Published online Mar 21, 2014. doi: 10.3748/wjg.v20.i11.2902
Revised: October 23, 2013
Accepted: January 14, 2014
Published online: March 21, 2014
Processing time: 175 Days and 9.3 Hours
Some direct-acting antiviral agents for hepatitis C virus (HCV), such as telaprevir and boceprevir have been available since 2011. It was reported that HCV NS5A is associated with interferon signaling related to HCV replication and hepatocarcinogenesis. HCV NS5A inhibitors efficiently inhibited HCV replication in vitro. Human studies showed that dual, triple and quad regimens with HCV NS5A inhibitors, such as daclatasvir and ledipasvir, in combination with other direct-acting antiviral agents against other regions of HCV with or without peginterferon/ribavirin, could efficiently inhibit HCV replication according to HCV genotypes. These combinations might be a powerful tool for “difficult-to-treat” HCV-infected patients. “First generation” HCV NS5A inhibitors such as daclatasvir, ledipasvir and ABT-267, which are now in phase III clinical trials, could result in resistance mutations. “Second generation” NS5A inhibitors such as GS-5816, ACH-3102, and MK-8742, have displayed improvements in the genetic barrier while maintaining potency. HCV NS5A inhibitors are safe at low concentrations, which make them attractive for use despite low genetic barriers, although, in fact, HCV NS5A inhibitors should be used with HCV NS3/4A inhibitors, HCV NS5B inhibitors or peginterferon plus ribavirin. This review article describes HCV NS5A inhibitor resistance mutations and recommends that HCV NS5A inhibitors be used in combination regimens potent enough to prevent the emergence of resistant variants.
Core tip: Hepatitis C virus (HCV) NS5A inhibitors such as daclatasvir and ledipasvir in combination with other direct-acting antiviral agents against other regions of HCV with or without peginterferon/ribavirin are becoming available for daily clinical practice. These inhibitors can induce resistance mutations more easily in HCV genotype 1a patients than in HCV genotype 1b patients. HCV NS5A inhibitors should be used in combination regimens potent enough to prevent the emergence of resistant mutants and attention should be paid to these mutants’ potential emergence.
- Citation: Nakamoto S, Kanda T, Wu S, Shirasawa H, Yokosuka O. Hepatitis C virus NS5A inhibitors and drug resistance mutations. World J Gastroenterol 2014; 20(11): 2902-2912
- URL: https://www.wjgnet.com/1007-9327/full/v20/i11/2902.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i11.2902
Hepatitis C virus (HCV) infection is a leading cause of hepatocellular carcinoma (HCC) in Japan[1-3] and is one of the major causes of end-stage liver disease, HCC and liver transplantation in the United States and Europe[4,5]. A sustained virological response, defined as undetectable HCV RNA at week 24 (SVR24) after stopping combination therapy with peginterferon plus ribavirin for 48 wk in HCV genotype 1 and for 24 wk in HCV genotype 2, is achieved in approximately 50% and 80% of patients, respectively[6-9].
HCV genomes are translated into a single open reading frame of approximately 3011 amino acids after HCV infects hepatocytes. This protein is made into structural (core, E1, E2 and p7) and nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A and NS5B) by HCV-encoding and cellular proteases[9,10]. HCV NS5A is a multifunctional phosphoprotein required for HCV RNA replication and virus assembly[11,12]. HCV NS5A has no known enzymatic activities and its precise role in the HCV life cycle is not yet fully understood; however, there have been several reports concerning the association between HCV NS5A, interferon signaling and hepatocarcinogenesis[13-21].
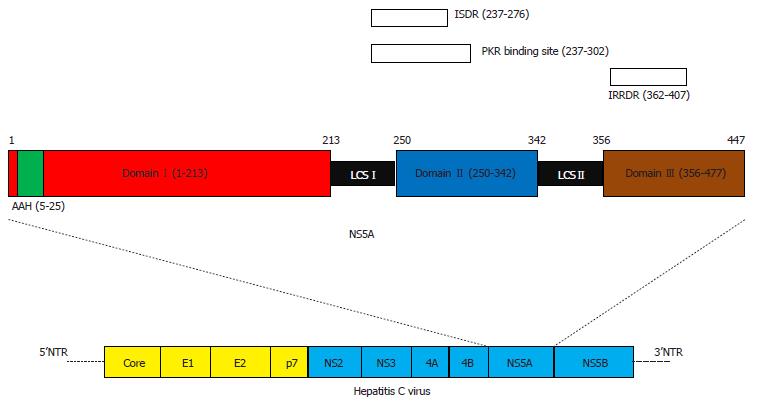
HCV NS5A is an approximately 447-amino acid protein with an N-terminal amphipathic alpha helix (amino acids 5-25) and 3 structural Domains (I, II and III; Figure 1)[22]. Domain I (amino acids 28-213) contains Zn2+-binding and RNA-binding motifs and has been crystallized as a dimer, which is essential for HCV replication[22-24]. Domains II (amino acids 250-342) and III (amino acids 356-447) appear unfolded[25]. Domain II has been linked to RNA replication, while Domain III is important for virus assembly. Both an interferon sensitivity-determining region and an interferon/ribavirin resistance-determining region exist in Domains II-III (Figure 1)[26-35].
Daclatasvir (DCV, formerly called BMS-790052) is a first-in-class HCV NS5A inhibitor with pmol/L potency and broad genotype coverage in vitro; DCV is currently the most studied among this class of inhibitors. The inhibitor was discovered through a screening hit with BMS-858, iminothiazolidinone, using a high-throughput cell-based HCV replicon assay. BMS-858 is a weak but specific inhibitor of HCV RNA replication for which resistance was mapped to the N-terminal of the HCV NS5A protein, indicating HCV NS5A protein as its target[36]. After chemical refinements to improve potency and HCV genotype coverage and the identification of symmetry as an important antiviral activity factor, DCV was identified as a candidate for clinical trials[36,37]. Its precise mode of action remains unclear, but DCV seems to bind with HCV NS5A at NS5A’s dimer interface based on current models of the HCV NS5A structure and analysis of drug resistant mutants[23,24]. DCV induces an alteration in the subcellular localization of HCV NS5A, which inhibits the formation and activation of HCV replication complexes[38], and DCV blocks intracellular HCV RNA synthesis, virus assembly and secretion[39]. HCV NS5A inhibitors are safe at low concentrations, which make them attractive for use despite low genetic barriers, although, in fact, HCV NS5A inhibitors should be used with HCV NS3/4A, NS5B inhibitors or peginterferon plus ribavirin. It seems to be relatively weak in HCV genotypes other than genotype 1b[40,41]. Many trials using direct acting antiviral agents (DAAs) including HCV NS5A inhibitors are currently underway, and the exact resistance profile is becoming apparent[42-49]. In this review, resistance-associated variants (RAVs) against HCV NS5A inhibitors and their correlations to clinical studies, as well as the difference in efficacy among HCV genotypes, are discussed.
Gao et al[36] reported that a chemical genetic approach identified an HCV NS5A inhibitor with a potent clinical effect. However, the in vitro resistance profile of DCV has been reported in detail, using the HCV replicon system, the HCV cell culture-adaptive virus system and human hepatocyte chimeric mice (Table 1)[36,40,50-53].
| EC50 | < 10 pmol/L | < 100 pmol/L | < 1 nmol/L | < 10 nmol/L | < 100 nmol/L | < 1 μmol/L | > 1 μmol/L |
| DCV | |||||||
| HCV GT | |||||||
| 1b | Wild (2.6 pmol/L) | R30E, H | 23F/93H | 31F, M, V/93H | Δ30/32L | ||
| L28M | L31F, V | 30Q/31F | 30Q/31M/93H | ||||
| L31M | P32L | 31V/58S | |||||
| R30Q | Y93H, N | 30H/31M | |||||
| 37L or 54H/93H | |||||||
| 23F/31F | |||||||
| 1a | Wild (6 pmol/L) | M28T | L31V | Q30E, K | 31V/93H | ||
| Q30H, R | Y93C, H | Y93N (> 500 nmol/L) | |||||
| L31M | 28T/30H | ||||||
| P32L | 30H/93H | ||||||
| H58D | 30R/93C | ||||||
| 30R/62D | |||||||
| 2-6 | GT2a (JFH1) | GT3a | GT2a (L31M) | GT2a (Y93H) | GT2a (F28S) | ||
| GT4a, 5a, 6a | GT2a (C92R) | GT2b (31M) | GT3a (Y93H) | ||||
| GT3a (A30K) | GT4a (L30I/Y93R) | ||||||
| GT3a (L31F) | |||||||
| GT4a (R30G) | |||||||
| GT4a (L30H) | |||||||
| ACH-3102 | |||||||
| HCV GT | |||||||
| 1b | Wild (7 pmol/L) | Y93H | P58S/Y93H | ||||
| L31V | 31V/93H | P58S/T64A/Y93H | |||||
| 1a | wild (20 pmol/L) | Q30R, E, K | Y93C | Y93H, N1 | |||
| Q30H | M28T | 28T/30H/93C1 | |||||
| L31M, V | P32L | ||||||
| H58D | |||||||
| 2-6 | GT2a (JFH1) | ||||||
| GT2a (L31M) | |||||||
| GT2b (31M) | |||||||
| GT3a, 4a, 5a, 6a |
Resistance mutations have been mapped to the N-terminal region of HCV NS5A (the first 100 amino acids within Domain I). The primary mutation sites are M28T, Q30E/H/R, L31M/V, P32L, and Y93C/H/N for HCV genotype 1a, and L31F/V, P32L, and Y93H/N for HCV genotype 1b. L23F, R30Q, and P58S in HCV genotype 1b acted as secondary resistance mutations. The genetic barrier to resistance is lower for HCV genotype 1a than for HCV genotype 1b (Table 1). As single mutations, Q30E and Y93N in HCV genotype 1a conferred the highest levels of resistance, while for HCV genotype 1b, DCV retained sub-nanomolar potency against all variants with single amino acid substitutions. However, a linked resistance mutation remarkably decreased inhibitor susceptibility for both HCV genotypes 1a and 1b. These substitutions also confer cross-resistance to other HCV NS5A inhibitors. DCV-resistant variants remained fully sensitive to interferon-alpha and other classes of DAAs such as HCV NS3/4A and HCV NS5B inhibitors. Pelosi et al[54] reported that combinations of double and triple inhibitors could generate resistance pathways that differ from those developed during HCV NS5A inhibitor monotherapy.
In HCV genotype 2a, HCV NS5A F28S, L31M, C92R, and Y93H were the major resistance mutations[55]. Residue 30 acted as a compensatory mutation, enhancing viral replication and modulating inhibitor sensitivity. The majority of HCV genotype 2a sequences stored in the European HCV database contain methionine at HCV NS5A residue 31[56], which showed 140-fold resistance [50% effective concentrations (EC50) 6.9 nmol/L] compared to the HCV genotype 2a strain JFH1 wild-type replicon. In HCV genotype 3a, DCV showed sub-nanomolar potency, with EC50 ranging from 120 to 870 pmol/L[57]. HCV NS5A residues 31 and 93 were also detected as sites for DCV-selected resistance in HCV genotype 3a. DCV retained pmol/L potency against the HCV genotype 4 replicon (EC50 7-13 pmol/L). HCV NS5A residue 30 was an important site for DCV-selected resistance; R30G and L30H conferred an EC50 of > 10 nmol/L. L30I-Y93R showed an EC50 of > 100 nmol/L[58].
In addition to DCV, ledipasvir (LDV, formerly GS-5885) and ABT-267 are entering phase III clinical study[59], and GSK-2336805 is currently in a phase II trial[60]. LDV has a similar potency and resistance profile to that of DCV. Y93N conferred the highest resistance (EC50 > 500 nmol/L) as a single mutation[61]. ABT-267 also exhibited pmol/L potency and broad HCV genotype coverage with a low barrier to resistance[59,62]. “Second generation” HCV NS5A inhibitors such as GS-5816, ACH-3102, and MK-8742, displayed improved potency against the resistant variants selected by the “first generation” HCV NS5A inhibitors, including HCV genotype 1a-Q30E and L31V[63-65]. ACH-3102 has sub-nanomolar potency to HCV genotype 1b-combined variants such as 31V-93H as well as many HCV genotype 1a single variants, while the HCV genotype 1a-Y93 variant confers high-level resistance (Table 1).
Analyses of the sequence variants in a 14-d DCV monotherapy study revealed a correlation between resistant variants emerging in vivo with DCV treatment and those observed in the in vitro HCV replicon system (major substitutions at residues 28, 30, 31, and 93 for HCV genotype 1a, and residues 31 and 93 for HCV genotype 1b)[41,66]. Generally, HCV genotype 1b responded better to DCV than HCV genotype 1a. The primary difference in the resistance patterns observed in vitro and in vivo was the increased complexity of linked variant combinations observed in clinical samples[67].
The influence of natural baseline polymorphisms at positions involved in drug resistance within the HCV genome has been reported (Table 2). In an HCV genotype 1a patient with Q30R, 14-d DCV treatment at 60 mg exhibited the maximal response with a 2.9 log decrease in HCV RNA, while the mean HCV decrease in this study group was 3.8 logIU/mL[66]. The natural prevalence of Q30R in HCV genotype 1a is reported to be 1% (Figure 2)[40,55-58,68-72]. Patients with high baseline HCV genotype 1a resistant variants Q30E or L31M responded poorly to LDV.
| HCV NS5A inhibitor | Mean maximal viral decline (log IU/mL) | Maximal viral decline in patients with RAVs at baseline (log IU/mL) | |
| HCV genotype 1a-infected | |||
| DCV | 3.8 (60 mg, 14 d) | Q30R (10%) | 2.9 |
| LDV | 3.1-3.3 (10-90 mg, 3 d) | Q30E/Q | 0.88 |
| L31M | 0.16 | ||
| Y93C (12%) | 1.6 | ||
| PPI-668 | 3.3 (80-160 mg, 3 d) | M28V (50%) | 3.7 |
| M28T (7%) | 2.8 | ||
| M28T (10%)/L31M (11%) | 3.6 | ||
| H58D (69%)/N (31%) | 2.2 | ||
| IDX719 | 3.2-3.6 (25-100 mg, 3 d) | M28M/V | 3.6 |
| ACH-3102 | 3.5-3.9 (50-300 mg, single dose) | M28V, T (2%-24%) | 3.4 to 4 |
| L31M (28%) | 3.4 | ||
| Y93C, D, H (2%-3%) | 4 to 4.6 | ||
| HCV genotype 1b-infected | |||
| DCV | 4.3 (10 mg, 14 d) | Q54H, N | > 4 |
| Q54H/Y93H | > 4 | ||
| LDV | 3.3 (10 mg, 3 d) | L31M | 2.09 |
| PPI-668 | 2.9 (40 mg, 3 d; only 1 patient) | R30Q | 2.9 |
| 3.8-4 (80-240 mg, 3 d) | L31M | 4 | |
| P58S | 3.8 | ||
| L28M (7%), R30Q (76%), L31M (20%) | 3.5 | ||
| R30Q/L31I/Y93H | 0.33 | ||
| IDX-719 | 3-4.3 (25-50 mg, 3 d) | R30Q/Y93H | 2.8 |
| HCV genotype 2a-infected | |||
| PPI-668 | 0.33 (160 mg, 3 d; only 1 patient) | F28L/A30K/L31M | 0.33 |
| IDX-719 | 2.0 (50-100 mg, 3 d) | L31M | 0.45 |
| L31L/M | 0.85 | ||
| HCV genotype 2b-infected | |||
| PPI-668 | 3.0 (160 mg, 3 d) | A30K | 0.48 |
| Y93H (7%) | 0.45 | ||
| Y93H | 0.25 | ||
In HCV genotype 1b, the natural prevalence of L31M or Y93H is 4%-8% according to the HCV database, and these variants are observed at a higher frequency than HCV genotype 1a variants. However, the resistance levels of HCV genotype 1b single variants are relatively low compared to those of HCV genotype 1a variants (Table 1). Low-level resistant variants such as R30Q and Q54H-Y93H in HCV genotype 1b responded well to DCV treatment, while the combined variants R30Q-L31I-Y93H responded poorly to PPI-668 (Table 2)[73].
Few studies have examined HCV genotype 2 patients. IDX-719 exhibited a mean maximal viral load reduction of 2 logIU/mL, while patients with a pre-existing resistance substitution (L31M in HCV NS5A at baseline) responded poorly (Table 2). Indeed, the HCV genotype 2a M31 variant was less sensitive to IDX-719 (EC50 1.8 nmol/L) compared to the HCV wild-type L31 replicon (EC50 0.024 nmol/L)[74]. In the HCV database, the most prevalent amino acid at residue 31 is methionine, indicating that HCV genotype 2a may respond poorly to DCV. In HCV genotype 3a patients, A30K or Y93H conferred high-level resistance to PPI-668 (Table 2). These data indicate that the in vitro resistance profile correlates with the in vivo HCV NS5A inhibitor monotherapy efficacy. As for the “second generation” HCV NS5A inhibitor ACH-3102, potency was not attenuated at least in patients having a minor prevalence of M28V, L31M or Y93 variants (approximately 30%)[75].
Long-term persistence of HCV NS5A resistance polymorphisms was observed following 14-d DCV monotherapy and preserved for up to 6 mo. Viral fitness, rather than DCV resistance, may determine which viral variants emerge as dominant in populations[67]. In 3-d monotherapy treatment of patients with LDV[61,76], HCV NS5A resistance polymorphisms, present at baseline or selected during LDV treatment, persisted in 100% and 50% of HCV genotype 1a- and 1b-infected patients, respectively, at 48 wk following treatment cessation. These data indicated that in contrast to HCV NS3 resistant variants to HCV NS3/4A inhibitors, those of HCV NS5A can fit well instead of HCV wild-type. The data also highlighted the need to use HCV NS5A inhibitor in combination with other DAAs or interferon to avoid producing drug-resistant virus. A baseline polymorphism with a minimal effect on DCV’s anti-HCV effect can affect the emergence of resistance. E62D at baseline did not contribute to DCV resistance; however, the linked variant, Q30R-E62D, conferred high-level resistance in vitro and is likely responsible for a viral breakthrough in vivo[77]. The pattern of resistant variants and the level of resistance observed varied depending on the selective pressure; 14-d treatment with low-dose DCV (1 mg) selected relatively low-level resistant variants, such as Q30R/H and M28T, while treatment with high-dose DCV (60 mg) selected high-level resistant variants, such as Q30E and Y93N, and linked variants, such as 28T-30H[66].
In vitro studies showed that DCV-resistant variants remained fully sensitive to other classes of DAAs, such as protease inhibitor and interferon. The COMMAND-1 study combining DCV with peginterferon-alpha and ribavirin revealed that SVR24 rates are lower in patients infected with HCV genotype 1a than in patients infected with HCV genotype 1b[78], which is consistent with the in vitro data. In a 24-wk dual-oral therapy with DCV and asunaprevir (ASV) in HCV genotype 1b-infected Japanese patients, 90.5% of null responders and 63.6% of patients ineligible for or intolerant of peginterferon-alpha and ribavirin achieved SVR24[79]. Of interest, many patients in this study with pre-existing resistance-associated HCV NS5A polymorphisms were cured of their chronic HCV infection[80]. The presence of DCV or ASV RAVs at baseline was no longer a strong predictor of treatment failure in HCV genotype 1b patients receiving dual therapy[44].
In line with these results, the AAI-447-011 US/French study demonstrated that a dual regimen in HCV genotype 1b null responders resulted in SVR12, defined as undetectable HCV RNA at week 12 after treatment cessation, in 78% with ASV 200 mg bid regimens and 65% with qid regimens. Among 8 patients with viral breakthrough, 5 had baseline HCV NS5A resistant variants[81]. At viral breakthrough, 7 patients had RAVs in both the HCV NS5A and HCV NS3 regions. In 1 relapsed patient, no baseline variants were detected. At relapse, both HCV NS5A and HCV NS3 RAVs were observed.
On the other hand, in an AAI-447-011 sentinel cohort, 6 of 9 HCV genotype 1a patients receiving dual therapy had viral breakthrough[82]. No baseline variants were detected and resistance mutations to both antiviral agents were found in all cases at the time of viral breakthrough. In these cases, the following high-level HCV NS5A-resistant variants were detected (3400- to > 330000-fold resistance) Y93N, C, L31V, Q30R-L31M[83]. In prior null responders receiving triple therapy (DCV, ASV, and ribavirin), 10/18 (56%) HCV genotype 1a patients experienced viral breakthrough, while 4/4 HCV genotype 1b patients achieved SVR4, defined as undetectable HCV RNA at week 4 after treatment cessation[81]. In difficult-to-cure subgroups, such as HCV genotype 1a or interferon-ineligible patients, a combination of only 2 DAAs with a low genetic barrier to resistance with or without ribavirin could be sub-optimal in terms of SVR rates and lead to selection of HCV NS5A RAVs[84,85].
A quad regimen of DCV, ASV, and peginterferon-alpha plus ribavirin has also been reported, resulting in 90%-95% SVR24 in HCV genotype 1a and 1b prior null responders. No viral breakthrough occurred. Two patients relapsed with no baseline variants being detected. At relapse, both NS3 and NS5A variants were observed. In 1 HCV genotype 1a patient, HCV NS3 (Q80Q/L) and HCV NS5A (L31M/L) variants were detected 8 h after treatment initiation. The quad regimen seems to be sufficient to suppress the emergence of high-level HCV NS5A resistance[82,83].
Karino et al[80] reported the long-term virologic characterization of patients failing to respond to ASV/DCV. In line with the results of monotherapy studies, DCV-resistant variants persisted through 48 wk post-treatment, whereas ASV-resistant substitutions were no longer detectable after 48 wk. The selection of durable resistant variant strains with high-level resistance might be problematic. These results indicated that, in difficult-to-treat patients, the optimal interferon-free regimen should combine a drug with potent antiviral activity (HCV NS3/4A inhibitor or HCV NS5A inhibitor) with a drug with a high genetic barrier to resistance (HCV NS5B polymerase inhibitor)[84].
While the ASV and DCV dual regimen only seems effective in limited patients such as HCV genotype 1b patients, high SVR has been achieved with broad genotype coverage by regimens combining 3 or more DAAs, or DAAs with a high barrier to resistance. A quad therapy regimen consisting of 12 wk of a ritonavir-boosted HCV NS3/4A inhibitor (ABT-450/r) plus an HCV NS5B polymerase inhibitor (ABT-333) and an HCV NS5A inhibitor (ABT-267) obtained 96% of SVR12 in untreated HCV genotype 1a patients and 89% of SVR12 in HCV genotype 1a null responders[86,87]. An interferon-free and ribavirin-free triple combination of DAAs (ASV, DCV, and the non-nucleoside polymerase inhibitor BMS-791325) resulted in more than 90% of SVR12 in untreated HCV genotype 1a and 1b patients[88]. A combination of DCV and a nucleotide analogue inhibitor of HCV RNA-dependent RNA polymerase, sofosbuvir, with or without ribavirin for 24 wk achieved SVR in 100% of untreated HCV genotype 1a and 1b patients (n = 44) as well as in HCV genotype 1a and 1b patients (n = 41) who failed to respond to prior treatment with telaprevir or boceprevir, peginterferon and ribavirin. SVR was achieved in 93% of patients infected with HCV genotypes 2 and 3 (n = 44)[89,90].
Currently, DCV is the most studied HCV NS5A inhibitor with pmol/L potency and HCV pan-genotype coverage. HCV NS5A-resistant variants exist naturally and emerge frequently after virologic response failure with suboptimal treatment including HCV NS5A inhibitors. Cross-resistance is observed to all HCV NS5A inhibitors currently entering the clinical trial stage, although promising “second generation” HCV NS5A inhibitors with improved potency are reported. Long-term clinical consequences of previously selected resistant variants are still unknown at this time. At present, patients should be treated according to the recommendations[91], which do not mention any HCV NS5A inhibitors. HCV NS5A inhibitors should be used in combination regimens potent enough to prevent the emergence of resistant variants. An algorithm of the current treatment options for HCV genotypes 1 and 2 is shown in Figure 3. In the near future, HCV NS5A inhibitors will play a critical role in interferon-free regimens in HCV genotype 1 patients.
P- Reviewers: Bock T, Enjoji M, Kapoor S, Liu XY, Wirth S S- Editor: Gou SX L- Editor: A E- Editor: Zhang DN
| 2. | Nakamoto S, Imazeki F, Fukai K, Fujiwara K, Arai M, Kanda T, Yonemitsu Y, Yokosuka O. Association between mutations in the core region of hepatitis C virus genotype 1 and hepatocellular carcinoma development. J Hepatol. 2010;52:72-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 3. | Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547-6549. [PubMed] |
| 4. | Di Bisceglie AM. Hepatitis C and hepatocellular carcinoma. Hepatology. 1997;26:34S-38S. [PubMed] |
| 5. | Pacholczyk M, Łągiewska B, Lisik W, Tronina O, Wasiak D, Cieciura T, Chmura A. Liver transplantation for HCV cirrhosis; cautious optimism after 10 years of experience. Ann Transplant. 2012;17:5-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 6. | Nakamoto S, Kanda T, Yonemitsu Y, Arai M, Fujiwara K, Fukai K, Kanai F, Imazeki F, Yokosuka O. Quantification of hepatitis C amino acid substitutions 70 and 91 in the core coding region by real-time amplification refractory mutation system reverse transcription-polymerase chain reaction. Scand J Gastroenterol. 2009;44:872-877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 7. | Kanda T, Imazeki F, Yonemitsu Y, Mikami S, Takada N, Nishino T, Takashi M, Tsubota A, Kato K, Sugiura N. Quantification of hepatitis C virus in patients treated with peginterferon-alfa 2a plus ribavirin treatment by COBAS TaqMan HCV test. J Viral Hepat. 2011;18:e292-e297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 8. | Kanda T, Imazeki F, Azemoto R, Yonemitsu Y, Mikami S, Kita K, Takashi M, Sunaga M, Wu S, Nakamoto S. Response to peginterferon-alfa 2b and ribavirin in Japanese patients with chronic hepatitis C genotype 2. Dig Dis Sci. 2011;56:3335-3342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 9. | Kanda T, Imazeki F, Yokosuka O. New antiviral therapies for chronic hepatitis C. Hepatol Int. 2010;4:548-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 10. | Watashi K, Shimotohno K. Chemical genetics approach to hepatitis C virus replication: cyclophilin as a target for anti-hepatitis C virus strategy. Rev Med Virol. 2007;17:245-252. [PubMed] |
| 11. | Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980-3986. [PubMed] |
| 12. | Macdonald A, Harris M. Hepatitis C virus NS5A: tales of a promiscuous protein. J Gen Virol. 2004;85:2485-2502. [PubMed] |
| 13. | Gale MJ, Korth MJ, Tang NM, Tan SL, Hopkins DA, Dever TE, Polyak SJ, Gretch DR, Katze MG. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217-227. [PubMed] |
| 14. | Gale M, Blakely CM, Kwieciszewski B, Tan SL, Dossett M, Tang NM, Korth MJ, Polyak SJ, Gretch DR, Katze MG. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208-5218. [PubMed] |
| 15. | Polyak SJ, Paschal DM, McArdle S, Gale MJ, Moradpour D, Gretch DR. Characterization of the effects of hepatitis C virus nonstructural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology. 1999;29:1262-1271. [PubMed] |
| 16. | Gale M, Kwieciszewski B, Dossett M, Nakao H, Katze MG. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J Virol. 1999;73:6506-6516. [PubMed] |
| 17. | Ghosh AK, Majumder M, Steele R, Meyer K, Ray R, Ray RB. Hepatitis C virus NS5A protein protects against TNF-alpha mediated apoptotic cell death. Virus Res. 2000;67:173-178. [PubMed] |
| 18. | Majumder M, Ghosh AK, Steele R, Ray R, Ray RB. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J Virol. 2001;75:1401-1407. [PubMed] |
| 19. | Pflugheber J, Fredericksen B, Sumpter R, Wang C, Ware F, Sodora DL, Gale M. Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc Natl Acad Sci USA. 2002;99:4650-4655. [PubMed] |
| 20. | Kanda T, Steele R, Ray R, Ray RB. Inhibition of intrahepatic gamma interferon production by hepatitis C virus nonstructural protein 5A in transgenic mice. J Virol. 2009;83:8463-8469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 21. | Tamura R, Kanda T, Imazeki F, Wu S, Nakamoto S, Tanaka T, Arai M, Fujiwara K, Saito K, Roger T. Hepatitis C Virus nonstructural 5A protein inhibits lipopolysaccharide-mediated apoptosis of hepatocytes by decreasing expression of Toll-like receptor 4. J Infect Dis. 2011;204:793-801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 22. | Tellinghuisen TL, Marcotrigiano J, Gorbalenya AE, Rice CM. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J Biol Chem. 2004;279:48576-48587. [PubMed] |
| 23. | Tellinghuisen TL, Marcotrigiano J, Rice CM. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature. 2005;435:374-379. [PubMed] |
| 24. | Love RA, Brodsky O, Hickey MJ, Wells PA, Cronin CN. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J Virol. 2009;83:4395-4403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 200] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 25. | Bartenschlager R, Lohmann V, Penin F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat Rev Microbiol. 2013;11:482-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 287] [Cited by in RCA: 292] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 26. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Invest. 1995;96:224-230. [PubMed] |
| 27. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77-81. [PubMed] |
| 28. | Kurosaki M, Enomoto N, Murakami T, Sakuma I, Asahina Y, Yamamoto C, Ikeda T, Tozuka S, Izumi N, Marumo F. Analysis of genotypes and amino acid residues 2209 to 2248 of the NS5A region of hepatitis C virus in relation to the response to interferon-beta therapy. Hepatology. 1997;25:750-753. [PubMed] |
| 29. | Penin F, Brass V, Appel N, Ramboarina S, Montserret R, Ficheux D, Blum HE, Bartenschlager R, Moradpour D. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J Biol Chem. 2004;279:40835-40843. [PubMed] |
| 30. | Kanda T, Yokosuka O, Imazeki F, Tanaka M, Shino Y, Shimada H, Tomonaga T, Nomura F, Nagao K, Ochiai T. Inhibition of subgenomic hepatitis C virus RNA in Huh-7 cells: ribavirin induces mutagenesis in HCV RNA. J Viral Hepat. 2004;11:479-487. [PubMed] |
| 31. | El-Shamy A, Nagano-Fujii M, Sasase N, Imoto S, Kim SR, Hotta H. Sequence variation in hepatitis C virus nonstructural protein 5A predicts clinical outcome of pegylated interferon/ribavirin combination therapy. Hepatology. 2008;48:38-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 110] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 32. | Verdegem D, Badillo A, Wieruszeski JM, Landrieu I, Leroy A, Bartenschlager R, Penin F, Lippens G, Hanoulle X. Domain 3 of NS5A protein from the hepatitis C virus has intrinsic alpha-helical propensity and is a substrate of cyclophilin A. J Biol Chem. 2011;286:20441-20454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 85] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 33. | El-Shamy A, Shoji I, Kim SR, Ide Y, Imoto S, Deng L, Yoon S, Fujisawa T, Tani S, Yano Y. Sequence heterogeneity in NS5A of hepatitis C virus genotypes 2a and 2b and clinical outcome of pegylated-interferon/ribavirin therapy. PLoS One. 2012;7:e30513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 34. | El-Shamy A, Shoji I, El-Akel W, Bilasy SE, Deng L, El-Raziky M, Jiang DP, Esmat G, Hotta H. NS5A sequence heterogeneity of hepatitis C virus genotype 4a predicts clinical outcome of pegylated-interferon-ribavirin therapy in Egyptian patients. J Clin Microbiol. 2012;50:3886-3892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 35. | Scheel TK, Prentoe J, Carlsen TH, Mikkelsen LS, Gottwein JM, Bukh J. Analysis of functional differences between hepatitis C virus NS5A of genotypes 1-7 in infectious cell culture systems. PLoS Pathog. 2012;8:e1002696. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 36. | Gao M, Nettles RE, Belema M, Snyder LB, Nguyen VN, Fridell RA, Serrano-Wu MH, Langley DR, Sun JH, O’Boyle DR. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature. 2010;465:96-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 773] [Cited by in RCA: 762] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 37. | Lemm JA, Leet JE, O’Boyle DR, Romine JL, Huang XS, Schroeder DR, Alberts J, Cantone JL, Sun JH, Nower PT. Discovery of potent hepatitis C virus NS5A inhibitors with dimeric structures. Antimicrob Agents Chemother. 2011;55:3795-3802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 38. | Lee C, Ma H, Hang JQ, Leveque V, Sklan EH, Elazar M, Klumpp K, Glenn JS. The hepatitis C virus NS5A inhibitor (BMS-790052) alters the subcellular localization of the NS5A non-structural viral protein. Virology. 2011;414:10-18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 39. | Guedj J, Dahari H, Rong L, Sansone ND, Nettles RE, Cotler SJ, Layden TJ, Uprichard SL, Perelson AS. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc Natl Acad Sci USA. 2013;110:3991-3996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 257] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 40. | Fridell RA, Qiu D, Wang C, Valera L, Gao M. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob Agents Chemother. 2010;54:3641-3650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 238] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 41. | Nettles RE, Gao M, Bifano M, Chung E, Persson A, Marbury TC, Goldwater R, DeMicco MP, Rodriguez-Torres M, Vutikullird A. Multiple ascending dose study of BMS-790052, a nonstructural protein 5A replication complex inhibitor, in patients infected with hepatitis C virus genotype 1. Hepatology. 2011;54:1956-1965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 155] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 42. | Pol S, Ghalib RH, Rustgi VK, Martorell C, Everson GT, Tatum HA, Hézode C, Lim JK, Bronowicki JP, Abrams GA. Daclatasvir for previously untreated chronic hepatitis C genotype-1 infection: a randomised, parallel-group, double-blind, placebo-controlled, dose-finding, phase 2a trial. Lancet Infect Dis. 2012;12:671-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 124] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 43. | Suk-Fong Lok A. HCV NS5A inhibitors in development. Clin Liver Dis. 2013;17:111-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 44. | Pawlotsky JM. NS5A inhibitors in the treatment of hepatitis C. J Hepatol. 2013;59:375-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 141] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 45. | Wyles DL. Antiviral resistance and the future landscape of hepatitis C virus infection therapy. J Infect Dis. 2013;207 Suppl 1:S33-S39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 75] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 46. | Vermehren J, Sarrazin C. The role of resistance in HCV treatment. Best Pract Res Clin Gastroenterol. 2012;26:487-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 47. | Yang PL, Gao M, Lin K, Liu Q, Villareal VA. Anti-HCV drugs in the pipeline. Curr Opin Virol. 2011;1:607-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 48. | Londoño MC, Lens S, Forns X. Interferon free regimens for the “difficult-to-treat”: are we there? J Hepatol. 2013;58:643-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 49. | Herbst DA, Reddy KR. NS5A inhibitor, daclatasvir, for the treatment of chronic hepatitis C virus infection. Expert Opin Investig Drugs. 2013;22:1337-1346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 50. | Scheel TK, Gottwein JM, Mikkelsen LS, Jensen TB, Bukh J. Recombinant HCV variants with NS5A from genotypes 1-7 have different sensitivities to an NS5A inhibitor but not interferon-α. Gastroenterology. 2011;140:1032-1042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 115] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 51. | Ramirez S, Li YP, Jensen SB, Pedersen J, Gottwein JM, Bukh J. Highly efficient infectious cell culture of three hepatitis C virus genotype 2b strains and sensitivity to lead protease, nonstructural protein 5A, and polymerase inhibitors. Hepatology. 2014;59:395-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 52. | Abe H, Hayes CN, Hiraga N, Imamura M, Tsuge M, Miki D, Takahashi S, Ochi H, Chayama K. A translational study of resistance emergence using sequential direct-acting antiviral agents for hepatitis C using ultra-deep sequencing. Am J Gastroenterol. 2013;108:1464-1472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 53. | Shi N, Hiraga N, Imamura M, Hayes CN, Zhang Y, Kosaka K, Okazaki A, Murakami E, Tsuge M, Abe H. Combination therapies with NS5A, NS3 and NS5B inhibitors on different genotypes of hepatitis C virus in human hepatocyte chimeric mice. Gut. 2013;62:1055-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 54. | Pelosi LA, Voss S, Liu M, Gao M, Lemm JA. Effect on hepatitis C virus replication of combinations of direct-acting antivirals, including NS5A inhibitor daclatasvir. Antimicrob Agents Chemother. 2012;56:5230-5239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 55. | Fridell RA, Qiu D, Valera L, Wang C, Rose RE, Gao M. Distinct functions of NS5A in hepatitis C virus RNA replication uncovered by studies with the NS5A inhibitor BMS-790052. J Virol. 2011;85:7312-7320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 120] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 56. | Combet C, Garnier N, Charavay C, Grando D, Crisan D, Lopez J, Dehne-Garcia A, Geourjon C, Bettler E, Hulo C. euHCVdb: the European hepatitis C virus database. Nucleic Acids Res. 2007;35:D363-D366. [PubMed] |
| 57. | Wang C, Valera L, Jia L, Kirk MJ, Gao M, Fridell RA. In vitro activity of daclatasvir on hepatitis C virus genotype 3 NS5A. Antimicrob Agents Chemother. 2013;57:611-613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 58. | Wang C, Jia L, Huang H, Qiu D, Valera L, Huang X, Sun JH, Nower PT, O’Boyle DR, Gao M. In vitro activity of BMS-790052 on hepatitis C virus genotype 4 NS5A. Antimicrob Agents Chemother. 2012;56:1588-1590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 59. | Lawitz E, Marbury T, Cambell A, Dumas E, Kapoor M, Pilot-Matias T, Krishnan P, Setze C, Xie W, Podsadecki T. Safety and antiviral acyivity of ABT-267, a novel NS5A inhibitor, during 3-day monotherapy: first study in HCV genotype-1 (GT-1)-infected treatment-naive subject. J Hepatol. 2012;56 suppl 2:S469-S470 [47th annual meeting EASL abstract]. |
| 60. | Wilfret DA, Walker J, Adkison KK, Jones LA, Lou Y, Gan J, Castellino S, Moseley CL, Horton J, de Serres M. Safety, tolerability, pharmacokinetics, and antiviral activity of GSK2336805, an inhibitor of hepatitis C virus (HCV) NS5A, in healthy subjects and subjects chronically infected with HCV genotype 1. Antimicrob Agents Chemother. 2013;57:5037-5044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 61. | Lawitz EJ, Gruener D, Hill JM, Marbury T, Moorehead L, Mathias A, Cheng G, Link JO, Wong KA, Mo H. A phase 1, randomized, placebo-controlled, 3-day, dose-ranging study of GS-5885, an NS5A inhibitor, in patients with genotype 1 hepatitis C. J Hepatol. 2012;57:24-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 131] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 62. | Gao M. Antiviral activity and resistance of HCV NS5A replication complex inhibitors. Curr Opin Virol. 2013;3:514-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 113] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 63. | Cheng G, Yu M, Peng B, Lee YJ, Trejo-Martin A, Gong R, Bush C, Worth A, Nash M, Chan K. GS-5816, a second generation HCV NS5A inhibitor with potent antiviral activity, broad genotypic coverage and a high resistance barrier. J Hepatol. 2013;58 suppl 1:S484-S485 [48th annual meeting EASL abstract]. |
| 64. | Yang G, Wiles J, Patel D, Zhao Y, Fabrycki J, Weinheimer S, Marlor C, Rivera J, Wang Q, Gadhachanda V. Preclinical characteristics of ACH-3102: a novel HCV NS5A inhibitor with improved potency against genotype-1a virus and variants resistant to 1st generation of NS5A inhibitors. J Hepatol. 2012;56 suppl 2:S330. [RCA] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 65. | Liu R, Kong R, Mann P, Ingravallo P, Zhai Y, Xia E, Ludmerer S, Kozlowski J, Coburn C. In vitro resistance analysis of HCV NS5A inhibitor: MK-8742 demonstrates increased potency against clinical resistance variants and higher resisitance barrier. J Hepatol. 2012;56 suppl 2:S334-S335. [DOI] [Full Text] |
| 66. | Fridell RA, Wang C, Sun JH, O’Boyle DR, Nower P, Valera L, Qiu D, Roberts S, Huang X, Kienzle B. Genotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlations. Hepatology. 2011;54:1924-1935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 197] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 67. | Wang C, Sun JH, O’Boyle DR, Nower P, Valera L, Roberts S, Fridell RA, Gao M. Persistence of resistant variants in hepatitis C virus-infected patients treated with the NS5A replication complex inhibitor daclatasvir. Antimicrob Agents Chemother. 2013;57:2054-2065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 68. | Bartels DJ, Sullivan JC, Zhang EZ, Tigges AM, Dorrian JL, De Meyer S, Takemoto D, Dondero E, Kwong AD, Picchio G. Hepatitis C virus variants with decreased sensitivity to direct-acting antivirals (DAAs) were rarely observed in DAA-naive patients prior to treatment. J Virol. 2013;87:1544-1553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 129] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 69. | Kuiken C, Hraber P, Thurmond J, Yusim K. The hepatitis C sequence database in Los Alamos. Nucleic Acids Res. 2008;36:D512-D516. [PubMed] |
| 70. | Hernandez D, Zhou N, Ueland J, Monikowski A, McPhee F. Natural prevalence of NS5A polymorphisms in subjects infected with hepatitis C virus genotype 3 and their effects on the antiviral activity of NS5A inhibitors. J Clin Virol. 2013;57:13-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 71. | Plaza Z, Soriano V, Vispo E, del Mar Gonzalez M, Barreiro P, Seclén E, Poveda E. Prevalence of natural polymorphisms at the HCV NS5A gene associated with resistance to daclatasvir, an NS5A inhibitor. Antivir Ther. 2012;17:921-926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 72. | Suzuki F, Sezaki H, Akuta N, Suzuki Y, Seko Y, Kawamura Y, Hosaka T, Kobayashi M, Saito S, Arase Y. Prevalence of hepatitis C virus variants resistant to NS3 protease inhibitors or the NS5A inhibitor (BMS-790052) in hepatitis patients with genotype 1b. J Clin Virol. 2012;54:352-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 97] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 73. | Lalezari JP, Farrell GC, Shah PS, Schwab C, Walsh D, Vig P, Brown NA, Ruby E, Halfon S, Colonno R. PPI-668, a potent new pan-genotypic HCV NS5A inhibitor: phase 1 efficacy and safety. Hepatology. 2012;56 Suppl:1065A. |
| 74. | McCarville JF, Seifer M, Standring DN, Mayers DL. IDX-06A-001 Investigator Team. Treatment-emergent variants following 3 days of monotherapy with IDX719, a potent, pan-genotypic NS5A inhibitor, in subjects infected with HCV genotypes 1-4. J Hepatol. 2013;58 suppl 1:S491-S492 [48th annual meeting EASL abstract]. |
| 75. | Yang G, Patel D, Zhao Y, Fabrycki J, Yang W, Podos S, Robison H, Robarge L, Kocinsky H, Deshpande M. Findings from clinical virology studies on ACH-3102 are consistent with preclinical observations on its improved potency against genotype-1a HCV and resistant variants. J Hepatol. 2013;58 suppl 1:S487-S488. [DOI] [Full Text] |
| 76. | Wong KA, Worth A, Martin R, Svarovskaia E, Brainard DM, Lawitz E, Miller MD, Mo H. Characterization of Hepatitis C virus resistance from a multiple-dose clinical trial of the novel NS5A inhibitor GS-5885. Antimicrob Agents Chemother. 2013;57:6333-6340. [PubMed] |
| 77. | Sun JH, O’Boyle Ii DR, Zhang Y, Wang C, Nower P, Valera L, Roberts S, Nettles RE, Fridell RA, Gao M. Impact of a baseline polymorphism on the emergence of resistance to the hepatitis C virus nonstructural protein 5A replication complex inhibitor, BMS-790052. Hepatology. 2012;55:1692-1699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 78. | Hezode C, Hirschfield GM, Ghesquiere W, Sievert W, Rodriguez-Torres M, Shafran SD, Thuluvath PJ, Tatum HA, Waked I, Esmat GE. Daclatasvir, an NS5A replication complex inhibitor, combined with peginterferon alfa-2a and ribavirin in treatment-naive hcv-genotype 1 or 4 patients: phase 2b COMMAND-1 SVR12 results. Hepatology. 2012;56 suppl:553A. |
| 79. | Suzuki Y, Ikeda K, Suzuki F, Toyota J, Karino Y, Chayama K, Kawakami Y, Ishikawa H, Watanabe H, Hu W. Dual oral therapy with daclatasvir and asunaprevir for patients with HCV genotype 1b infection and limited treatment options. J Hepatol. 2013;58:655-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 198] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 80. | Karino Y, Toyota J, Ikeda K, Suzuki F, Chayama K, Kawakami Y, Ishikawa H, Watanabe H, Hernandez D, Yu F. Characterization of virologic escape in hepatitis C virus genotype 1b patients treated with the direct-acting antivirals daclatasvir and asunaprevir. J Hepatol. 2013;58:646-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 144] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 81. | Lok AS, Gardiner DF, Hezode C, Lawitz E, Bourliere M, Everson GT, Marcellin P, Rodriguez-Torres M, Pol S, Serfaty L. Sustained virologic response in chronic HCV genotype (GT) 1-infected null responders with combination of daclatasvir (DCV; NS5A inhibitor) and asunaprevir (ASV; NS3 inhibitor) with or without peginterferon alfa-2a/ribavirin (PEG/RBV). Hepatology. 2012;56 suppl:230A. |
| 82. | Lok AS, Gardiner DF, Lawitz E, Martorell C, Everson GT, Ghalib R, Reindollar R, Rustgi V, McPhee F, Wind-Rotolo M. Preliminary study of two antiviral agents for hepatitis C genotype 1. N Engl J Med. 2012;366:216-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 476] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
| 83. | Hernandez D, Yu F, Ueland J, Wang C, Huang H, Gardiner D, Fridell R, Gao M. Characterization of virologic escape in hcv genotype 1 null responders receiving a combination of the NS3 protease inhibitor BMS-650032 and NS5A inhibitor BMS-790052. J Hepatol. 2011;54 suppl 1:S28-S29. [DOI] [Full Text] |
| 84. | Aghemo A, De Francesco R. New horizons in hepatitis C antiviral therapy with direct-acting antivirals. Hepatology. 2013;58:428-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 128] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 85. | Aghemo A, Colombo M. Selection of resistant-associated variants to the NS5A inhibitor daclatasvir: revenge of the hepatitis C virus. Gastroenterology. 2013;145:247-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 86. | Lange CM, Zeuzem S. Perspectives and challenges of interferon-free therapy for chronic hepatitis C. J Hepatol. 2013;58:583-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 87. | Kowdley K, Lawitz E, Poordad F, Cohen DE, Nelson DR, Zeuzem S, Everson GT, Kwo PY, Foster GR, Sulkowski MS. A 12-week Interferon-free Treatment Regimen With ABT-450/r, ABT 267, ABT-333, and Ribavirin Achieves SVR12 Rates (Observed Data) of 99% in Treatment-naive Patients and 93% in Prior Null Responders With HCV Genotype 1 Infection. Hepatology. 2012;56:1515A-1516A. |
| 88. | Everson GT, Sims KD, Rodriguez-Torres , Hezode C, Lawitz E, Bourliere M, Loustaud-Ratti V, Rustgi VK, Schwartz H, Tatum HA. An Interferon-Free, Ribavirin-Free 12-Week Regimen of Daclatasvir (DCV), Asunaprevir (ASV), and BMS-791325 Yielded SVR4 of 94% in Treatment-Naive Patients with Genotype (GT) 1 Chronic Hepatitis C Virus (HCV) Infection. Hepatology. 2012;56:1517A-1518A. |
| 89. | Sulkowski M, Gardiner D, Rodriguez-Torres M, Reddy KR, Hassanein T, Jacobson IM, Lawitz E, Lok AS, Hinestrosa F, Thuluvath PJ. High Rate of Sustained Virologic Response With the All-Oral Combination of Daclatasvir (NS5A Inhibitor) Plus Sofosbuvir (Nucleotide NS5B Inhibitor), With or Without Ribavirin, in Treatment-Naive Patients Chronically Infected With HCV GT 1, 2, or 3. Hepatology. 2012;56:1516A-1517A. |
| 90. | Sulkowski MS, Gardiner DF, Rodriguez-Torres M, Reddy KR, Hassanein T, Jacobson IM, Lawitz E, Lok AS, Hinestrosa F, Thuluvath PJ, Schwartz H, Nelson DR, Eley T, Wind-Rotolo M, Huang SP, Gao M, McPhee F, Sherman D, Hindes R, Symonds WT, Pasquinelli C, Grasela DM, AI444040 Study Group. Sustained Virologic Response With Daclatasvir Plus Sofosbuvir Ribavirin (RBV) in Chronic HCV Genotype (GT) 1-Infected Patients Who Previously Failed Telaprevir (TVR) or Boceprevir (BOC). J Hepatol. 2013;58:S570. |
| 91. | Omata M, Kanda T, Yu ML, Yokosuka O, Lim SG, Jafri W, Tateishi R, S . Hamid S, Chuang WL, Chutaputti A, Wei L, Sollano J, Sarin SK, Kao JH, W. McCaughan G. APASL consensus statements and management algorithms for hepatitis C virus infection. Hepatol Int. 2012;6:409-435. [RCA] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 142] [Article Influence: 10.9] [Reference Citation Analysis (0)] |