, murine models, clinical samples as tissue, serum or faeces, for CRC biomarker discovery, discussing their advantages and disadvantages, and finally summarize the candidate CRC markers most frequently identified.
- Citation: Álvarez-Chaver P, Otero-Estévez O, Páez de la Cadena M, Rodríguez-Berrocal FJ, Martínez-Zorzano VS. Proteomics for discovery of candidate colorectal cancer biomarkers. World J Gastroenterol 2014; 20(14): 3804-3824
- URL: https://www.wjgnet.com/1007-9327/full/v20/i14/3804.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i14.3804
Colorectal cancer (CRC) is the second most frequently diagnosed cancer and the second most common cause of cancer-related deaths in Europe and other Western countries[1]. This is mainly due to the lack of well-validated and clinically useful biomarkers with adequate sensitivity and specificity to detect this disease at early stages. Over the last two decades CRC survival rates have barely changed, with more than 50% of the patients having regional or distant metastasis at the time of presentation[2].
However, CRC is potentially curable if detected early before the development of metastasis. After the surgical resection of a tumour that is still localized to the colon or rectum (Duke’s stage A) patients have a 5-year survival rate of more than 90%. Contrarily, patients with Duke’s stage D cancer, where the tumour has spread to other organs, have a 5-year survival rate of less than 10%[2]. Nowadays it is well known that the pathogenesis of CRC is a progressive accumulation of mutations in multiple genes such as APC, KRAS and p53[3]. CRC development is a multi-step process that usually spans about 5-10 years, which offers a period of several years to detect the tumour in an early stage and to interfere with the natural course of the disease[4].
Early detection of CRC can therefore significantly reduce the mortality for this malignancy. However, current screening methods including faecal occult blood test (FOBT), sigmoidoscopy, colonoscopy and virtual computed tomography scanning either lack the required sensitivity and specificity or are costly and invasive[5]. Some biomarkers such as the circulating carcinoembryonic antigen (CEA) levels and tumour-associated gene mutations have only shown some prognostic or predictive value. In particular, the KRAS mutation has been proposed as a marker of probable failure of epidermal growth factor receptor (EGFR)-targeted therapy[6]. In patients with metastasis, for whom no curative options remain, therapies include the combination of traditional chemotherapy with the use of new drugs. There is therefore an urgent need for developing new screening tests and identifying new biomarkers to diagnose, predict, and monitor the progress of CRC, and eventually find more efficient drug targets for this disease.
Following the genomics revolution, recent technological advancements allow the proteomic analyses of complex protein mixtures. Proteins, not only genes, are responsible for the phenotypes of cells, therefore it is impossible to elucidate mechanisms of disease solely by studying the genome. Proteomics is the large-scale study of proteins to comprehensively map biological processes such as the molecular mechanisms of carcinogenesis[7]. The proteome is more complex than the genome due to alternative transcription initiation, alternative splicing, RNA editing, proteolytic processing and post-translational modifications (phosphorylation, glycosylation, etc.), among others. Therefore, the knowledge of the human proteome is an extraordinary challenge. Proteomics can be defined as the discipline that includes the set of methodologies used for the large-scale study of a proteome, i.e. the set of proteins in an organism, a cell or any biological system, in a given time and under certain conditions. It should be noted that proteomics does not focus exclusively on the identification and quantification of these proteins, but also in the study of their location, their modifications, their interactions and their functions.
Proteomic studies generate large protein databases and an expanding list of candidate protein markers that are differentially expressed in CRC patients, identified using two-dimensional electrophoresis (2-DE) and two-dimensional differential in-gel electrophoresis (2D-DIGE) techniques[8]. As an alternative to 2-DE and 2D-DIGE, proteomic studies have also employed the technique of direct expression profiling of tumour and normal tissue by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS) or by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS)[9]. With these approaches, the reproducible spectra profiles of tumour and normal tissue are used to generate classification models. More recently, the technique of choice is the LC-MS/MS (liquid chromatography-tandem mass spectrometry), as it is more rapid and sensitive. With this methodology, commonly referred to as shotgun analysis, proteins from a complex mixture are collectively in-solution digested and the resulting complex mixture of peptides is separated by high-performance liquid chromatography (HPLC). Then, chromatographic fractions are introduced directly into a sensitive tandem mass spectrometer capable of isolating and fragmenting individual peptides. Protein identification is performed at the level of peptide fragmentation patterns acquired during tandem MS, which are indicative of the amino acid sequence.
Briefly, in this review we provide an overview of recent reports describing the different proteomic techniques used and the diverse biological samples employed for the discovery of new candidate protein markers for CRC.
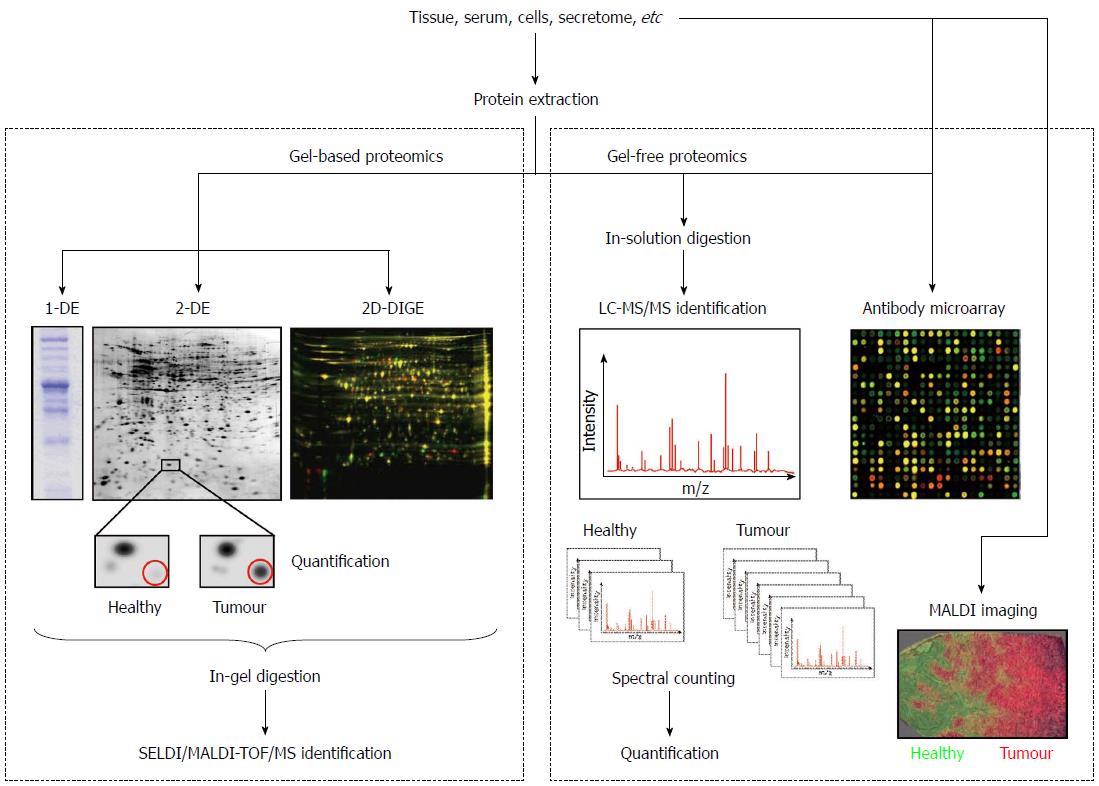
The analysis of the proteome changes between normal and diseased samples is known as comparative proteomics and is fundamental for the discovery of candidate cancer biomarkers. This area of proteomics employs the techniques described below and the workflows outlined in Figure 1.
For many proteomic applications, 1-D electrophoresis (1-DE) is the method of choice to resolve protein mixtures. Proteins are solubilised in sodium dodecyl sulphate (SDS) and then separated on the basis of their molecular weight (MW). This technique is simple to perform, is reproducible, and can be used to resolve proteins with molecular masses ranging between 10 and 300 kDa[10]. Due to its limited resolving power, the most common application of 1-DE is the characterization of proteins after a purification procedure[11]. Nowadays, it is also often employed to carry out the digestion of samples to be analysed by LC-MS/MS because it is more effective than in-solution digestion. For example, Lim et al[12] employed SDS polyacrylamide gel electrophoresis (PAGE) to overcome the limitation of two-dimensional (2-D) electrophoresis for resolving extreme acidic, basic, or membrane proteins. In their study, the protein bands were subjected to in-gel digestion and protein analysis was performed using electrospray ionization (ESI) ion trap mass spectrometer. Among the differentially expressed proteins they identified low abundant proteins and proteins with extreme pH, which were previously not detected in 2-D gels. In another study, a lectin affinity-based approach followed by the same proteomic strategy (SDS-PAGE coupled to LC-MS/MS) was employed to detect differentially expressed secreted proteins in the secretome (conditioned media) of cultured paired normal and CRC tissues. EGF-containing fibulin-like extracellular matrix protein 2 (EFEMP2) was found up-regulated and was further validated by immunohistochemistry (IHC) at tissue level and enzyme-linked immunosorbent assay (ELISA) at serum level. The expression level of EFEMP2 was dramatically increased in CRC patients, even at early stages. Moreover, the diagnostic accuracy of EFEMP2 was superior to that of CEA, with an area under the receiver operating characteristic curve of 0.923 and 0.728, respectively. These authors concluded that EFEMP2 is a promising serum biomarker for the early detection of CRC[13].
Although 2-DE is commonly described as a laborious technique with low throughput that requires a relatively large amount of sample, its capability to separate thousands of proteins in a single analysis made it a major player in the profiling of protein expression in cancer and the core technology for protein separation prior MS characterization. In fact, this procedure offers a good resolution because it combines two types of gel electrophoresis techniques: isoelectric point (pI)-based protein separation by isoelectric focusing (IEF) and MW separation by SDS-PAGE.
Traditionally, a comparative proteomic study involves the extraction of the protein content from the samples (tissue, serum, cells, etc.) in an appropriate lysis buffer, the separation of samples on 2-D gels, the staining of gels with a protein stain such as Coomassie brilliant blue, silver nitrate or SYPRO, the acquisition of images and the matching of protein spots using a statistical package[14] such as Melanie, PDQuest or Progenesis Samespots. These analyses generate two master 2D maps, one for the healthy samples and another one for the pathological samples. Subsequent analyses with the same software compare the two maps to detect proteins which are present in greater or lesser quantities in one of the samples. Then, differentially expressed proteins are excised and subjected to in-gel digestion with trypsin for MS identification in protein databases.
Using 2-DE, often referred to as gel-based proteomics, protein isoforms and variants expressed by a biological system (tissue, cells, etc) may be displayed, allowing the visualization of different phenotypes[15,16]. However, it is clear that classical 2-DE has several limitations. For example, proteins that are low-abundant, or have a MW lower than 10 kDa or higher than 150 kDa, as well as those with very basic pI values, are seldom detected using conventional gels. Moreover, insolubility precludes the penetration inside the gels of hydrophobic or membrane-associated proteins that are of special interest in the biomarker discovery field. However, since its development in the 1970s as the first approach to separate complex protein mixtures, the role of 2-DE in the proteomics workflow has been well preserved due to the emergence of new methodologies and protocols that overcome its limitations. In 1975 the first buffer for protein solubilisation was introduced, but the proposed procedure was not very suitable for membrane-associated, alkaline or hydrophobic proteins separation[14]. To perform a complete protein solubilisation several additives must be included in the buffers, such as chaotropes (urea, thiourea) to prevent protein aggregation, detergents (Triton x 100 or CHAPS) to increase the solubility of certain proteins, and reducing agents such as dithiothreitol (DTT) to reduce disulphide bonds. After disulphide links reduction, the newly produced free sulphidryl groups must be protected by alkylation, being iodoacetamide (IAA) the alkylating reagent most compatible with 2-DE. Moreover, sample solubilisation can be improved by procedures such as agitation and ultra-sonication[17].
Using gel-based proteomics, many studies have been carried out in order to find new CRC biomarkers. Among the most recent we can highlight the work of Chen et al[18] who found an overexpression of alpha-enolase, the heat shock protein HSP27 and macrophage migration inhibitory factor (MIF) in tumour tissue of CRC patients with low preoperative serum CEA. They corroborated that serum alpha-enolase and MIF were significantly overexpressed in those patients, improving the diagnosis of primary CRC when combined with the determination of preoperative CEA levels. Other examples of 2D gel-based discovery studies that have yielded novel candidate CRC serum markers include S100A8 and S100A9[19], and desmin[20].
Besides comparisons between normal and tumour-derived tissues, several 2D gel studies have analysed metastatic and non-metastatic CRC tissues and have validated candidates for CRC markers using IHC and functional analyses in cell lines and mouse models. Zhao et al[21] performed a comparative proteomic analysis to show that Rho GDP-dissociation inhibitor (RhoGDI) is markedly up-regulated in metastatic CRC, validating the result by Western blot in tissue and cells and by IHC in 126 pathologically characterized CRC cases. RhoGDI was shown to correlate with tumour invasion, lymph node metastasis and clinical stage. Authors also demonstrated that the transfection of the RhoGDI gene in HT29 cells with low levels of this inhibitor resulted in an increase in cell proliferation and motility in vitro. In another similar study, the same authors showed that gene transfection-mediated overexpression of LIM and SH3 domain protein 1 (LASP-1) in SW480 human colon adenocarcinoma cells resulted in an aggressive phenotype of cancer cells and promoted cancer growth and metastasis[22]. More recently, using 2D serum proteome analysis combined with MS, transthyretin (TTR) was also identified by these authors as a specific marker of CRC metastasis[11]. Other CRC tumour markers, which have been widely studied by our group through the use of 2D technology, are the proteins clusterin[23] and nucleoside diphosphate kinase A (NDK A)[24].
As we have mentioned above, many proteomic studies from the past 10 years have focused on the comparison of colorectal tumour and adjacent normal mucosa tissues. These analyses predominantly employed 2D gel separation of total tissue lysates which limit the loading amount of sample, restricting the analysis to the more abundant proteins that mask minor proteins that could be interesting as possible CRC marker candidates. In an attempt to identify less abundant CRC proteins, few studies have combined more targeted approaches with 2-DE, including studies focusing on membrane proteins[25,26] or basic proteins[27,28]. Despite these targeted attempts, the analyses still are largely limited to abundant proteins that are found overexpressed in several tumours, such as structural proteins, glycolytic enzymes or heat shock proteins. In serum 2D analysis, the previous depletion of albumin and IgG, which account for more than 60% of the total serum proteins, may result in the loss of potentially important proteins bounded to them. Therefore, sample preparation improvements like sonication before the depletion and desalting steps allowed the detection of valuable, low abundant proteins in serum of CRC patients[11]. Other improvements of 2-DE were the introduction of new gels which can separate proteins with extreme pI values and/or make use of narrow range pH gradients for increased resolution, as well as the use of improved pre-fractionation strategies. Consequently, 2-DE remains as a pivotal methodology for the display of an extensive image of a complex mixture of proteins.
An exciting advance in 2-DE, which improved the speed and reproducibility of conventional 2-DE, was introduced by Unlü et al[29]. This technology is called differential in-gel electrophoresis (DIGE). Basically, different protein samples (healthy vs pathological, for example) are labelled on lysine side chains with succinimidyl esters of propyl-Cy3 and methyl-Cy5, two fluorophores that emit light at different wavelengths (Figure 1). An internal standard is prepared by pooling equal amounts of samples, labelled with a third dye (Cy2). The protein samples are mixed prior to separation and loaded onto the 2D gel together. After electrophoresis, the 2-D pattern is visualized by imaging the gel with a fluorescence scanner by sequential excitation of the fluorescent dyes. Three images are obtained, which are combined to identify pattern differences. Because the samples run together, differences in gel preparation, running conditions and local gel structure are eliminated, making this technique of great utility for biomarker studies. However, 2D-DIGE also has its drawbacks: fluorescent labels are less sensitive than both SYPRO dyes and silver staining, proteins differ in their labelling efficiency, and the technique is relatively expensive compared to silver or Coomassie staining of gels.
Using 2D-DIGE, proteome analysis of membrane fractions in colorectal carcinomas revealed several proteins with an altered expression[26]. Among them, annexins (A2, A4, A5 and A7), lamin B, calponin 1 and voltage-dependent anion channel (VDAC) were analysed by IHC using tissue microarrays. Authors proposed annexin A2, annexin A4 and VDAC as candidate markers for colorectal cancer diagnosis and, presumably, also for therapy. More recently, Ma et al[30] used 2D-DIGE coupled with MS to screen for biomarker candidates in the serum proteome of CRC patients and healthy donors. They identified and validated transaldolase 1 and thyroid receptor interactor as CRC-associated serum biomarkers. Sawhney et al[31] demonstrated the compatibility between the subcellular fractionation by laser microdissection (LMD) of human colon tissue and 2D-DIGE. They observed a greater coverage of proteins from very small amounts of micro-dissected material. Sugihara et al[32] compared the proteome of normal colorectal epithelial tissues with that of the tumour in 59 CRC patients using 2D-DIGE. They found a higher expression of 110 protein spots and focused on the adenoma polyposis coli-binding protein (EB1). This protein was originally discovered as a binding protein of the tumour suppressor gene product APC, and had been associated with poor prognosis in several malignancies but not in CRC. Immunohistochemical analysis of 132 CRC cases revealed that EB1 was overexpressed in tumour cells and was correlated with poor prognosis. Therefore, they proposed EB1 as a candidate biomarker and therapeutic target for CRC. In another study, the proteomic analysis of six paired normal and CRC tissues by 2D-DIGE and MALDI-TOF-MS showed two markedly down-regulated proteins, which were identified as cytoplasmic carbonic anhydrase I and II and whose changes were further validated by IHC and Western blot. The down-regulation of these enzymes is an early event in colorectal carcinogenesis, but is not correlated with lymph node metastasis[33]. More recently, Zhou et al[34] showed that overexpression of carbonic anhydrase II remarkably suppressed tumour cell growth both in vitro and in vivo. Using the Caco-2 cell line, an in vitro model to study colorectal carcinogenesis, our research group identified the translationally-controlled tumour protein (TCTP) and the transforming growth factor-β-induced protein ig-h3 (TGFβ1p), among others, as candidate biomarkers for CRC[35]. Grandjean et al[36] used the new methodology of sequential immunoaffinity depletion-differential in gel electrophoresis (SID-DIGE), that allowed the efficient screening of sera for the identification of autoantibodies as candidate biomarkers. The identification of autoantibodies is based on the characterization of tumour-associated antigens against which they are directed. Among the 25 tumour-associated antigens identified, 7 were also detected using the conventional SERPA (serological proteome analysis) technique, validating their new approach. The identification of the additional 18 autoantibodies proved the potential of this new method.
Using protein microarrays, the simultaneous analysis of different proteins is performed in one single experiment, allowing the study of the protein identity, quantity, interaction and function. There are two types of protein arrays: forward-phase protein arrays and reverse-phase protein arrays.
For forward-phase protein microarrays (also known as capture arrays), the elements of the array are capture molecules (antibodies, proteins, nucleotides or aptamers), each binding specifically to a particular protein. Antibody arrays are the most common, and use antibodies immobilized on a solid surface or membrane to specifically interact with the proteins of interest. For the detection, samples can be labelled with different fluorophores like in 2D-DIGE (Figure 1), allowing two possible samples to undergo the same treatment for comparison[37]. This technology has started to be implemented extensively in cancer research. For example, Ellmark et al[38] prepared a cell suspension from a colorectal tumour containing a mixed population of cells which was captured on an antibody microarray. Cancer cells were detected using a fluorescently tagged antibody for carcinoembryonic antigen (CEA-Alexa647) or epithelial cell adhesion molecule (EpCAM-Alexa488). Using this multiplexing procedure, authors found a differential expression of CD45, CD71 and CEA in cancer cells, among others proteins.
In a reverse-phase protein microarray, the samples are immobilized on the surface or membrane and antibodies are then be applied to the array to detect specific epitopes, protein sequences or structures[37]. For example, Oliveira et al[39] used tissue microarrays and found that NDK A protein expression was higher in tumour tissue of CRC patients than in adjacent non-neoplastic mucosa. In another study, serum CRC biomarker candidates Apolipoprotein AI (Apo A1) and C9 complement component (C9) were selected by liquid chromatography (LC) and MS, and then validated using a reverse-phase protein microarray[40].
Currently, antibody microarrays are attracting considerable attention in cancer biomarker discovery. Several aspects of microarray technology make it well suited to cancer research because of the low-volume requirements and its multiplexed detection capability that make optimal use of precious clinical samples. These assays are rapid and highly amenable to automation, which makes them ideal for biomarker studies[41]. For a review of array-based detection of serum autoantibodies in CRC the reader is referred to Tan et al[42]. In addition, the equipment of SPR (Surface Plasmon Resonance) is now available for the analysis of protein microarrays, allowing the study of interactions and the identification by MS of ligands of interest. A protein array variant vastly used in the search for new biomarkers for CCR is the technique named Surface-Enhanced Laser Desorption/Ionization Time-Of-Flight Mass Spectrometry (SELDI-TOF-MS). It is a widely used technology platform for biomarker discovery in tissue, plasma and serum, though it is most commonly associated with the development of serum-based markers. SELDI-TOF-MS combines two powerful technologies, chromatography and MS, and consists of solid supports or chips made of aluminium or stainless steel coated with specific chromatographic surfaces, including reverse phase, anionic exchange, cationic exchange and immobilised metal affinity surface. In the case of serum samples, the prefractionation technique called ProteoMiner™ (Bio-Rad Laboratories, Hercules, CA, United States) is widely used prior to the analysis by SELDI-TOF-MS due to the presence of a wide range of protein concentrations.
For the protein identification by MS two strategies can be pursued. On one hand the so-called peptide mass fingerprinting (PMF) and on the other hand, the sequencing or tandem mass spectrometry (MS/MS). In both cases it is necessary to digest the protein, and its fragments (peptides) should pass to a gas state. At the end of the 1980s, two revolutionary methods for the ionization of peptides and proteins were developed: the electrospray ionization (ESI) and the matrix-assisted laser desorption/ionization (MALDI), for which the authors received the Nobel Prize in Chemistry in 2002[43].
Less than five years ago, the strategy used in most of the proteomic studies aimed at the search of new tumour markers was firstly the separation of the proteins in the sample using 2-DE or 2D-DIGE to compare the proteome between healthy and tumour tissues and, secondly, the removal of the spots of interest from the gel for the subsequent identification by SELDI/MALDI-TOF-MS[44]. However, as an alternative to 2-DE and 2D-DIGE, most of the proteomic studies in the last three years have employed another technique of MS to carry out studies of differential expression between samples from healthy individuals and patients: the liquid-chromatography coupled to tandem mass spectrometry (LC-MS/MS). This methodology allows performing a previous separation of the peptides typically in a reverse phase chromatographic column. As the LC equipment is usually connected on-line to the mass spectrometer, the fractions obtained after the chromatography enter successively in the mass analyser, allowing the one by one slicing of the peptides present in the sample. This analysis can be performed with digested bands from a 1-D gel, with digested spots from a 2-D gel or with a complex mixture of proteins not previously separated so that the peptides from different proteins are mixed after digestion. This latter type of analysis is known as shot-gun proteomics or multidimensional protein identification technology (MudPIT). However, prior to the MS analysis, it is necessary to perform two independent chromatographic separations in different conditions to resolve the complexity of the sample. In addition, mass spectrometers used for this type of studies have great resolution and sensitivity, obtaining a much higher number of proteins identified compared to that obtained with classical proteomic studies based on 2-DE or 2D-DIGE[45]. Several authors have described the advantages of this technology in comparison with gel-based proteomics. For example, the proteasome activator complex subunit 3 (PSME3), an intracellular CRC-associated protein, was identified in a 2D gel-based study comparing normal and cancer tissues. This candidate was then selected for follow-up analysis by immunoblotting[46]. One year later, in a second study of the same authors, PSME3 was validated as a novel CRC serum marker using MS[47]. This new study detailed the technical advantages of the mass spectrometry-based approach for relative quantification of protein abundance, as compared to the traditional image analysis approach. Importantly, PSME3 up-regulation would have been missed by image analysis because this protein was masked by another co-migrating high-abundant protein, annexin A4. As another interesting example, Thierolf et al[48] analysed the same paired normal and CRC tissues by 2-DE and 2D-LC-MS/MS, showing the complementarity of these two different strategies since a set of proteins were uniquely identified by each one. Among the identified proteins S100A12 was validated in serum, and authors concluded that this protein was more sensitive for the detection of CRC patients than CEA.
Another MS approach, the MALDI-TOF-MS, is most commonly used to discriminate tumour from normal tissue and in some cases can sub-classify disease. For example, Liao et al[49] evaluated the potential value of this approach to classify various clinic-pathological features in CRC. They found 73 protein peaks with a higher expression in tumours than in adjacent normal mucosa tissues in the mass range of 1800-16000 Da. Using “leave-one-out” cross validation algorithms for tumour spectra they correctly classified poorly, well and moderately differentiated tumours. Similar analyses of normal mucosa spectra correctly predicted disease recurrence, disease-free survival and metastasis. More recently, Fan et al[50] employed a well-defined technology platform called ClinProt (Bruker Daltonics, Germany), based on magnetic bead purification of peptides and MALDI-TOF-MS. They successfully detected 61 short peptides, from 1 to 18 kDa, which were differentially expressed in the serum of patients with CRC, concluding that this peptidome pattern may provide an alternative for CRC diagnosis or may help in tailoring the use of chemotherapy to each patient. MALDI is also a method of ionization used for analysing the molecular content of tissue sections, including formalin-fixed and paraffin-embedded tissue samples, which are the standard embedding techniques used in clinical routines. This technique is named imaging mass spectrometry (MALDI-IMS), commonly known as MALDI imaging. It has emerged in the last few years as a useful tool for the molecular classification of tissue samples regarding disease stage, risk stratification and therapy response, as well as for the identification of disease biomarkers[51]. Recently, this approach was employed to evaluate fresh frozen sections of CRC tissue and adjacent healthy mucosa, offering novel insights into tumour micro-environmental biochemistry[52].
As mentioned above, SELDI-TOF-MS is a widely used technology platform for biomarker discovery. It can be described as a type of MALDI-TOF-MS where the sample matrix, known as protein chip, has an active role in the sample purification as well as in the desorption/ionization step[53].
Several peptide-profiling studies using serum of CRC patients have reported the combination of SELDI-TOF-MS with bioinformatics in order to perform pattern diagnostics. For an overview of these studies, readers can refer to the work of Gemoll et al[54]. As an example, Liu et al[55] determined a set of two protein peaks that had the ability to distinguish patients with different stages of CRC from healthy subjects, with a sensitivity of 95.0% and a specificity of 94.9%. More recently, Helgason et al[56] identified 13 candidate biomarkers for CRC (m/z 2.0-31.9 kDa) and 2 peaks (m/z 2022 and 28100 Da) that changed during chemotherapy in accordance with patient response to the treatment. Nevertheless, when human serum or plasma is studied with this technique, its sensitivity is restricted due to the wide dynamic range of serum protein concentrations. In this context, sample pre-fractionation targeting the low abundant proteins may help to overcome these limitations. Therefore, the combination of ProteoMiner™ (Bio-Rad Laboratories, Hercules, CA, United States) pre-fractionation and SELDI based protein profiling is suitable for large-scale serum proteome profiling studies yielding reliable and reproducible results[57].
In tissue studies, only Melle et al[58] have reported the detection of differences between normal mucosa and colorectal carcinoma by SELDI. In 2005 they found that pituitary adenylate cyclase activating polypeptide precursor (PACAP), heterogeneous nuclear ribonucleoproteína A1 (HNRNP A1), flavin reductase, calgizzarin (S100A11), nucleoside diphosphate kinase B (NDK B), cyclophilin A and smooth muscle protein 22-alpha showed significantly differential abundance between colorectal tumour tissue and adjacent normal mucosa. In another study published a year later, these authors validated the differential expression of the calgizzarin (S100A11) by immunological techniques[59]. Regarding other clinical material, Ward et al[60] used urine of CRC patients as sample to detect proteomic changes in MALDI and SELDI spectra. They found a number of changes in peak intensity significantly associated with colon cancer and these, in conjunction with class prediction models, yielded a diagnostic sensitivity of 78% and a specificity of 87% (values higher than those obtained with serum CEA).
Although the quantification in gel-based approaches as 2-DE and 2D-DIGE is very accurate and sensitive, the relative high amount of protein sample necessary for protein identification, as well as the multiple experimental steps required, are the major disadvantages of these techniques. Due to these drawbacks and as a consequence of the technical improvements in the fields of chromatography and mass spectrometry, novel MS-based quantification strategies have been developed, allowing high throughput proteome analyses complementary to gel-based approaches, leading to a higher proteome coverage.
Current MS-based strategies for protein quantification can be divided into two main groups: strategies based on labelling a specific amino acid residue and the so-called label-free proteomics. In all labelling strategies the first and maybe the most critical step is to modify the molecular mass of a specific amino acid so it can be distinguished from its unlabelled counterpart in the detection phase. This can be done in various ways: in one approach stable isotope labelling is used without changing the chemical identity of the amino acid, as in the case of introducing stable isotopes of 2H, 13C, 15N and 18O within various functional groups. In a second approach, chemical modification with or without stable isotope labelling is used. Alkylating Cys is an example of the first case, while guanidination which transfers C-terminal Lys to homoarginine is an example of the latter[61]. Basically, proteins or peptides in one of the samples are modified with an isotope tag. The two samples are then mixed before being processed and analysed by MS. Although the physical characteristics of the peptides remain the same, the masses of the isotopically labelled peptides are proportionally shifted in the spectrum, and the ratio between the intensities of the differentially labelled peptides peaks then permits accurate relative quantitation of the proteins. Stable isotopes can be incorporated into proteins or peptides using different techniques: O18 proteolytic labelling, isotope-coded affinity tags (ICAT), isotope-coded protein labelling (ICPL), iTRAQ (isobaric Tags for Relative and Absolute Quantification), or SILAC (Stable Isotope Labelling with Amino acids in cell Culture), where stable isotopes are incorporated into growing cells. The advantage of labelling strategies is that in the same experiment several samples can be analysed and very small changes of expression can be detected. For example, Kim et al[62] combined 2-DE with ICAT and found a set of five proteins (VCP, TPM2, ITLN1, TAGLN and FABP1) differentially expressed in the tumour tissue of CRC patients, with value for the prognosis of the disease. Using the iTRAQ-based quantitative proteomics approach Ghosh et al[63] validated the role of calcyclin binding protein (CacyBP) in promoting colorectal cancer metastasis.
The goal in the search for new CRC biomarkers is not the identification of proteins with small changes of expression; instead, large variations in their expression are desirable. Therefore, proteomic techniques based on LC-MS/MS without prior marking of the samples (label-free proteomics) are frequently used, despite these are time-consuming since the samples are analysed one by one. Using this approach with the insoluble fractions from tissues from CRC patients, Yang et al[64] found four proteins (KRT5, JUP, TUBB, and COL6A1) that gave specific network information for CRC. Their panel of novel markers proposed as candidate targets for treatment was further validated by Western-blotting. By label-free quantitative mass spectrometry and protein microarray, Matsubara et al[65] reported that adipophilin is a plasma biomarker potentially useful for the detection of early-stage CRC, showing improved diagnostic performance.
A novel label-free quantitative proteomics technique is the so-called SRM (Selected Reaction Monitoring). It is an MS/MS method, which consists in selecting a specific precursor ion or peptide that will later be divided, allowing the selection of one of its product ions. This detection is used to make a relative or absolute quantification of the peptide and, by extension, of the protein to which it belongs in the analysed sample. Although this approach can be used without making a prior labelling, a recent study combined this technique with ICAT to compare healthy mucosa and tumour tissues of CRC patients, identifying more than 1000 proteins overexpressed in tumours, related to endocytosis, mitochondrial dysfunction and various cell signalling pathways[66]. Other MS-based strategy for protein quantification is label-free spectral counting. Considering that the detected spectral counts are correlated with the abundance of corresponding proteins, Yao et al[13] compared and identified proteins in the secretome of paired normal and CRC cultured tissues before and after a lectin capture method. After lectin capture the percentage of spectral counts of secreted proteins was significantly increased from 45% to 80% for the conditioned medium of normal tissues, and from 50% to 85% for the CRC ones, indicating that secreted proteins were effectively enriched by the lectin affinity based approach.
The first critical issue in the proteomic analysis for CRC biomarker discovery is the selection of the sample. Different samples can be used for searching candidate protein markers, including clinical samples such as serum, tissue or faeces from patients, as well as other biological samples like cell lines or animal models. Below, we will briefly discuss the advantages and disadvantages of these different types of samples, and summarize the putative CRC markers most frequently identified.
Blood is the most suitable sample used to identify biomarkers for the screening or diagnosis of CRC due to its availability and non-invasive collection. However, biomarker detection in serum or plasma has some drawbacks. One is the fact that in these fluids a heterogeneous mixture of proteins derived from different tissues is found, making it difficult to attribute a differentially expressed protein to a tissue-specific disease. This limitation can result in the identification of putative protein markers that are not specific for CRC. Furthermore, in serum a small number of major proteins (e.g., albumin) are highly concentrated and mask other less abundant proteins that could be interesting as biomarkers. One strategy to overcome this problem is to remove the serum most abundant proteins by using commercial affinity columns designed specifically for this purpose[67].
Despite these shortcomings, in the last years several proteomic studies have been published focusing on the search of serum proteins to discriminate CRC patients from healthy individuals. Interestingly, different authors including our own group identified the same differentially expressed proteins using a variety of proteomic tools[18,19,40,68-74]. Among those proteins we found apolipoproteins, cathepsins, alpha-1 antitrypsin, alpha-1-acid glycoprotein types 1 and 2, transferrin, beta-2 microglobulin, complement components C3 and C9, gelsolin, heat shock proteins as HSP60, transthyretin, defensins (also known as human neutrophil peptides), alpha-enolase, S100 A calcium binding proteins, as well as the inhibitor of metalloproteases TIMP-1. This latter marker is known to be aberrantly glycosylated in patients with CRC[75].
Recently, the application of new proteomic technologies allowed the identification of other candidate proteins such as adipophilin, also known as perilipin-2 or adipose differentiation-related protein[65], and kininogen-1[76], for the early detection of CRC. Although in some cases these putative markers show good diagnostic parameters, in general it has been demonstrated that a single protein marker is not enough to get high sensitivity and specificity values for CRC diagnosis. Therefore, in order to reach a greater diagnostic accuracy it can be more useful to use a panel of several markers. In fact, Brock et al[77] have described that the protein panel alpha-1-acid glycoprotein 1, gelsolin, C3, C9, hyaluronic acid binding protein 2 (HABP2) and serum amiloide A2 (SAA2) yielded a sensitivity of 94% and a specificity of 83% to differentiate CRC patients from healthy donors.
Looking for the identification of metastasis-associated proteins, the serum of healthy individuals vs patients with metastatic CRC were compared. Specific proteins were identified in the patients, including mitogen activated protein kinase activated protein kinase 3 (MAPKAPK3), activin A receptor IIB (ACVR2B), Pim-1 oncogene (PIM-1), v-src sarcoma viral oncogene homolog (SRC) and fibroblast growth factor receptor-4 (FGFR4)[78]. Other authors have compared serum samples from CRC patients with lymph node or liver metastasis and those from patients without recurrence or metastasis for at least 3 years. In these studies the protein transthyretin (TTR) was identified as a candidate lymph node metastasis marker in CRC[11], whereas a panel of 8 peptides identified as fragments of alpha-fetoprotein, complement C4-A, fibrinogen alpha, eukaryotic peptide chain release factor GTP-binding subunit ERF3B, and angiotensinogen, demonstrated promising value for predicting liver metastasis in patients who underwent radical resection of CRC[79].
The analysis of tissue samples obtained from CRC patients allows comparing the protein profile between tumour and the adjacent healthy mucosa, particularly useful in the discovery of prognostic tumour markers. This approach has several advantages over serum or plasma analysis. The first one is that not all the proteins altered in the tumour, and therefore marker candidates, are secreted to the blood from the tumour cells. Therefore, the concentration of a putative marker is higher in tumour tissue than in blood. In addition, since tumour samples are analysed, there is no doubt that the altered proteins identified originate in the tumour itself. However, proteomic studies in tumours have also some disadvantages. First of all, the availability of this type of sample is limited. Moreover, tumours are heterogeneously composed of neoplastic cells besides the surrounding stromal cells. As an example of this latter drawback, various studies have described alterations of the protein vimentin in colorectal tumours[80-82] even though this protein is not expressed in epithelial cells, only in stromal cells and lymphocytes. Therefore, the identification of specific biomarkers would require the isolation of the tumour epithelial cells. However, because of the small size of tissue biopsies and other technical reasons, this separation is usually not carried out. In consequence, some proteins overexpressed in tumours are also increased in inflammation and this lack of specificity may prevent its use in clinical practice.
An overview of the proteins with altered expression in colorectal tumours identified by different authors comparing matched normal and tumour tissues reveals that many of them are abundant proteins such as structural and cytoskeleton proteins, annexins, chaperones or glycolytic enzymes which are also modified in other cancers[83]. In order to detect less abundant proteins, an approach that will be discussed later in this review is the isolation of subcellular fractions and the characterization of the corresponding sub-proteomes.
Among the proteins differentially expressed in colorectal tumours and consistently described in literature as candidate markers we can highlight translationally controlled tumour protein (TCTP)[21,46,84], NDK A[19,20,46,84] and S100A9[19,85], all of them overexpressed in tumours, as well as selenium-binding protein (SELENBP1 protein)[19,85] and carbonic anhydrase 1 (CA-1)[27,85,86] which are less expressed in the tumour tissue. Noticeably, in some cases the analysis of whole tumour lysates has led to the identification of marker candidates which have been subsequently validated in tissue or serum samples from a different cohort of CRC patients. Among these interesting biomarkers are proteins from the S100A family such as S100A8, S100A9[19] and S100A12[48], ribonucleoprotein HNRNPA1[87], the enzyme nicotinamide N-methyl transferase (NNMT)[46], proteasome activator complex subunit 3 (PSME3)[47], desmin[20] and olfactomedin-4 (OLF4). This latter protein is overexpressed not only in adenocarcinomas at early stages, but also in adenomas[88].
Besides the comparisons between healthy mucosa and tumour tissue, other authors have analysed metastatic and non-metastatic CRC tissues. Kang et al[89] compared the primary tumour tissues from 14 CRC patients with and without hepatic metastasis. They found a differential protein cluster, consisting of 17 proteins from the PI3K/AKT pathway which was validated by Western blot. A similar approach was performed by Zhao et al[21] who detected that Rho GDP-dissociation inhibitor alpha is markedly up-regulated in metastatic CRC.
A different approach to analyse whole tumour proteins has been recently published by Wiśniewski et al[90]. In this study, using formalin-fixed paraffin-embedded tissues, they were able to identify about 10000 proteins, many of them previously described as candidate CRC markers. This method offers a great advantage for the identification of prognostic markers since it allows retrospective studies analysing archival paraffin-embedded samples from patients who have been clinically followed-up for several years.
Stool samples provide important advantages over other clinical samples in the search for CRC markers such as the availability and the non-invasive collection of the sample. Furthermore, tumour-derived proteins may be more abundant in faeces than in blood. However, one drawback of the proteomic analysis of stool samples is the proteolysis caused by the gut micro-biota, leading to protein degradation.
In faeces of CRC patients the following proteins have been identified: defensins, S100A calcium binding proteins (including calprotectin), haemoglobin, haptoglobin, alpha-1 antitrypsin, lactoferrin, CEA, dipeptidyl peptidases I and IV, cadherin 17, SELENBP1, pyruvate kinase type M2 (M2PK), metalloprotease 9 (MMP9) and its inhibitor TIMP1[91]. Although some of these proteins are related with inflammation, resulting inespecific for CRC, others have been previously reported as CRC-associated proteins and have been tested as candidate CRC screening biomarkers. Interestingly, tumour pyruvate kinase type M2 showed good sensitivity for the detection of tumours (85%) but not for adenomas (28%)[92].
Moreover, it has been demonstrated that a marker panel containing the proteins S100A12, TIMP1 and haemoglobin-haptoglobin showed higher sensitivity than faecal occult blood, and identified 74% of the patients at early stages of tumour development[93]. More recently, Ang et al[94] developed a multiplex analysis for 40 human proteins on faecal samples from eight CRC patient and seven healthy volunteers. These authors identified 24 proteins consistently found in all samples and nine proteins (alpha-1 antitrypsin, alpha 1-acid glycoprotein, complement C3, fibrinogen, haptoglobin, hemoglobin alpha, hemoglobin beta, myeloblastin and transferrin) detected only in the CRC patients. The relatively high abundance of these nine candidate markers in the CRC patient samples indicate that they could be clearly differentiated from the healthy controls.
Cell lines derived from colon tumours offer several advantages over clinical samples for biomarker identification. Cell lines are homogeneous cell populations, they are easy to work with and their availability is almost unlimited. Besides, it is easier to obtain subcellular fractions such as plasma membrane, nucleus, secretome (the conditioned media in which the cells grow), and exosomes from cell cultures. Thereby, cellular proteome complexity is reduced, simplifying the identification of less abundant proteins. Nevertheless, the use of cell cultures has some limitations. Almost all available human colon cell lines come from tumours, whereas cell lines derived from normal mucosa epithelial cells do not exist. This fact prevents the study of the proteome changes associated to the CRC malignant transformation sequence: normal mucosa-adenoma-adenocarcinoma. In addition, cell cultures do not exactly represent the in vivo situation since they lack features of an in situ tumour such as the interactions among tumoral cells, stromal cells and the immune system. For this reason, it is mandatory that the candidate biomarkers discovered in cell lines are eventually validated in clinical samples from CRC patients[67].
The human colon adenocarcinoma Caco-2 cell line is an accepted in vitro model to study colorectal tumorigenesis since Caco-2 cells seems to lose their carcinogenic phenotype during the differentiation process. Taking into account this advantage, several authors have applied different proteomic technologies to compare the whole cell lysates before and after differentiation of Caco-2 cells. Along the differentiation process, the proteins glutathione S-transferase alpha 1 (GSTA1), annexin A4 (ANXA4), villin 1 (VIL1), galectin (LGALS3) and phosphoglycerate kinase 1 (PGK1) were up-regulated, whereas the proteins CDC2, PCNA and HNRNPH3 were down-regulated[95,96]. Fanayan et al[97] analysed cell lysates of other human colon cancer cell lines (LIM1215, LIM1899 and LIM2405) that were selected to represent a wide range of pathological states of colorectal cancer. They identified both cancer-associated proteins with differential expression patterns, as well as protein networks and pathways which appear to be de-regulated in these cell lines. Examples of candidate markers include mortalin, nucleophosmin, ezrin, alpha and beta forms of spectrin, exportin, the carcinoembryonic antigen family, EGFR and met-proto-oncogene (MET).
In order to identify metastasis-associated proteins, some studies have compared two cell lines with different metastatic potential as SW-480, derived from a Dukes’ stage B colon carcinoma and SW-620, derived from a lymph node metastasis of the same patient. Ghosh et al[98] analysed the whole cell proteome profiles of these two isogenic colorectal cancer cell lines and identified 147 proteins significantly altered in the metastatic cell. Up-regulated proteins in the SW620 cell line included stathmin, villin, myosin 10 and myristoylated alanine-rich C-kinase substrate (MARCKS), whereas down-regulated proteins included those related to cytoskeletal signalling (type 1 cytoskeletal 13, type II cytoskeletal 5, tubulin beta 2A/2B, actin, and several acting binding proteins), annexin A1 and proteins with roles in cellular adhesion including beta-catenin and neural cell adhesion molecule1 (NCAM1). However, a beta-catenin degrading protein, calcyclin-binding protein (CacyBP), was found up-regulated in SW620 cells, suggesting the possible involvement of CacyBP in CRC metastasis through the alteration of beta-catenin-mediated cellular adhesion.
In recent years, the proteomic analysis of the secretome of human colon adenocarcinoma cell lines has been described as a useful strategy for the detection of candidate blood-based markers. This is due because the analysis of a cell-specific secretome limits the contamination by the major proteins of the human serum, favouring the identification of tissue-specific proteins[99]. However, as Malard et al[100] pointed out, the analysis of human cell line secretomes by proteomics techniques presents some drawbacks. First, proteins secreted in culture media are highly diluted. Moreover, changes in the physiology of the cells may occur upon a change of medium, as most of secretome studies are performed with the basal medium without the addition of fetal calf serum.
Secretomes comprise protein released through different mechanisms, including secreted membrane vesicles (exosomes) able to transfer information to target cells. Searching for CRC-specific markers, Wu et al[101] studied the secretome of 21 cancer cell lines from 12 different cancer types, and found that collapsin response mediator protein-2 (CRMP-2) was exclusively detected in the colon cell lines Colo205 and SW480. This protein was eventually validated in serum, demonstrating its value for discriminating CRC patients from healthy donors. Aiming to find secreted metastasis-associated proteins, other studies have compared the secretome of SW-480 and SW-620 cells. Among the proteins differentially expressed between these two cell lines the proteins alpha 5 and alpha 6 integrins, peroxiredoxins 2 and 6, soluble E-cadherin, growth/differentiation factor GDF15 and trefoil factor 3 (TFF3) have been found[71,102]. Noticeably, the serum levels of GDF15 and TFF3 were significantly increased in CRC patients with lymph node metastasis as compared to patients without metastasis or healthy donors[71]. Besides, another study showed that serum levels of soluble E-cadherin reflect the disease status of CRC patients, although validation in a larger cohort would be required to confirm this finding[103].
As we have previously mentioned, the analysis of whole cell lysates can yield good candidate markers, although the complexity of such a sample may hamper biomarker discovery. An alternative for reducing sample complexity of tissue biopsies or cell lysates is to perform a previous subcellular fractionation of the sample and then to analyse the sub-proteome of different subcellular fractions. In this regard, Yang et al[64] focused on the identification of proteins in the insoluble fraction of biopsies from 13 CRC patients and found a panel of four proteins (KRT5, JUP, TUBB and COL6A1) as candidate CRC biomarkers. Other authors have analysed the nuclear matrix proteome comparing adenomas and adenocarcinomas since nuclear phenotypic alterations and chromosomal instability are a hallmark of tumour cells[104]. Therefore, it is expected that among the differentially expressed proteins identified in that study (MNDA, HNRPH2, DNASE1, NUP62, NUP88), some meaningful tumour markers could be eventually validated in larger cohorts.
A subcellular fraction of particular interest that has been studied by several researchers including our own group is the membrane fraction. Membrane-associated proteins account for approximately 30% of human proteome and play essential roles in cell-cell and cell-extracellular matrix interactions as well as in signal transduction. Most of the biomarkers currently used in clinical practice (e.g., CEA) and about 70% of all known therapeutic targets are membrane proteins. However, because of the difficulties to isolate and analyse these proteins, mainly due to their hydrophobic character, there are only few proteomic studies analysing membrane-associated proteins in CRC patients. Comparing paired tumours and healthy mucosa samples the overexpression of several membrane proteins including CEA, claudin-3 and the A1 antigen of the histocompatibility complex has been reported in tumours[105]. Another membrane protein identified as a possible biomarker is STOML2 (stomatin-like protein 2), which was found increased in tumours and also detected in serum of CRC patients at early stages of the disease[106]. Other authors have identified candidate markers for metastasis when comparing the cell surface proteomes of the low metastatic cell line KM12C and the high metastatic KM12SM cells. A number of cell signalling, integrins and other cell adhesion proteins (cadherin 17, junction plakoglobin) were described among the most de-regulated proteins[107].
Our research group has analysed the sub-proteomes of membrane and soluble fractions of tumours, and compared them with those from adjacent healthy mucosa. Among the de-regulated membrane proteins we found cytoskeleton proteins, chaperones and two isoforms of the calcium binding protein S100A6[81]. Regarding the soluble fraction a panel of four putative biomarker proteins were identified, 14-3-3 protein zeta/delta, parkinson protein 7 (DJ-1), retinoblastoma binding protein 4 (RBBP-4) and nucleoside diphosphate kinase A (NDK A), differentially expressed in normal and tumour tissues. Furthermore, NDK A was detected in serum and preliminary results indicated an increased concentration in CRC patients. Thus NDK A is an interesting candidate serum biomarker for CRC[24].
Exosomes are 40-100-nm diameter membrane vesicles of endocytic origin that are released from most cell types and circulate in body fluids, including blood. Therefore, analysing the protein profile of these vesicles released by tumour cells can be a useful method to identify markers for the diagnosis of CRC. Indeed, the proteome analysis of exosomes derived from tumoral colon cell lines has led to the identification of the candidate markers claudin-3, Ephrin-B1 and galectin 4[108], cadherin-17, CEA, EGFR, glycoprotein A33 and EpCAM (epithelial cell surface adhesion molecule), among others[109]. Recently, the exosome protein profiles of SW480 and SW620 cell lines were compared to identify metastatic factors and signalling molecules fundamental for tumour progression. A major finding was the selective enrichment of metastatic factors (MET, S100A8, S100A9, TNC) and signal transduction molecules (EFNB2, JAG1, SRC, TNIK) in exosomes derived from the metastatic cells[110].
Animal models can be an alternative to clinical samples for the discovery of biomarkers since proteomic studies using inbred strains of mice avoid genetic and physiological variations among individuals. Unfortunately, there are only a small number of mouse models that show genetic alterations that lead to CRC. Among such murine models are the Apc (Min/+) mice, which are mutant in the APC gene and can develop tumours in the small intestine and the colon. The analyses of serum and tumours alterations of the proteins catepsins B and D, DJ-1, clusterin and S100A9 have been described[111]. In another study, among a total of 52 de-regulated proteins identified when comparing tumours and adjacent healthy tissue, a co-expressed gene network linked to innate immunity and inflammation was found in tumours. The network included cathelicidin antimicrobial peptide (CAMP), Toll-like receptors, IL-8 and triggering receptor expressed on myeloid cells 1 (TREM1)[112]. More recently, serum proteins from tumour-bearing Apc (Min/+) mice were quantitatively compared to tumour-free wild type mice via in anima metabolic labelling and LTQ Orbitrap mass spectrometer. Among the 40 differentially expressed proteins identified, MGAM, ITIH3 and F5 were validated in neoplastic colonic tissues from the mutant mice. These proteins provided a set of candidate biomarkers for future validation in humans[113].
An overview of recent reports focused on the different proteomic techniques and samples used for the discovery of candidate protein markers for CRC is provided in Table 1.
| Gene | Protein name | Sample | Potential clinical value | Proteomic technique | Validation | Ref. |
| YWHAZ | 14-3-3 protein zeta/delta | Tissue | Tissue biomarker | 2-DE, LC-MS/MS | WB | [24,46] |
| HLA-A1 | HLA class I histocompatibility antigen A-1 | Membrane fraction of tissue | Tissue biomarker | 1-DE, iTRAQ labelling, LC-MS/MS | WB | [105] |
| ACVR2B | Activin receptor type-2B | Serum | Diagnosis | FP-protein microarray | WB, RP-tissue microarray, ELISA | [78] |
| PLIN2 | Adipophilin | Serum | Diagnosis | Label-free quantitative LC-MS/MS | WB, RP-microarray, IHC | [65] |
| SPTBN1 | Beta-spectrin | Cell lines | Metastasis | 1-DE, LC-MS/MS | RNA-Seq | [97] |
| A1AT | Alpha-1 antitrypsin | Serum, stool | Diagnosis | SELDI-TOF-MS, 1-DE, MudPIT, SRM | - | [69,94] |
| ORM1 | Alpha-1-acid glycoprotein type 1 | Serum, stool | Diagnosis | 1-DE, MudPIT, SRM | SRM | [77,94] |
| ORM2 | Alpha-1-acid glycoprotein type 2 | Serum, stool | Diagnosis | iTRAQ labelling, LC-MS/MS, 1-DE, MudPIT, SRM | WB, ELISA | [73,94] |
| ENO1 | Alpha-enolase | Tissue, validated in serum | Diagnosis | 2-DE, 2D-DIGE, LC-MS/MS, MALDI-TOF-MS | WB | [18,19,46] |
| AFP | Alpha-fetoprotein | Serum | Liver metastasis from CRC | MALDI-TOF-MS, LC-MS/MS | - | [79] |
| AGT | Angiotensinogen | Serum | Liver metastasis from CRC | MALDI-TOF-MS, LC-MS/MS | - | [79] |
| ANXA2 | Annexin A2 | Membrane fractions from colorectal carcinoma | Diagnosis and presumably therapy | 2D-DIGE, MALDI-TOF-MS | RP-tissue microarray | [25] |
| ANXA4 | Annexin A4 | Membrane fractions from colorectal carcinoma | Diagnosis and presumably therapy | 2D-DIGE, MALDI-TOF-MS | RP-tissue microarray | [25,80] |
| APOA1 | Apolipoprotein A1 | Serum | Diagnosis | 2-DE, 2D-DIGE, MALDI-TOF-MS, LC-MS/MS | WB, RP-protein microarray, RP-tissue microarray | [40,70,80] |
| CTNNB1 | Beta-catenin | Cell lines | Metastasis | 1-DE, iTRAQ labelling, LC-MALDI-TOF-MS, LC-MS/MS | WB, RP-tissue microarray, flow cytometry, confocal microscopy | [98,107] |
| CACYBP | Calcyclin binding protein | Cell lines | Metastasis | iTRAQ labelling, LC-MALDI-TOF-MS | WB | [63,98] |
| CDH17 | Cadherin 17 | Membrane fractions from cell lines, exosomes, stool | Metastasis | 1-DE, LC-MS/MS | WB, RP-tissue microarray, flow cytometry, confocal microscopy | [91,107,109] |
| CEACAM5 | Carcinoembryonic antigen | Cell lines, membrane fraction from tissue, exosomes, stool | Prognosis | Protein microarray, iTRAQ labelling, 1-DE, LC-MS/MS | WB, RP-tissue microarray, flow cytometry, confocal microscopy, RNA-Seq | [38,91,97,105,107,109] |
| CAMP | Cathelicidin antimicrobial peptide | Tissue from animal model | Candidate tissue biomarker | 2-DE, MudPIT, LC-MS/MS | - | [112] |
| CTSB | Cathepsin B | Animal model | Candidate diagnosis biomarker | MudPIT, LC-MS/MS | WB, Ab microarray, IHC | [111] |
| CTSD | Cathepsin D | Tissue, animal model | Candidate diagnosis biomarker | 2D-DIGE, MudPIT, LC-MS/MS | WB, Ab microarray, IHC | [80,111] |
| CDC2 | Cell division control protein 2 homolog | Cell lines | Candidate tissue biomarker | 2-DE, MALDI-TOF-MS | WB | [95] |
| CLDN3 | Claudin-3 | Membrane fraction from tissue, exosomes | Tissue biomarker | 1-DE, iTRAQ labelling, LC-MS/MS | WB | [105,108] |
| PTPRC | Cluster of differentiation 45 | Tumor tissue | Diagnosis | Protein microarray | - | [38] |
| TFRC | Cluster of differentiation 71 | Tumor tissue | Diagnosis | Protein microarray | - | [38] |
| CLU | Clusterin | Serum, animal model | Diagnosis | 2-DE, MALDI-TOF-MS, MudPIT, LC-MS/MS | WB, antibody microarray, IHC | [23,70,111] |
| F5 | Coagulation factor V | Animal model | Candidate serum biomarker | Metabolic labelling, LC-MALDI-TOF-MS, SRM, LC-MS-MS | - | [113] |
| COL6A1 | Collagen, type VI α-1 chain | Tissue | Prognosis | LC-MS/MS | WB, IHC | [64] |
| DPYSL2 | Collapsin response mediator protein-2 | Secretome from cell lines, validated in serum | Diagnosis | 1-DE, MALDI-TOF-MS | WB, IHC, ELISA | [101] |
| C3 | Complement C3 | Serum, stool | Diagnosis | SELDI-TOF-MS, 1-DE, MudPIT, SRM, LC-MS/MS | SRM | [69,74,77,94] |
| C4A | Complement C4-A | Serum | Liver metastasis from CRC | MALDI-TOF MS, LC-MS/MS | - | [79] |
| C9 | Complement C9 | Serum | Diagnosis early stage | LC-MS/MS, SRM | RP-protein microarray, SRM | [40,77] |
| PPIA | Cyclophilin A | Tissue | Tissue biomarker | 2-DE, SELDI-TOF-MS | IHC | [58] |
| CAI | Cytoplasmic carbonic anhydrase I | Tissue | Tissue biomarker | 2D-DIGE, MALDI-TOF-MS | WB, IHC | [27,33,84,86] |
| CAII | Cytoplasmic carbonic anhydrase II | Tissue | Tissue biomarker | 2D-DIGE, MALDI-TOF-MS | WB, IHC | [33] |
| DEFA1 | Defensin-1 | Tissue, serum, stool | Diagnosis | SELDI-TOF-MS, MALDI-TOF-MS | - | [68,91] |
| DEFA2 | Defensin-2 | Tissue, serum | Diagnosis | SELDI-TOF-MS, MALDI-TOF-MS | - | [68] |
| DEFA3 | Defensin-3 | Tissue, serum, stool | Diagnosis | SELDI-TOF-MS, MALDI-TOF-MS | - | [68,91] |
| DES | Desmin | Tissue, validated in serum | Diagnosis and prognosis | 2-DE, MALDI-TOF-MS | WB, IHC | [20] |
| MAPRE1 | The adenomatous polyposis coli-binding protein | Tissue | Prognosis | 2D-DIGE | IHC | [32] |
| CDH1 | E-cadherin | Secretome from cell lines , serum | Diagnosis | 2D-DIGE, MALDI-TOF-MS, LC-MS/MS | WB | [102,103] |
| EFEMP2 | EGF-containing fibulin-like extracellular matrix protein 2 | Secretome of cultured fresh normal and CRC tissues | Diagnosis | 1-DE, LC-MS/MS | WB, IHC | [13] |
| EFNB2 | Ephrin-B2 | Exosomes | Metastasis | 1-DE, LC-MS/MS | - | [108,110] |
| EGFR | Epidermal growth factor receptor | Cell lines, exosomes | Candidate diagnosis biomarker | 1-DE, LC-MS/MS | WB, RP-tissue microarray, flow cytometry, confocal microscopy, RNA-Seq | [97,107,109] |
| EPCAM | Epithelial cell surface adhesion molecule | Exosomes | Candidate diagnosis biomarker | LC-MS/MS | - | [109] |
| GSPT2 | Eukaryotic peptide chain release factor GTP-binding subunit | Serum | Liver metastasis from CRC | MALDI-TOF-MS, LC-MS/MS | - | [79] |
| XPO4 | Exportin | Cell lines | Tissue biomarker | 1-DE, LC-MS/MS | RNA-Seq | [97] |
| EZR | Ezrin | Cell lines | Tissue biomarker | 1-DE, LC-MS/MS | RNA-Seq | [97] |
| FABP1 | Fatty acid-binding protein 1 | Tissue | Prognosis | 2D-DIGE, ICAT, LC-MS/MS | WB, IHC | [62] |
| FGFR4 | Fibroblast growth factor receptor 4 | Serum | Diagnosis | FP-protein microarray | - | [78] |
| FGA | Fibrinogen alpha | Serum, stool | Liver metastasis from CRC, diagnosis | MALDI-TOF MS, LC-MS/MS, 1-DE, MudPIT, SRM | - | [79,94] |
| BLVRB | Flavin reductase | Tissue | Tissue biomarker | 2-DE, SELDI-TOF-MS | IHC | [58] |
| LGALS3 | Galectin 3 | Cell lines | Tissue biomarker | 2-DE, MALDI-TOF-MS | WB | [95] |
| LGALS4 | Galectin 4 | Exosomes | Tissue biomarker | 1-DE, LC-MS/MS | - | [108] |
| GSN | Gelsolin | Tissue, serum | Diagnosis | 2D-DIGE, MALDI-TOF-MS, SRM, LC-MS/MS | WB, IHC | [19,77] |
| GSTA1 | Glutathione S-transferase alpha-1 | Cell lines | Tissue biomarker | 2-DE, MALDI-TOF-MS, LC-MS/MS | WB | [95,96] |
| GPA33 | Glycoprotein A33 | Exosomes | Candidate diagnosis biomarker | Immunoaffinity capture microbeads and LC-MS/MS | - | [109] |
| GDF15 | Growth/differentiation factor 15 | Secretome from cell lines, validated in serum | Metastasis | LC-MS/MS | WB, ELISA | [71] |
| HBG | Haemoglobin | Stool | Diagnosis | 1-DE, MudPIT, SRM | Immunoassay | [93,94] |
| HSPA9 | Heat shock 70 kDa protein 9 (mortalin) | Cell lines | Tissue biomarker | 1-DE, LC-MS/MS | RNA-Seq | [97] |
| HSPB1 | Heat shock protein 27 | Tissue, validated in serum | Diagnosis | 2-DE, LC-MS/MS | - | [18] |
| HSPD1 | Heat shock protein 60 | Serum | Diagnosis of late stage CRC, prognosis? | 2D-DIGE, MALDI-TOF-MS | WB, ELISA, Tissue microarray | [72] |
| HNRNPH2 | Heterogeneous nuclear ribonucleoprotein H2 | Nuclear fraction from tissue | Tissue biomarker | 1-DE, MALDI-TOF-MS, LC-MS/MS | - | [104] |
| HNRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 | Tissue, validated in serum | Diagnosis and prognosis | 2-DE , MALDI-TOF-MS, SELDI-TOF-MS | IHC | [58,87] |
| HNRNPH3 | Heterogeneous nuclear ribonucleoprotein H3 | Cell lines | Tissue biomarker | 2-DE, MALDI-TOF-MS | WB | [95] |
| HABP2 | Hyaluronan-binding protein 2 | Serum | Diagnosis | SRM, LC-MS/MS | - | [77] |
| ITLN1 | Intelectin-1 | Tissue | Prognosis | 2D-DIGE, ICAT, LC-MS/MS | WB, IHC | [62] |
| ITIH3 | Inter-alpha-trypsin inhibitor heavy chain 3 | Serum from animal model | Serum biomarker | Metabolic labelling, LC-MALDI-TOF-MS, SRM, LC-MS-MS | - | [113] |
| IL8 | Interleukin 8 | Animal model | Candidate tissue biomarker | 2-DE, MudPIT, LC-MS/MS | - | [112] |
| JAG1 | Jagged 1 protein | Exosomes | Metastasis | 1-DE, LC-MS/MS | - | [110] |
| JUP | Junction plakoglobin | Membrane fraction from cell lines, tissue | Diagnosis, prognosis and metastasis | 1-DE, LC-MS/MS | WB, RP-tissue microarray, flow cytometry, confocal microscopy | [64,107] |
| KRT5 | Keratin 5 | Tissue, cell lines | Prognosis | LC-MS/MS, 2-DE, iTRAQ labelling, LC-MALDI-TOF-MS | WB, IHC | [64,98] |
| KNG1 | Kininogen-1 | Serum, tissue | Diagnosis | MB-WCX, MALDI-TOF-MS | ELISA, IHC | [76] |
| MIF | Macrophage migration inhibitory factor | Tissue, validated in serum | Diagnosis | 2-DE, LC-MS/MS | - | [18] |
| MGAM | Maltase-glucoamylase | Serum from animal model | Candidate serum biomarker | Metabolic labelling, LC-MALDI-TOF-MS, SRM, LC-MS-MS | - | [113] |
| MAPKAPK3 | MAP kinase-activated protein kinase 3 | Serum | Diagnosis | FP-protein microarray | WB, RP-tissue microarray, ELISA | [78] |
| MET | Met-proto-oncogen | Cell lines, exosomes | Metastasis | 1-DE, LC-MS/MS | RNA-Seq | [97,110] |
| PRTN3 | Myeloblastin | Stool | Diagnosis | 1-DE, MudPIT, SRM | Immunoassay | [94] |
| MNDA | Myeloid cell nuclear differentiation antigen | Nuclear fraction from tissue | Tissue biomarker | na1-DE, MALDI-TOF-MS, LC-MS/MS | - | [104] |
| MYH10 | Myosin 10 | Cell lines | Candidate tissue biomarker | iTRAQ labelling, LC-MALDI-TOF-MS | WB | [98] |
| MARCKS | Myristoylated alanine-rich C-kinase substrate | Cell lines | Candidate tissue biomarker | iTRAQ labelling, LC-MALDI-TOF-MS | WB | [98] |
| NCAM1 | Neural cell adhesion molecule 1 | Cell lines | Candidate tissue biomarker | iTRAQ labelling, LC-MALDI-TOF-MS | WB | [98] |
| NNMT | Nicotinamide N-methyltransferase | Tissue, validated in serum | Diagnosis | 2-DE, MALDI-TOF-MS | WB, ELISA | [46] |
| NUP62 | Nuclear pore complex 62 | Nuclear fraction from tissue | Tissue biomarker | 1-DE, MALDI-TOF-MS, LC-MS/MS | - | [104] |
| NUP88 | Nuclear pore complex 88 | Nuclear fraction from tissue | Tissue biomarker | 1-DE, MALDI-TOF-MS, LC-MS/MS | - | [104] |
| NPM1 | Nucleophosmin | Cell lines | Candidate tissue biomarker | 1-DE, LC-MS/MS | - | [97] |
| NME1 | Nucleoside diphosphate kinase A | Tissue, cell lines, serum | Diagnosis | 2-DE, 2D-DIGE, RP-tissue microarray, MALDI-TOF-MS, LC-MS/MS | WB, IHC | [4,19,20,24,39,46,84] |
| NME2 | Nucleoside diphosphate kinase B | Tissue | Tissue biomarker | 2-DE, SELDI-TOF-MS | IHC | [58] |
| OLFM4 | Olfactomedin-4 | Secreted proteins from tissue | Tissue biomarker | OFFGEL, iTRAQ labelling, MALDI-TOF-MS | IHC | [88] |
| PARK7 | Parkinson protein 7 | Tissue, animal model | Tissue biomarker | 2-DE, LC-MS/MS | WB, IHC | [24,111] |
| PRDX2 | Peroxiredoxins 2 | Cell lines | Candidate tissue biomarker | 2D-DIGE, MALDI-TOF-MS, LC-MS/MS | WB | [102] |
| PRDX6 | Peroxiredoxins 6 | Cell lines | Candidate tissue biomarker | 2D-DIGE, MALDI-TOF-MS, LC-MS/MS | WB | [102] |
| PGK1 | Phosphoglycerate kinase I | Cell lines | Candidate tissue biomarker | 2-DE, MALDI-TOF-MS | WB | [95] |
| ADCYAP1 | Pituitary adenylate cyclase-activating polypeptide | Tissue | Tissue biomarker | 2-DE, SELDI-TOF-MS | IHC | [58] |
| PCNA | Proliferating cell nuclear antigen | Cell lines | Candidate tissue biomarker | 2-DE, MALDI-TOF-MS | WB | [95] |
| PSME3 | Proteasome activator complex subunit 3 | Tissue, validated in serum | Diagnosis and prognosis | 2-DE, MALDI-TOF-MS | IHC | [46,47] |
| RBBP4 | Retinoblastoma binding protein 4 | Cell line, tissue | Candidate tissue biomarker | 2-DE, LC-MS/MS | WB | [24] |
| ARHGDI | Rho GDP-dissociation inhibitor | Tissue, cell lines | Metastasis | 2-DE, MALDI-TOF-MS | [21] | |
| S100A11 | S100-A11 calcium binding protein | Tissue | Tissue biomarker | 2-DE, SELDI-TOF-MS | IHC, protein microarray | [58,59] |
| S100A12 | S100-A12 calcium binding protein | Tissue, stool, validated in serum | Diagnosis | 2-DE, MALDI-TOF-MS, LC-MS/MS | Immunoassay | [48,93] |
| S100A6 | S100-A6 calcium binding protein | Membrane fraction from tissue | Tissue biomarker | 1-DE, 2-DE, MALDI-TOF-MS, LC-MS/MS | - | [81,110] |
| S100A8 | S100-A8 calcium binding protein | Tissue, cell lines, exosomes, validated in serum, stool | Diagnosis | 1-DE, 2D-DIGE, MALDI-TOF-MS, LC-MS/MS | WB, IHC | [19,84,91,110] |
| S100A9 | S100-A9 calcium binding protein | Tissue, cell lines, exosomes, animal model, validated in serum, stool | Diagnosis | 1-DE, 2D-DIGE, MALDI-TOF-MS, LC-MS/MS | WB, IHC | [19,84,85,91,110] |
| SELENBP1 | Selenium binding protein 1 | Tissue, cell lines, serum | Prognosis | 2D-DIGE, MALDI-TOF/TOF, and MALDI-TOF-MS | WB | [19,84,86] |
| PIM1 | Serine/threonine-protein kinase Pim-1 | Serum | Diagnosis | FP-protein microarray | WB, RP-tissue microarray, ELISA | [78] |
| SAA2 | Serum amyloid A-2 protein | Serum | Diagnosis | 1-DE, MudPIT, LC-MS/MS | SRM | [77] |
| TAGLN | Smooth muscle protein 22-alpha | Tissue | Tissue biomarker | 2-DE, SELDI-TOF-MS | IHC | [58] |
| SPTAN1 | Spectrin | Cell lines | Candidate tissue biomarker | 1-DE, LC-MS/MS | RNA-seq | [97] |
| SRC | Proto-oncogene tyrosine-protein kinase Src | Serum, exosomes | Diagnosis, metastasis | FP-protein microarray, 1-DE, LC-MS/MS | WB, RP-tissue microarray, ELISA | [78,110] |
| STMN1 | Stathmin | Cell lines | Metastasis | iTRAQ, LC-MALDI-TOF-MS | WB | [98] |
| STOML2 | Stomatin-like protein 2 | Tissue, serum | Diagnosis and prognosis | LC-MS/MS | ELISA | [106] |
| TNC | Tenascin C | Exosomes | Metastasis | 1-DE, LC-MS/MS | - | [110] |
| TRIP | Thryroid receptor interactor | Serum | Diagnosis | 2D-DIGE, MALDI-TOF MS | - | [30] |
| TIMP1 | TIMP metallopeptidase inhibitor 1 | Serum | Diagnosis | Lectin Fractionation, MALDI-MS-MS | SRM | [75] |
| TLRs | Toll-like receptors | Animal model | Candidate tissue biomarker | 2-DE, MudPIT, LC-MS/MS | - | [112] |
| TNIK | TRAF2 and NCK-interacting protein kinase | Exosomes | Metastasis | 1-DE, LC-MS/MS | - | [110] |
| TALDO1 | Transaldolase 1 | Serum | Diagnosis | 2D-DIGE, MALDI-TOF MS | - | [30] |
| TFRC | Transferrin | Serum, stool | Diagnosis | SELDI-TOF-MS, 1-DE, MudPIT, SRM | - | [69,94] |
| TGFBI | Transforming growth factor β-induced protein ig-h3 | Cell lines | Candidate serum biomarker | 2-DE, 2D-DIGE, MALDI-TOF-MS | - | [35,46] |
| TAGLN | Transgelin | Tissue | Prognosis | 2D-DIGE, ICAT, LC-MS/MS | WB, IHC | [62] |
| TPT1 | Translationally controlled tumour protein | Secretome from cell lines, tissue | Candidate serum biomarker | 2D-DIGE, MALDI-TOF-MS | - | [35,84] |
| TTR | Transthyretin | Serum | Metastasis | 2-DE, MALDI-TOF MS | ELISA | [11] |
| TFF3 | Trefoil factor 3 | Secretome from cell lines, tissue, validated in serum | Metastasis | LC-MS/MS | WB, ELISA | [71] |
| TREM1 | Triggering receptor expressed on myeloid cells 1 | Animal model | Candidate tissue biomarker | 2-DE, MudPIT, LC-MS/MS | - | [112] |
| TPM2 | Tropomyosin 2 (beta chain) | Tissue | Prognosis | 2D-DIGE, ICAT, LC-MS/MS | WB, IHC | [62] |
| TUBB | Tubulin beta chain | Tissue, cell lines | Diagnosis and prognosis | 2DE, iTRAQ labelling, LC-MALDI-TOF-MS, LC-MS/MS, | WB, IHC | [64,98] |
| PKM | Pyruvate kinase type M2 | Tissue, stool | Diagnosis | 2D-DIGE, MALDI-TOF-MS | - | [19,92] |
| VCP | Valosin-containing protein | Tissue | Prognosis | 2D-DIGE, ICAT, LC-MS/MS | WB, IHC | [62] |
| VIL1 | Villin 1 | Tissue, cell lines | Metastasis | iTRAQ labelling, 2-DE, 2D-DIGE, LC-MALDI-TOF-MS, MALDI-TOF-MS | WB | [19,95,98] |
| VDAC1 | Voltage-dependent anion-selective channel protein 1 | Membrane fraction from colorectal carcinoma | Diagnosis and presumably therapy | 2D-DIGE, MALDI-TOF MS | RP-tissue microarray | [25] |
Many proteomic studies have identified CRC-associated proteins, but little work has been done to validate these candidate biomarkers. Therefore, it would be worthwhile to collect samples from larger cohorts of healthy individuals and CRC patients and to analyse their peptidome and proteome in depth to fully assess the biomarker potential of proteomic changes. Only this validation will then allow transferring novel biomarkers into clinical use for a better detection and treatment of CRC.
Nowadays, antibody-based methods (Western blotting, IHC, microarrays and particularly ELISA) are the most widely used for quantitative biomarker measurement. Nevertheless, antibody specificity is often lacking and considerable care must be taken in the validation of the signals observed. Insufficient samples, often of poor quality, are a key problem, as well as a lack of suitable antibodies or ELISA kits. Additionally, developing a validated ELISA is expensive and time-consuming. This fact, together with the recent advances in MS technology, is further stimulating the development of quantitative MS technologies for protein and peptide biomarker analysis. Multiple reaction monitoring (MRM, also known as selective reaction monitoring, SRM) is rapidly becoming the method of choice[94]. With this technique tens of protein candidates can be followed up in one LC-MS/MS analysis without the use of antibodies, and employing only microliters of sample though it cannot be still performed routinely. Nowadays, MALDI-TOF-MS for peptide profiling acquisition may be also applied as an alternative method for diagnosis of CRC[50]. Moreover, it is expected that combining several markers for CRC and applying multivariate analyses will significantly improve their diagnostic performance[114-116].
Future work should also be directed to evaluate the functional protein interaction networks derived from the proteomics data, in order to elucidate the molecular mechanisms of CRC carcinogenesis. Recently, Jimenez et al[83] reported a summary of differently expressed proteins in clinical proteomics studies comparing colorectal healthy and cancer tissues. Authors also analysed these proteins using the STRING tool (http://string.embl.de/) to visualize the protein-protein interaction networks. At the heart of the network they found a cluster of five up-regulated and well-connected proteins in CRC (enolase 1, glyceraldehyde-3-phosphate-dehydrogenase, pyruvate kinase isoenzymes M1/M2, fructose-bisphosphate aldolase A and transketolase), all involved in glycolysis. Glycolytic activity is increased in almost all cancers to provide cells with enough energy for proliferation, a phenomenon called “Warburg effect”[117]. Other molecular and cellular functions associated with the up-regulated proteins include “Cell Death”, “Cell-To-Cell Signalling and Interaction” and “Cellular Assembly and Organization”[83].
The discovery of novel biomarkers in CRC is crucial for the early detection of the disease, the characterization of the disease progression and the prediction to therapy response. Recent advances in proteomic technologies allow large-scale analysis of proteins which can be applied for discovering aberrant protein profiles of clinical and other biological samples related to CRC. A first critical issue in the search for candidate markers is the selection of the sample set. Tumour tissue is the most direct approach for biomarkers discovery as they are most likely present in cancer tissues at higher concentration than in serum or plasma. In the next step, protein candidate markers must be assessed by a targeted assay and validated in large independent cohorts. Only this validation will allow the translation of the candidate CRC markers to the clinical practice. Finally, it is now generally accepted that a combination of proteins rather than a single individual candidate can better discriminate CRC patients from controls, or determine the prognosis of the patients, implying the advantage of using proteins involved in different physio-pathological pathways. In conclusion, there has been a significant amount of research for the identification of CRC biomarkers using proteomic techniques; however, these are yet to yield stronger candidates. A single unique biomarker, of sufficient sensitivity and specificity, may not be a realistic goal, but instead a panel of markers may be a useful way to overcome the difficulties imposed by inter-individual variability.
P- Reviewers: Armengaud J, Bozdayi AM, Kadiyska YK S- Editor: Ma YJ L- Editor: A E- Editor: Ma S
| 1. | Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374-1403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3526] [Cited by in RCA: 3653] [Article Influence: 304.4] [Reference Citation Analysis (2)] |
| 2. | Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9215] [Cited by in RCA: 9855] [Article Influence: 821.3] [Reference Citation Analysis (4)] |
| 3. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8087] [Cited by in RCA: 8001] [Article Influence: 228.6] [Reference Citation Analysis (1)] |
| 4. | Søreide K, Nedrebø BS, Knapp JC, Glomsaker TB, Søreide JA, Kørner H. Evolving molecular classification by genomic and proteomic biomarkers in colorectal cancer: potential implications for the surgical oncologist. Surg Oncol. 2009;18:31-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 5. | de Wijkerslooth TR, Bossuyt PM, Dekker E. Strategies in screening for colon carcinoma. Neth J Med. 2011;69:112-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 6. | Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9:489-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 496] [Cited by in RCA: 506] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 7. | Wilkins MR, Sanchez JC, Gooley AA, Appel RD, Humphery-Smith I, Hochstrasser DF, Williams KL. Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng Rev. 1996;13:19-50. [PubMed] |
| 8. | Derijks-Engwegen JY, Cats A, Smits ME, Schellens JH, Beijnen JH. Improving colorectal cancer management: the potential of proteomics. Biomark Med. 2008;2:253-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 9. | Palmblad M, Tiss A, Cramer R. Mass spectrometry in clinical proteomics - from the present to the future. Proteomics Clin Appl. 2009;3:6-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 10. | Graves PR, Haystead TA. Molecular biologist’s guide to proteomics. Microbiol Mol Biol Rev. 2002;66:39-63; table of contents. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 357] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 11. | Zhao L, Liu Y, Sun X, Peng K, Ding Y. Serum proteome analysis for profiling protein markers associated with lymph node metastasis in colorectal carcinoma. J Comp Pathol. 2011;144:187-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 12. | Lim SR, Gooi BH, Singh M, Gam LH. Analysis of differentially expressed proteins in colorectal cancer using hydroxyapatite column and SDS-PAGE. Appl Biochem Biotechnol. 2011;165:1211-1224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 13. | Yao L, Lao W, Zhang Y, Tang X, Hu X, He C, Hu X, Xu LX. Identification of EFEMP2 as a serum biomarker for the early detection of colorectal cancer with lectin affinity capture assisted secretome analysis of cultured fresh tissues. J Proteome Res. 2012;11:3281-3294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 14. | Görg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;4:3665-3685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1331] [Cited by in RCA: 1184] [Article Influence: 56.4] [Reference Citation Analysis (0)] |
| 15. | Rodríguez-Piñeiro AM, Álvarez-Chaver P, Martínez-Zorzano VS, Rodríguez-Berrocal FJ, Páez de la Cadena M. Relevance of protein isoforms in proteomic studies. Curr Proteomics. 2007;4:235-252. [RCA] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 16. | O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007-4021. [PubMed] |
| 17. | Xavier T, Ganesan TS, Menon KN. A simple and efficient method for processing of cell lysates for two-dimensional gel electrophoresis. Electrophoresis. 2010;31:2429-2435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 18. | Chen WT, Chang SC, Ke TW, Chiang HC, Tsai FJ, Lo WY. Identification of biomarkers to improve diagnostic sensitivity of sporadic colorectal cancer in patients with low preoperative serum carcinoembryonic antigen by clinical proteomic analysis. Clin Chim Acta. 2011;412:636-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (1)] |
| 19. | Kim HJ, Kang HJ, Lee H, Lee ST, Yu MH, Kim H, Lee C. Identification of S100A8 and S100A9 as serological markers for colorectal cancer. J Proteome Res. 2009;8:1368-1379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 114] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 20. | Ma Y, Peng J, Liu W, Zhang P, Huang L, Gao B, Shen T, Zhou Y, Chen H, Chu Z. Proteomics identification of desmin as a potential oncofetal diagnostic and prognostic biomarker in colorectal cancer. Mol Cell Proteomics. 2009;8:1878-1890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 84] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 21. | Zhao L, Wang H, Li J, Liu Y, Ding Y. Overexpression of Rho GDP-dissociation inhibitor alpha is associated with tumor progression and poor prognosis of colorectal cancer. J Proteome Res. 2008;7:3994-4003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 62] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 22. | Zhao L, Wang H, Liu C, Liu Y, Wang X, Wang S, Sun X, Li J, Deng Y, Jiang Y. Promotion of colorectal cancer growth and metastasis by the LIM and SH3 domain protein 1. Gut. 2010;59:1226-1235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 110] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 23. | Rodríguez-Piñeiro AM, de la Cadena MP, López-Saco A, Rodríguez-Berrocal FJ. Differential expression of serum clusterin isoforms in colorectal cancer. Mol Cell Proteomics. 2006;5:1647-1657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 24. | Alvarez-Chaver P, Rodríguez-Piñeiro AM, Rodríguez-Berrocal FJ, García-Lorenzo A, Páez de la Cadena M, Martínez-Zorzano VS. Selection of putative colorectal cancer markers by applying PCA on the soluble proteome of tumors: NDK A as a promising candidate. J Proteomics. 2011;74:874-886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 25. | Alfonso P, Cañamero M, Fernández-Carbonié F, Núñez A, Casal JI. Proteome analysis of membrane fractions in colorectal carcinomas by using 2D-DIGE saturation labeling. J Proteome Res. 2008;7:4247-4255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 26. | Yeoh LC, Loh CK, Gooi BH, Singh M, Gam LH. Hydrophobic protein in colorectal cancer in relation to tumor stages and grades. World J Gastroenterol. 2010;16:2754-2763. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 6] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 27. | Bi X, Lin Q, Foo TW, Joshi S, You T, Shen HM, Ong CN, Cheah PY, Eu KW, Hew CL. Proteomic analysis of colorectal cancer reveals alterations in metabolic pathways: mechanism of tumorigenesis. Mol Cell Proteomics. 2006;5:1119-1130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 121] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 28. | Tanaka S, Sakai A, Kimura K, Yoshida H, Fushitani H, Ogata A, Miyamoto A, Fukushima M, Wada A, Tanigawa N. Proteomic analysis of the basic proteins in 5-fluorouracil resistance of human colon cancer cell line using the radical-free and highly reducing method of two-dimensional polyacrylamide gel electrophoresis. Int J Oncol. 2008;33:361-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 29. | Unlü M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18:2071-2077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1671] [Cited by in RCA: 1435] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 30. | Ma Y, Peng J, Huang L, Liu W, Zhang P, Qin H. Searching for serum tumor markers for colorectal cancer using a 2-D DIGE approach. Electrophoresis. 2009;30:2591-2599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 31. | Sawhney S, Stubbs R, Hood K. Reproducibility, sensitivity and compatibility of the ProteoExtract subcellular fractionation kit with saturation labeling of laser microdissected tissues. Proteomics. 2009;9:4087-4092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 32. | Sugihara Y, Taniguchi H, Kushima R, Tsuda H, Kubota D, Ichikawa H, Sakamoto K, Nakamura Y, Tomonaga T, Fujita S. Proteomic-based identification of the APC-binding protein EB1 as a candidate of novel tissue biomarker and therapeutic target for colorectal cancer. J Proteomics. 2012;75:5342-5355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 33. | Wang N, Chen Y, Han Y, Zhao Y, Liu Y, Guo K, Jiang Y. Proteomic analysis shows down-regulations of cytoplasmic carbonic anhydrases, CAI and CAII, are early events of colorectal carcinogenesis but are not correlated with lymph node metastasis. Tumori. 2012;98:783-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 34. | Zhou R, Huang W, Yao Y, Wang Y, Li Z, Shao B, Zhong J, Tang M, Liang S, Zhao X. CA II, a potential biomarker by proteomic analysis, exerts significant inhibitory effect on the growth of colorectal cancer cells. Int J Oncol. 2013;43:611-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 35. | García-Lorenzo A, Rodríguez-Piñeiro AM, Rodríguez-Berrocal FJ, Cadena MP, Martínez-Zorzano VS. Changes on the Caco-2 Secretome through Differentiation Analyzed by 2-D Differential In-Gel Electrophoresis (DIGE). Int J Mol Sci. 2012;13:14401-14420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 36. | Grandjean M, Dieu M, Raes M, Feron O. A new method combining sequential immunoaffinity depletion and differential in gel electrophoresis to identify autoantibodies as cancer biomarkers. J Immunol Methods. 2013;396:23-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 37. | Lin JC. Protein microarrays for cancer diagnostics and therapy. Med Princ Pract. 2010;19:247-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 38. | Ellmark P, Belov L, Huang P, Lee CS, Solomon MJ, Morgan DK, Christopherson RI. Multiplex detection of surface molecules on colorectal cancers. Proteomics. 2006;6:1791-1802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 51] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 39. | Oliveira LA, Artigiani-Neto R, Waisberg DR, Fernandes LC, Lima Fde O, Waisberg J. NM23 protein expression in colorectal carcinoma using TMA (tissue microarray): association with metastases and survival. Arq Gastroenterol. 2010;47:361-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 40. | Murakoshi Y, Honda K, Sasazuki S, Ono M, Negishi A, Matsubara J, Sakuma T, Kuwabara H, Nakamori S, Sata N. Plasma biomarker discovery and validation for colorectal cancer by quantitative shotgun mass spectrometry and protein microarray. Cancer Sci. 2011;102:630-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 41. | Hamdan M, Righetti PG. Proteomics in cancer research. Hoboken, New Jersey: John Wiley & Sons, Inc 2005; 69-126. |
| 42. | Tan HT, Low J, Lim SG, Chung MC. Serum autoantibodies as biomarkers for early cancer detection. FEBS J. 2009;276:6880-6904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 243] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 43. | Cañas B, López-Ferrer D, Ramos-Fernández A, Camafeita E, Calvo E. Mass spectrometry technologies for proteomics. Brief Funct Genomic Proteomic. 2006;4:295-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 71] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 44. | Barderas R, Babel I, Casal JI. Colorectal cancer proteomics, molecular characterization and biomarker discovery. Proteomics Clin Appl. 2010;4:159-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 45. | de Wit M, Fijneman RJ, Verheul HM, Meijer GA, Jimenez CR. Proteomics in colorectal cancer translational research: biomarker discovery for clinical applications. Clin Biochem. 2013;46:466-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 75] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 46. | Roessler M, Rollinger W, Palme S, Hagmann ML, Berndt P, Engel AM, Schneidinger B, Pfeffer M, Andres H, Karl J. Identification of nicotinamide N-methyltransferase as a novel serum tumor marker for colorectal cancer. Clin Cancer Res. 2005;11:6550-6557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 162] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 47. | Roessler M, Rollinger W, Mantovani-Endl L, Hagmann ML, Palme S, Berndt P, Engel AM, Pfeffer M, Karl J, Bodenmüller H. Identification of PSME3 as a novel serum tumor marker for colorectal cancer by combining two-dimensional polyacrylamide gel electrophoresis with a strictly mass spectrometry-based approach for data analysis. Mol Cell Proteomics. 2006;5:2092-2101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 118] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 48. | Thierolf M, Hagmann ML, Pfeffer M, Berntenis N, Wild N, Roeßler M, Palme S, Karl J, Bodenmüller H, Rüschoff J. Towards a comprehensive proteome of normal and malignant human colon tissue by 2-D-LC-ESI-MS and 2-DE proteomics and identification of S100A12 as potential cancer biomarker. Proteomics Clin Appl. 2008;2:11-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 49. | Liao CC, Ward N, Marsh S, Arulampalam T, Norton JD. Mass spectrometry protein expression profiles in colorectal cancer tissue associated with clinico-pathological features of disease. BMC Cancer. 2010;10:410. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 50. | Fan NJ, Gao CF, Wang XL, Zhao G, Liu QY, Zhang YY, Cheng BG. Serum peptidome patterns of colorectal cancer based on magnetic bead separation and MALDI-TOF mass spectrometry analysis. J Biomed Biotechnol. 2012;2012:985020. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 51. | Balluff B, Schöne C, Höfler H, Walch A. MALDI imaging mass spectrometry for direct tissue analysis: technological advancements and recent applications. Histochem Cell Biol. 2011;136:227-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 52. | Mirnezami R, Spagou K, Vorkas PA, Lewis MR, Kinross J, Want E, Shion H, Goldin RD, Darzi A, Takats Z. Chemical mapping of the colorectal cancer microenvironment via MALDI imaging mass spectrometry (MALDI-MSI) reveals novel cancer-associated field effects. Mol Oncol. 2014;8:39-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 96] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 53. | Merchant M, Weinberger SR. Recent advancements in surface-enhanced laser desorption/ionization-time of flight-mass spectrometry. Electrophoresis. 2000;21:1164-1177. [PubMed] |
| 54. | Gemoll T, Roblick UJ, Auer G, Jörnvall H, Habermann JK. SELDI-TOF serum proteomics and colorectal cancer: a current overview. Arch Physiol Biochem. 2010;116:188-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 55. | Liu XP, Shen J, Li ZF, Yan L, Gu J. A serum proteomic pattern for the detection of colorectal adenocarcinoma using surface enhanced laser desorption and ionization mass spectrometry. Cancer Invest. 2006;24:747-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 44] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 56. | Helgason HH, Engwegen JY, Zapatka M, Vincent A, Cats A, Boot H, Beijnen JH, Schellens JH. Identification of serum proteins as prognostic and predictive markers of colorectal cancer using surface enhanced laser desorption ionization-time of flight mass spectrometry. Oncol Rep. 2010;24:57-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 57. | Sihlbom C, Kanmert I, Bahr Hv, Davidsson P. Evaluation of the combination of bead technology with SELDI-TOF-MS and 2-D DIGE for detection of plasma proteins. J Proteome Res. 2008;7:4191-4198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 60] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 58. | Melle C, Osterloh D, Ernst G, Schimmel B, Bleul A, von Eggeling F. Identification of proteins from colorectal cancer tissue by two-dimensional gel electrophoresis and SELDI mass spectrometry. Int J Mol Med. 2005;16:11-17. [PubMed] |
| 59. | Melle C, Ernst G, Schimmel B, Bleul A, Mothes H, Kaufmann R, Settmacher U, Von Eggeling F. Different expression of calgizzarin (S100A11) in normal colonic epithelium, adenoma and colorectal carcinoma. Int J Oncol. 2006;28:195-200. [PubMed] |
| 60. | Ward DG, Nyangoma S, Joy H, Hamilton E, Wei W, Tselepis C, Steven N, Wakelam MJ, Johnson PJ, Ismail T. Proteomic profiling of urine for the detection of colon cancer. Proteome Sci. 2008;6:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 54] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 61. | Hamdan M, Righetti PG. Current strategies for protein quantification. Hoboken, New Jersey: John Wiley & Sons Inc 2005; 69-126. |
| 62. | Kim HJ, Kang UB, Lee H, Jung JH, Lee ST, Yu MH, Kim H, Lee C. Profiling of differentially expressed proteins in stage IV colorectal cancers with good and poor outcomes. J Proteomics. 2012;75:2983-2997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 63. | Ghosh D, Li Z, Tan XF, Lim TK, Mao Y, Lin Q. iTRAQ based quantitative proteomics approach validated the role of calcyclin binding protein (CacyBP) in promoting colorectal cancer metastasis. Mol Cell Proteomics. 2013;12:1865-1880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 64. | Yang HY, Kwon J, Park HR, Kwon SO, Park YK, Kim HS, Chung YJ, Chang YJ, Choi HI, Chung KJ. Comparative proteomic analysis for the insoluble fractions of colorectal cancer patients. J Proteomics. 2012;75:3639-3653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 65. | Matsubara J, Honda K, Ono M, Sekine S, Tanaka Y, Kobayashi M, Jung G, Sakuma T, Nakamori S, Sata N. Identification of adipophilin as a potential plasma biomarker for colorectal cancer using label-free quantitative mass spectrometry and protein microarray. Cancer Epidemiol Biomarkers Prev. 2011;20:2195-2203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 66. | Kang UB, Yeom J, Kim HJ, Kim H, Lee C. Expression profiling of more than 3500 proteins of MSS-type colorectal cancer by stable isotope labeling and mass spectrometry. J Proteomics. 2012;75:3050-3062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 67. | Nibbe RK, Chance MR. Approaches to biomarkers in human colorectal cancer: looking back, to go forward. Biomark Med. 2009;3:385-396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 68. | Albrethsen J, Bøgebo R, Gammeltoft S, Olsen J, Winther B, Raskov H. Upregulated expression of human neutrophil peptides 1, 2 and 3 (HNP 1-3) in colon cancer serum and tumours: a biomarker study. BMC Cancer. 2005;5:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 121] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 69. | Ward DG, Suggett N, Cheng Y, Wei W, Johnson H, Billingham LJ, Ismail T, Wakelam MJ, Johnson PJ, Martin A. Identification of serum biomarkers for colon cancer by proteomic analysis. Br J Cancer. 2006;94:1898-1905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 163] [Article Influence: 8.6] [Reference Citation Analysis (1)] |
| 70. | Rodríguez-Piñeiro AM, Ayude D, Rodríguez-Berrocal FJ, Páez de la Cadena M. Concanavalin A chromatography coupled to two-dimensional gel electrophoresis improves protein expression studies of the serum proteome. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;803:337-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 71. | Xue H, Lü B, Zhang J, Wu M, Huang Q, Wu Q, Sheng H, Wu D, Hu J, Lai M. Identification of serum biomarkers for colorectal cancer metastasis using a differential secretome approach. J Proteome Res. 2010;9:545-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 140] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 72. | Hamelin C, Cornut E, Poirier F, Pons S, Beaulieu C, Charrier JP, Haïdous H, Cotte E, Lambert C, Piard F. Identification and verification of heat shock protein 60 as a potential serum marker for colorectal cancer. FEBS J. 2011;278:4845-4859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 73. | Zhang X, Xiao Z, Liu X, Du L, Wang L, Wang S, Zheng N, Zheng G, Li W, Zhang X. The potential role of ORM2 in the development of colorectal cancer. PLoS One. 2012;7:e31868. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 74. | Zhu D, Wang J, Ren L, Li Y, Xu B, Wei Y, Zhong Y, Yu X, Zhai S, Xu J. Serum proteomic profiling for the early diagnosis of colorectal cancer. J Cell Biochem. 2013;114:448-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 75. | Ahn YH, Kim KH, Shin PM, Ji ES, Kim H, Yoo JS. Identification of low-abundance cancer biomarker candidate TIMP1 from serum with lectin fractionation and peptide affinity enrichment by ultrahigh-resolution mass spectrometry. Anal Chem. 2012;84:1425-1431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 76. | Wang J, Wang X, Lin S, Chen C, Wang C, Ma Q, Jiang B. Identification of kininogen-1 as a serum biomarker for the early detection of advanced colorectal adenoma and colorectal cancer. PLoS One. 2013;8:e70519. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 77. | Brock R, Xiong B, Li L, Vanbogelen RA, Christman L. A multiplex serum protein assay for determining the probability of colorectal cancer. Am J Cancer Res. 2012;2:598-605. [PubMed] |
| 78. | Babel I, Barderas R, Díaz-Uriarte R, Martínez-Torrecuadrada JL, Sánchez-Carbayo M, Casal JI. Identification of tumor-associated autoantigens for the diagnosis of colorectal cancer in serum using high density protein microarrays. Mol Cell Proteomics. 2009;8:2382-2395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 137] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 79. | Zhu D, Zhong Y, Wu H, Ye L, Wang J, Li Y, Wei Y, Ren L, Xu B, Xu J. Predicting metachronous liver metastasis from colorectal cancer using serum proteomic fingerprinting. J Surg Res. 2013;184:861-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 80. | Alfonso P, Núñez A, Madoz-Gurpide J, Lombardia L, Sánchez L, Casal JI. Proteomic expression analysis of colorectal cancer by two-dimensional differential gel electrophoresis. Proteomics. 2005;5:2602-2611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 151] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 81. | Alvarez-Chaver P, Rodríguez-Piñeiro AM, Rodríguez-Berrocal FJ, Martínez-Zorzano VS, Páez de la Cadena M. Identification of hydrophobic proteins as biomarker candidates for colorectal cancer. Int J Biochem Cell Biol. 2007;39:529-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 82. | Watanabe M, Takemasa I, Kawaguchi N, Miyake M, Nishimura N, Matsubara T, Matsuo E, Sekimoto M, Nagai K, Matsuura N. An application of the 2-nitrobenzenesulfenyl method to proteomic profiling of human colorectal carcinoma: A novel approach for biomarker discovery. Proteomics Clin Appl. 2008;2:925-935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 83. | Jimenez CR, Knol JC, Meijer GA, Fijneman RJ. Proteomics of colorectal cancer: overview of discovery studies and identification of commonly identified cancer-associated proteins and candidate CRC serum markers. J Proteomics. 2010;73:1873-1895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 85] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 84. | Friedman DB, Hill S, Keller JW, Merchant NB, Levy SE, Coffey RJ, Caprioli RM. Proteome analysis of human colon cancer by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics. 2004;4:793-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 275] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 85. | Conrotto P, Roesli C, Rybak J, Kischel P, Waltregny D, Neri D, Castronovo V. Identification of new accessible tumor antigens in human colon cancer by ex vivo protein biotinylation and comparative mass spectrometry analysis. Int J Cancer. 2008;123:2856-2864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 54] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 86. | Kim H, Kang HJ, You KT, Kim SH, Lee KY, Kim TI, Kim C, Song SY, Kim HJ, Lee C. Suppression of human selenium-binding protein 1 is a late event in colorectal carcinogenesis and is associated with poor survival. Proteomics. 2006;6:3466-3476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 88] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 87. | Ma YL, Peng JY, Zhang P, Huang L, Liu WJ, Shen TY, Chen HQ, Zhou YK, Zhang M, Chu ZX. Heterogeneous nuclear ribonucleoprotein A1 is identified as a potential biomarker for colorectal cancer based on differential proteomics technology. J Proteome Res. 2009;8:4525-4535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 88. | Besson D, Pavageau AH, Valo I, Bourreau A, Bélanger A, Eymerit-Morin C, Moulière A, Chassevent A, Boisdron-Celle M, Morel A. A quantitative proteomic approach of the different stages of colorectal cancer establishes OLFM4 as a new nonmetastatic tumor marker. Mol Cell Proteomics. 2011;10:M111.009712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 89. | Kang B, Hao C, Wang H, Zhang J, Xing R, Shao J, Li W, Xu N, Lu Y, Liu S. Evaluation of hepatic-metastasis risk of colorectal cancer upon the protein signature of PI3K/AKT pathway. J Proteome Res. 2008;7:3507-3515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 90. | Wiśniewski JR, Duś K, Mann M. Proteomic workflow for analysis of archival formalin-fixed and paraffin-embedded clinical samples to a depth of 10 000 proteins. Proteomics Clin Appl. 2013;7:225-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 113] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 91. | Ang CS, Phung J, Nice EC. The discovery and validation of colorectal cancer biomarkers. Biomed Chromatogr. 2011;25:82-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 92. | Mulder SA, van Leerdam ME, van Vuuren AJ, Francke J, van Toorenenbergen AW, Kuipers EJ, Ouwendijk RJ. Tumor pyruvate kinase isoenzyme type M2 and immunochemical fecal occult blood test: performance in screening for colorectal cancer. Eur J Gastroenterol Hepatol. 2007;19:878-882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 93. | Karl J, Wild N, Tacke M, Andres H, Garczarek U, Rollinger W, Zolg W. Improved diagnosis of colorectal cancer using a combination of fecal occult blood and novel fecal protein markers. Clin Gastroenterol Hepatol. 2008;6:1122-1128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 94. | Ang CS, Rothacker J, Patsiouras H, Gibbs P, Burgess AW, Nice EC. Use of multiple reaction monitoring for multiplex analysis of colorectal cancer-associated proteins in human feces. Electrophoresis. 2011;32:1926-1938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 95. | Buhrke T, Lengler I, Lampen A. Analysis of proteomic changes induced upon cellular differentiation of the human intestinal cell line Caco-2. Dev Growth Differ. 2011;53:411-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 96. | Stierum R, Gaspari M, Dommels Y, Ouatas T, Pluk H, Jespersen S, Vogels J, Verhoeckx K, Groten J, van Ommen B. Proteome analysis reveals novel proteins associated with proliferation and differentiation of the colorectal cancer cell line Caco-2. Biochim Biophys Acta. 2003;1650:73-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 91] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 97. | Fanayan S, Smith JT, Lee LY, Yan F, Snyder M, Hancock WS, Nice E. Proteogenomic analysis of human colon carcinoma cell lines LIM1215, LIM1899, and LIM2405. J Proteome Res. 2013;12:1732-1742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 98. | Ghosh D, Yu H, Tan XF, Lim TK, Zubaidah RM, Tan HT, Chung MC, Lin Q. Identification of key players for colorectal cancer metastasis by iTRAQ quantitative proteomics profiling of isogenic SW480 and SW620 cell lines. J Proteome Res. 2011;10:4373-4387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 99. | Lin Q, Tan HT, Lim HS, Chung MC. Sieving through the cancer secretome. Biochim Biophys Acta. 2013;1834:2360-2371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 100. | Malard V, Chardan L, Roussi S, Darolles C, Sage N, Gaillard JC, Armengaud J. Analytical constraints for the analysis of human cell line secretomes by shotgun proteomics. J Proteomics. 2012;75:1043-1054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 101. | Wu CC, Chen HC, Chen SJ, Liu HP, Hsieh YY, Yu CJ, Tang R, Hsieh LL, Yu JS, Chang YS. Identification of collapsin response mediator protein-2 as a potential marker of colorectal carcinoma by comparative analysis of cancer cell secretomes. Proteomics. 2008;8:316-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 102. | Diehl HC, Stühler K, Klein-Scory S, Volmer MW, Schöneck A, Bieling C, Schmiegel W, Meyer HE, Schwarte-Waldhoff I. A catalogue of proteins released by colorectal cancer cells in vitro as an alternative source for biomarker discovery. Proteomics Clin Appl. 2007;1:47-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 103. | Weiss JV, Klein-Scory S, Kübler S, Reinacher-Schick A, Stricker I, Schmiegel W, Schwarte-Waldhoff I. Soluble E-cadherin as a serum biomarker candidate: elevated levels in patients with late-stage colorectal carcinoma and FAP. Int J Cancer. 2011;128:1384-1392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 104. | Albrethsen J, Knol JC, Piersma SR, Pham TV, de Wit M, Mongera S, Carvalho B, Verheul HM, Fijneman RJ, Meijer GA. Subnuclear proteomics in colorectal cancer: identification of proteins enriched in the nuclear matrix fraction and regulation in adenoma to carcinoma progression. Mol Cell Proteomics. 2010;9:988-1005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 105. | Chen JS, Chen KT, Fan CW, Han CL, Chen YJ, Yu JS, Chang YS, Chien CW, Wu CP, Hung RP. Comparison of membrane fraction proteomic profiles of normal and cancerous human colorectal tissues with gel-assisted digestion and iTRAQ labeling mass spectrometry. FEBS J. 2010;277:3028-3038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 49] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 106. | Han CL, Chen JS, Chan EC, Wu CP, Yu KH, Chen KT, Tsou CC, Tsai CF, Chien CW, Kuo YB. An informatics-assisted label-free approach for personalized tissue membrane proteomics: case study on colorectal cancer. Mol Cell Proteomics. 2011;10:M110.003087. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 107. | Luque-García JL, Martínez-Torrecuadrada JL, Epifano C, Cañamero M, Babel I, Casal JI. Differential protein expression on the cell surface of colorectal cancer cells associated to tumor metastasis. Proteomics. 2010;10:940-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 75] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 108. | Choi DS, Lee JM, Park GW, Lim HW, Bang JY, Kim YK, Kwon KH, Kwon HJ, Kim KP, Gho YS. Proteomic analysis of microvesicles derived from human colorectal cancer cells. J Proteome Res. 2007;6:4646-4655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 159] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 109. | Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics. 2010;9:197-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 415] [Cited by in RCA: 454] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 110. | Ji H, Greening DW, Barnes TW, Lim JW, Tauro BJ, Rai A, Xu R, Adda C, Mathivanan S, Zhao W. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics. 2013;13:1672-1686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 279] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 111. | Hung KE, Faca V, Song K, Sarracino DA, Richard LG, Krastins B, Forrester S, Porter A, Kunin A, Mahmood U. Comprehensive proteome analysis of an Apc mouse model uncovers proteins associated with intestinal tumorigenesis. Cancer Prev Res (Phila). 2009;2:224-233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 112. | Zhu W, Fang C, Gramatikoff K, Niemeyer CC, Smith JW. Proteins and an inflammatory network expressed in colon tumors. J Proteome Res. 2011;10:2129-2139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 113. | Ivancic MM, Huttlin EL, Chen X, Pleiman JK, Irving AA, Hegeman AD, Dove WF, Sussman MR. Candidate serum biomarkers for early intestinal cancer using 15N metabolic labeling and quantitative proteomics in the ApcMin/+ mouse. J Proteome Res. 2013;12:4152-4166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 114. | Duffy MJ, van Dalen A, Haglund C, Hansson L, Holinski-Feder E, Klapdor R, Lamerz R, Peltomaki P, Sturgeon C, Topolcan O. Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur J Cancer. 2007;43:1348-1360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 342] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 115. | Roblick UJ, Bader FG, Lenander C, Hellman U, Zimmermann K, Becker S, Ost A, Alaiya A, Bruch HP, Keller R. Undifferentiated pelvic adenocarcinomas: diagnostic potential of protein profiling and multivariate analysis. Int J Colorectal Dis. 2008;23:483-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 116. | Rodríguez-Piñeiro AM, Álvarez-Chaver P, Rodríguez-Berrocal FJ, Páez de la Cadena M, Martínez-Zorzano VS. Multivariate methods aid in pinpointing promising tumor marker candidates from colorectal biopsies. J Integr OMICS. 2012;2:24-30. [RCA] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 117. | Warburg O. On the origin of cancer cells. Science. 1956;123:309-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9117] [Cited by in RCA: 9909] [Article Influence: 143.6] [Reference Citation Analysis (0)] |