Published online Mar 21, 2014. doi: 10.3748/wjg.v20.i11.2785
Revised: December 3, 2013
Accepted: January 14, 2014
Published online: March 21, 2014
Processing time: 171 Days and 4.8 Hours
Hepatitis C virus (HCV) causes a clinically important disease affecting 3% of the world population. HCV is a single-stranded, positive-sense RNA virus belonging to the genus Hepacivirus within the Flaviviridae family. The virus establishes a chronic infection in the face of an active host oxidative defence, thus adaptation to oxidative stress is key to virus survival. Being a small RNA virus with a limited genomic capacity, we speculate that HCV deploys a different strategy to evade host oxidative defence. Instead of counteracting oxidative stress, it utilizes oxidative stress to facilitate its own survival. Translation is the first step in the replication of a plus strand RNA virus so it would make sense if the virus can exploit the host oxidative defence in facilitating this very first step. This is particularly true when HCV utilizes an internal ribosome entry site element in translation, which is distinctive from that of cap-dependent translation of the vast majority of cellular genes, thus allowing selective translation of genes under conditions when global protein synthesis is compromised. Indeed, we were the first to show that HCV translation was stimulated by an important pro-oxidant-hydrogen peroxide in hepatocytes, suggesting that HCV is able to adapt to and utilize the host anti-viral response to facilitate its own translation thus allowing the virus to thrive under oxidative stress condition to establish chronicity. Understanding how HCV translation is regulated under oxidative stress condition will advance our knowledge on how HCV establishes chronicity. As chronicity is the initiator step in disease progression this will eventually lead to a better understanding of pathogenicity, which is particularly relevant to the development of anti-virals and improved treatments of HCV patients using anti-oxidants.
Core tip: Oxidative stress inhibits canonical translation, however, emerging evidence suggests that oxidative stress can actually stimulate alternative translation from select internal ribosome entry site (IRES) elements including that involved in redox regulation and in persistent virus infection e.g., human immunodeficiency virus and hepatitis C virus (HCV). We postulate a novel role of oxidative stress-activated IRES-mediated translation in redox homeostasis and virus persistence. In the case of HCV, we explore the idea that HCV exploits oxidative stress to activate its own translation as a novel means of evading the host oxidative defence to establish chronicity.
- Citation: Chan SW. Establishment of chronic hepatitis C virus infection: Translational evasion of oxidative defence. World J Gastroenterol 2014; 20(11): 2785-2800
- URL: https://www.wjgnet.com/1007-9327/full/v20/i11/2785.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i11.2785
Hepatitis C virus (HCV) causes a clinically important disease affecting 3% of the world population[1]. About 75% of the infection will develop into chronic hepatitis, which can then progress into fibrosis, cirrhosis and hepatocellular carcinoma. A vaccine is not available. Current interferon (IFN) treatments are expensive, with numerous side effects and are particularly ineffective against the predominant genotype 1 in America and European countries[2]. The newly approved protease inhibitors likely promote the emergence of drug resistant mutants, owing to the high mutation rate of the HCV genome[2]. Thus there is a pressing need for alternative HCV therapies. HCV establishes a chronic infection in the face of an active immune response and the host oxidative defence. A number of mechanisms have been proposed to account for evasion of the antibody and cellular immunity and the natural killer, IFN and Toll-like receptor innate immunity[3-5]. However, little is known about how the virus can survive in a highly oxidative environment given that oxidative stress is such a prominent clinical feature associated with hepatitis C infection[6-12]. Adaptation to oxidative stress is key to virus survival. We postulate that adaptation can be at the level of translation as HCV uses an internal ribosome entry site (IRES) element for translation, distinctive from that of cellular translation[13]. Indeed, we were the first to show that translation from the HCV IRES was stimulated by an important pro-oxidant-hydrogen peroxide (H2O2) in hepatocytes, suggesting that HCV is able to adapt to and utilize host anti-viral response to facilitate its own translation thus allowing the virus to thrive under oxidative stress condition to establish chronicity[14]. Anti-oxidants are now in clinical trials in the treatment of HCV patients[15-17]. Understanding the mechanisms of how HCV evades host oxidative defence at the translational level may help shape the formulation of improved and new anti-oxidant treatments for HCV.
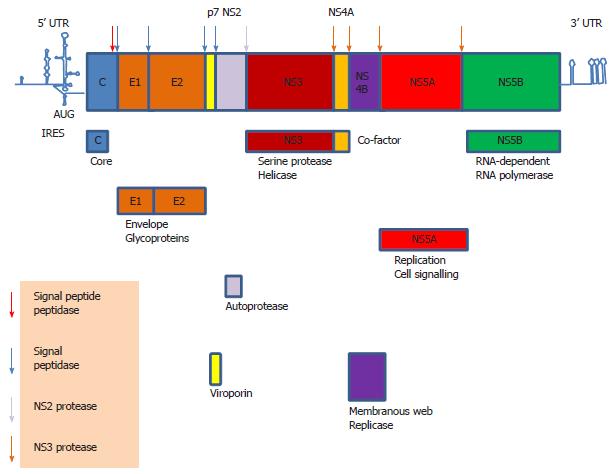
HCV is a Hepacivirus belonging to the family Flaviviridae[18]. As a single-stranded, positive-sense RNA virus, translation is the first step in the life cycle of HCV upon infection of a susceptible cell. Its 5’ untranslated region (UTR) contains an IRES element used to translate the 9.6 kb RNA genome into a single polypeptide which is then cleaved by the host and viral proteases into structural proteins core, envelopes E1 and E2, and non-structural (NS) proteins p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B (Figure 1)[19]. The RNA polymerase, NS5B, then catalyzes replication of the viral genome. The genome of HCV undergoes a high mutation rate giving rise to genetic variants, thus HCV is divided into genotypes and sub-types and is populated as “quasispecies”[20,21]. A “quasispecies” is a cloud of diverse, genetically linked mutants that function cooperatively and behave as a unit for natural selection[22]. Thus, a population of mutants with similar fitness values will out-compete those with a broad range of fitness values even though the latter includes mutants of high fitness values. This constitutes the basis of “the survival of the flattest” in “quasispecies” theory in contrast to Darwinian “the survival of the fittest”[23]. However, there is much debate on whether HCV or any RNA virus ever exists as a “quasispecies” in evolutionary term as the mutation rate of HCV is never high enough to lead to “quasispecies” dynamics[23]. Nevertheless, this “quasispecies”/intra-host variants phenomenon has great impact on virus persistence, pathogenesis, anti-viral treatment and vaccine design. However, different regions of the genome exhibit different degrees of sequence variability, with the envelope E2 region being the most variable harbouring hypervariable regions and the 5’ UTR the most conserved[21,24,25]. Thus, targeting 5’ UTR may be a solution to solve the problem of sequence variability in anti-viral therapies[26,27].
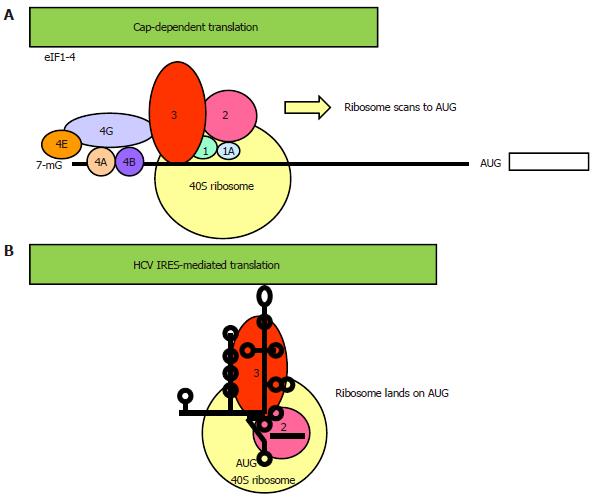
The vast majority of proteins is synthesized by a process known as cap-dependent translation, so named because it requires a 7-methyl guanosine (m7G) cap-structure at the 5’ end of the mRNA[28]. Translation is initiated when the cap is bound by the cap-binding complex eukaryotic initiation factor (eIF) 4F, which consists of eIF4E, eIF4A and eIF4G (Figure 2A). eIF4E is the cap-binding protein. eIF4A is a helicase, its unwinding activity is promoted by another initiation factor, eIF4B. eIF4G is the scaffold protein, which functions to recruit the 40S ribosomal subunit-eIF3-eIF2 pre-initiation complex to the 5’ end of the mRNA via protein-protein interaction between eIF4G and eIF3. The ribosomal complex, primed by eIF1/1A, then scans a short distance (50-100 nucleotides) to (usually) the nearest AUG triplet within a favorable (Kozak) sequence context to initiate translation[29].
IRES mediates an alternative form of translation distinctive from that of cap-dependent translation of the vast majority of cellular genes, thus allowing selective translation of genes under conditions when global protein synthesis is compromised e.g., virus infection, stress[30-32]. IRES translation is an important strategy employed by a subset of virus, mainly that of RNA viruses belonging to the Picornaviridae family, to continue viral protein synthesis during host translational shut off. IRESs found in cellular mRNAs mainly serve the function of regulating cellular processes such as apoptosis, differentiation, angiogenesis, thus their activity is usually tightly regulated and many are only responsive to stress. Studies on viral IRESs suggest that the IRES element forms a direct landing pad for the ribosome, therefore, the secondary and tertiary structures of the IRES are important for its activity (Figure 2B)[33-36]. As a result, the viral IRES element normally spans a considerably longer 5’ UTR that folds into a higher order structure and is interspersed with multiple AUG triplets[37]. However, short sequence motif rather than secondary structure is important in most cellular IRES activity[38,39]. As short as a 9-nucleotide sequence from the 5’ UTR of the cellular gene Gtx exhibited IRES activity[40,41]. In yeast and Drosophila, strong IRES activity was associated with weak secondary structure[42]. Because IRES-mediated translation is independent of a cap many canonical eIFs are dispensable, however, the requirement for canonical eIFs varies greatly amongst IRESs, ranging from the dependence of the entire set of eIFs in the hepatitis A virus (a picornavirus unrelated to HCV) IRES to none of them in the cricket paralysis virus IRES[31,35,37]. Another characteristic of IRES translation is that it is regulated by a diverse group of proteins known as IRES trans-acting factors (ITAFs)[43].
Each IRES has a unique set of ITAFs, even within the same group of IRES that shares primary sequence and secondary structure[44]. On the other hand, IRES of diverse origins can share common ITAFs[45]. Many of these ITAFs are RNA chaperone proteins. Most of them facilitate IRES-mediated translation although some are negatively regulating. Common ITAFs include the La autoantigen, polypyrimidine tract binding protein (PTB), heterogeneous nuclear ribonucleoproteins (hnRNPs), poly r(C) binding protein (PCBP), Upstream of N-ras (unr), death-associated protein 5 (DAP5) and the embryonic lethal abnormal vision/protein (ELAV/HuR)[30,46-52].
ITAF modification by stress signals is an important aspect in the regulation of IRES activity under stress conditions, using mechanisms such as nuclear-cytoplasmic shuttling, protein cleavage, phosphorylation and increased protein expression[53-58]. Many of the ITAFs are abundant nuclear proteins, thus nuclear-cytoplasmic shuttling presents an effective means of a fast response[59]. hnRNP A1 shuttled to the cytoplasm during osmotic shock to downregulate translation from the X-linked inhibitor of apoptosis protein (XIAP) IRES but upregulate translation from the fibroblast growth factor-2 IRES[53]. hnRNP A1 also shuttled to the cytoplasm in rhinovirus-2-infected and UVC-irradiated cells to enhance translation from the rhinovirus IRES but limit translation from the apoptotic peptidase activating factor 1 IRES[60]. Proteolysis also plays an important part in regulating IRES activity, either by directly conferring novel function to the truncated protein or by causing protein shuttling after the removal of the nuclear localization signal (NLS), or both. Caspase cleavage of DAP5 during endoplasmic reticulum (ER) stress released an active fragment with a novel ITAF function to activate the cellular inhibitor of apoptosis protein (HIAP2) IRES[54]. Cleavage of the La protein and PTB by the poliovirus serine protease released truncated fragments devoid of NLS to shuttle to the cytoplasm to either activate or repress the poliovirus IRES[55,56]. Phosphorylation of ITAFs is also commonly used to modify ITAF function. Phosphorylation of the hnRNP C protein during differentiation stimulated translation from the IRES of c-sis[57]. In some cases, over-expression of ITAF is sufficient to promote IRES translation. Elevated expression of unr stimulated translation from the IRES of PITSLRE cyclin-dependent protein kinase during G2/M phase of the cell cycle[58].
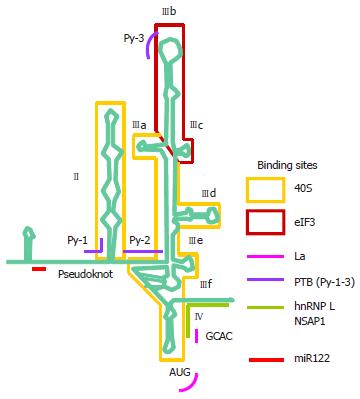
The HCV 5’ UTR is divided into four stem-loop domains (Figure 3)[61]. The IRES element is made up of approximately 340 nucleotides and spans domain II and extends into the core-coding region encompassing a double pseudoknot fold[13,62-64]. The HCV IRES has the second simplest requirement of eIFs, only needing eIF2 and eIF3. In reminiscent of the prokaryotic ribosomal binding of the Shine-Dalgarno sequence, the HCV IRES can directly recruit the 40S ribosome before association with eIF2 and eIF3 to form the 43S pre-initiation complex[35,63,65]. Structural and biochemical studies have indicated an unusually vast binding site for the ribosome encompassing domains II, III and IV and conformational changes in both the 40S ribosomal subunit and the IRES have been observed upon their interaction[66]. Binding between the IRES and the ribosome is thought to be mediated via ribosomal proteins although the role of RNA-RNA interaction cannot be excluded[67-69]. eIF3 can bind both the ribosome and junction domain IIIabc and domain IIIb of the IRES, thus playing a significant role in stabilizing the ribosome-eIF2 binary complex[70]. The ternary complex eIF2α-GTP-the tRNA for the first methionine (MettRNAi) does not directly bind the IRES, rather it forms a ternary complex with eIF3 and the 40S ribosomal subunit to position MettRNAi directly onto the AUG codon in the P-site of the ribosome[70].
A number of putative ITAFs for the HCV IRES have been identified, including La, PTB, hnRNP D, hnRNP L, HuR, the NS1-associated protein 1 and miR-122 (Figure 3)[71-82]. There is evidence for a critical role of the La autoantigen in IRES translation, by binding to and altering the conformation of the IRES to orchestrate assembly of the ribosomal complex[82,83]. La is one of the best known ITAFs and is pivotal in mediating translation from a number of IRESs[72,82,84-89]. It normally functions in RNA metabolism e.g., nascent polIII transcript processing and RNP assembly but is co-opted as an ITAF in IRES translation[90]. Pathologically, La is an autoantigen in a number of autoimmune diseases such as lupus and Sjögren’s syndrome[91]. The binding site for the La autoantigen has been mapped to the initiator AUG and the adjacent GCAC motif although additional binding sites may exist[88,92]. The binding sites for some other ITAFs have also been mapped[61,92]. Together with that of the ribosomal subunit and eIF3, these binding sites offer attractive targets for antiviral intervention because of (1) the highly conserved nature of the IRES[93]; (2) the distinctive mode of translation making it likely to produce an anti-viral with a high therapeutic index[94]; and (3) the use of cellular targets making it less ready to select for resistant mutants.
HCV IRES activity can also be modulated by the viral proteins core, NS2/3, NS3 and NS5A and the 3’ UTR[95-100]. A long range interaction between the IRES and the 3’ UTR is thought to be essential for IRES activity[101]. However, it is still unclear what constitutes the bona fide ITAFs and how they regulate HCV IRES activity, in particular under stress.
Accumulation of reactive oxygen species (ROS) and the generation of oxidative stress are implicated in the development of a number of inflammatory diseases, including viral hepatitis[102]. Chronic hepatitis C patients present elevated blood and hepatic levels of pro-oxidants, reduced anti-oxidants levels, iron overload with increased lipid peroxidation, decreased hepatic glutathione and increased oxidative DNA damage[103-108]. Proteomic and microarray analysis of liver biopsies revealed increased oxidative stress in hepatitis C samples[109,110].
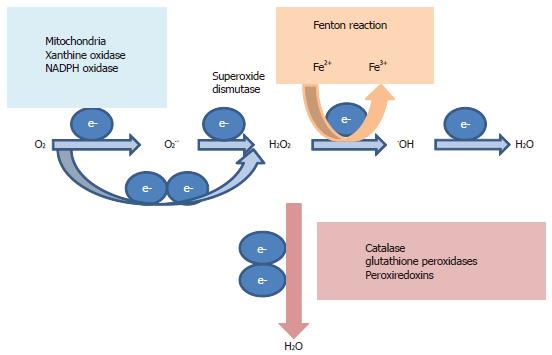
Important ROS include superoxide anion O2.-, H2O2 and hydroxyl radical .OH. ROS exist in every cell as part of the by-products of active respiration in the mitochondria (Figure 4)[111]. ROS are harmful to cells as they will cause oxidative damage to intracellular macromolecules and are eliminated by anti-oxidant enzymes such as superoxide dismutase, catalase and the glutathione system to maintain redox balance (Figure 4). A low level of ROS, in particular H2O2, is however, important mediator of cellular signal transduction pathways[111,112]. A high level of ROS is important in fighting infections[113]. Immune recognition of infected cells triggers the release of ROS from sequestered phagocytes and activated macrophages. Endogenous ROS are also produced as a direct result of hepatitis C viral replication and interactions of a number of hepatitis C viral proteins with the host cell, as evidenced by studying infected cultured cells and ectopically expressed viral proteins (core, NS3 or NS5A) in cultured hepatocytes, monocytes and isolated mitochondria[6-12,114,115]. This is supported by data from in vivo studies. Transgenic mice carrying the structural proteins exhibited elevated levels of ROS and were more susceptible to oxidant injury[10]. Infection of a SCID/Alb/uPA chimeric mouse (mouse with chimeric human and mouse liver) also revealed increased oxidative stress in infected hepatocytes[116]. It has recently been shown that the NAD(P)H oxidases, Nox1 and Nox4, are two of the endogenous ROS sources in HCV-infected cultured cells and liver samples[117,118].
ROS are lethal to pathogens. How do pathogens counteract the damaging effects of ROS? Bacteria do it at the transcriptional level as they normally do. Some bacteria such as Escherichia coli and Salmonella typhimurium can sense and counteract oxidative stress by inducing transcription of response genes from the OxyR regulon[119,120]. Viruses, being obligatory intracellular parasites with limited genomic capacity, exploit different strategies to suit their life style. The poxvirus molluscum contagiosum virus has a large DNA genome thus is capable of encoding their own anti-oxidant protein to become resistant to the cytotoxic effect of H2O2[121,122]. HCV is a small RNA virus with a limited genomic capacity. There is no evidence that HCV encodes an anti-oxidant protein. This has led us to speculate that HCV deploys a different strategy to evade the host oxidative defence. Instead of counteracting oxidative stress, it utilizes oxidative stress to facilitate its own survival. This would be advantageous to the virus because it persists as a chronic infection. Precedence can be found in human immunodeficiency virus (HIV), which also establishes a chronic infection and has been associated with increased oxidative stress in HIV patients[123]. HIV replication was facilitated by ROS via activation of the transcription factors nuclear factor-kappa B and hypoxia inducible factor 1 alpha to stimulate gene expression from the HIV long terminal repeat[124-126]. The effect of ROS on HCV replication is inconclusive, as opposing results were obtained from laboratory studies (most likely due to the use of different pro-oxidants and HCV expression systems) although some clinical studies and anti-oxidants trials do support a stimulatory role of ROS on HCV replication[17,127-137]. Translation is the first step in the replication of a plus strand RNA virus so it would make sense if the virus can exploit the host oxidative defence in facilitating this very first step. Indeed, we have previously shown that H2O2 stimulates translation from the HCV IRES[14].
Amongst viruses, HCV and HIV infections are commonly associated with elevated oxidative stress in patients, meaning that the viruses are continuously exposed to oxidative stress[123]. Coincidentally, they both cause chronic infections and translation from their IRESs is both upregulated by H2O2, suggesting that the viruses can adapt to and utilize oxidative stress to their own advantage[14,138]. Amongst cellular IRESs, translation from the IRESs of nuclear factor erythroid-2 related factor 2 (Nrf2) and ferritin is stimulated by pro-oxidants[139-141]. Coincidentally, these proteins are both involved in restoring redox balance, suggesting that upregulation of IRES translation could be a homeostatic response to oxidative stress. Nrf2 is the coordinator of the anti-oxidant response to oxidative stress and translation from its IRES was stimulated by H2O2[139,141]. Ferritin sequesters excess iron from catalyzing the Fenton reaction that leads to the production of free radicals and translation from the ferritin IRES was activated by iron (Figure 4)[140]. A protective response to oxidative stress was also mediated by IRES translation in a pathological setting of ischaemic insults[142]. A rapid rise in the level of H2O2 damaged the neuron but at the same time, conferred neuroprotection to ischaemic insults by stimulating translation from the Sp1 IRES. Altogether these results suggest that one of the adaptive responses to oxidative stress could be at the level of translation and that an IRES is being deployed to achieve this. It is interesting to see whether viruses co-opted a homeostatic cellular IRES in their counter-defence against oxidative stress or vice versa.
Will all viruses possessing an IRES element be capable of taking advantage of oxidative stress to increase their replication rate? IRES is present in all members of the Picornaviridae family including poliovirus, rhinovirus (common cold), encephalomyocarditis virus, foot-and-mouth disease virus and hepatitis A virus[143]. Picornavirus IRESs are divided into Type I-V based on structural and functional similarity[144]. Type IV IRES is grouped with IRESs from the two genera Hepacivirus and Pestivirus of the Flaviviridae family (here known as HCV-like IRES), leading to the speculation that HCV acquired an IRES element from picornavirus in the distant past by recombination[143,145]. This may explain why IRES is not a common feature of the Flaviviridae family and is absent from the genus Flavivirus. IRESs have also been found in some retroviruses and DNA viruses establishing chronic/latent infections such as HIV and Kaposi’s sarcoma-associated herpesvirus[138,146]. It is interesting to see whether responsiveness to oxidative stress is a function preserved in all HCV-like IRESs regardless of whether they establish an acute or chronic infection or it is a function evolved with persistent infection.
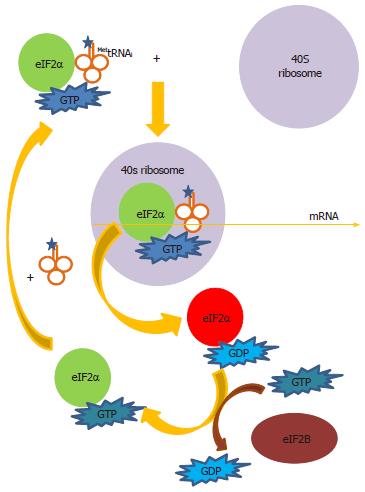
How then could an IRES facilitate an adaptive oxidative response? All protein synthesis relies on eIF2α to deliver MettRNAi to the 40S ribosomal subunit by forming a ternary complex eIF2α-GTP-MettRNAi (Figure 5)[147]. Following hydrolysis of GTP to GDP, the eIF2α-GDP complex leaves the ribosome. GDP is converted to GTP in an exchange reaction catalyzed by the exchanger eIF2B, allowing eIF2α-GTP to be recycled for more complex formation with MettRNAi to continue translation initiation. Phosphorylation of eIF2α at Serine-51 (Ser-51) inhibits eIF2B, thus arresting protein synthesis at the step of GDP-GTP exchange. To date four mammalian eIF2α kinases have been known. They are the RNA-activated protein kinase (PKR), PKR-like ER eIF2α kinase (PERK), heme-regulated inhibitor of translation (HRI) and the mammalian homologue of yeast eIF2α kinase general control nonderepressible 2 (GCN2)[148-151]. These kinases share similarity in their kinase domains but differ in their regulatory domains allowing them to respond to distinct stress stimuli whilst phosphorylating eIF2α at the identical residue Ser-51. PKR is specifically activated by ds-RNA during virus infections; PERK is specifically activated by ER stress; HRI is specifically activated by heme deficiency in erythroid cells primarily involved in the regulation of haemoglobin synthesis whereas GCN2 is specifically activated by amino acid starvation. We have shown that H2O2 is also a stress signal to induce phosphorylation of eIF2α although currently we do not have evidence to suggest which of the four mammalian kinases is operating in our system[14]. All of the four kinases have been shown to be the eIF2α kinase under different oxidative stress conditions and in different cell types: HRI in arsenite-induced oxidative stress; GCN2 in UV-irradiation-induced oxidative stress and PKR and PERK in H2O2-stimulated osteoblastoma and HEK293 cells[152-156]. It is equally possible that oxidative stress-induced eIF2α phosphorylation is a result of inhibition of a phosphatase rather than activation of a kinase[157].
Little is known of how MettRNAi is delivered to maintain IRES translation under oxidative stress condition, when eIF2α is phosphorylated. So the question will be under this condition what is used to deliver MettRNAi? Although many eIFs are dispensable for IRES-mediated translation, almost all still rely on eIF2α to deliver MettRNAi, thus are sensitive to the inhibitory effect of phospho-eIF2α. The HCV IRES is no exception. Under non-stressed condition, translation from the HCV IRES is still dependent on eIF2α to deliver MettRNAi[158-160]. However, some IRESs can evade this critical step of translational control and allow them to maintain translation under conditions that would otherwise inhibit protein synthesis. First, instead of downregulation by phospho-eIF2α, translation from select viral and cellular IRESs is actually upregulated by phospho-eIF2α[161-163]. The exact mechanism of how phospho-eIF2α upregulates select IRES translation is unclear. Regarding HCV, there is no evidence that translation from the HCV IRES is upregulated by phospho-eIF2α, either under stress conditions that induce phosphorylation of eIF2α or by ectopic expression of a phospho-mimetic eIF2α-SD (substitution of Ser-51 with Aspartate-51 which mimics the structure of phospho-eIF2α)[14,164,165]. Secondly, a minority of IRESs does not require any eIFs for translation. The cricket paralysis virus intergenic IRES simply folds to mimic the function of MettRNAi[166]. The HCV IRES can also operate without eIF. However, this “factor-less” translation was performed under in vitro condition, using a non-physiological high concentration of Mg2+[167]. It is not known whether the HCV IRES can operate in an eIF-less mode of translation in vivo. Thirdly, IRES translation can switch from eIF2-dependent to eIF2-independent mode of translation under stress conditions or during virus infections that induce phosphorylation of eIF2α. Translation from the poliovirus IRES during early phase of infection was dependent on eIF2α but was independent of eIF2α during late phase of infection and this eIF2-independence was assisted by the viral 2A protease[168]. HCV infection also induces phosphorylation of eIF2α[169]. Translation from the HCV, classical swine fever virus (CSFV) and the cellular XIAP IRESs was resistant to the inhibitory effect of eIF2α phosphorylation by switching from eIF2-dependent to eIF2-independent mode of translation, using alternative eIF such as eIF5B, eIF2A or eIF2D/ligatin to deliver MettRNAi[158-160,170-173]. It remains to be seen which mechanisms operate to deliver MettRNAi in IRES translation under oxidative stress condition.
However, the use of an eIF2-independent mode of translation simply allows translation to operate at a lower efficiency when the more efficient canonical eIF2α-dependent pathway is inhibited[158,160]. The HCV IRES behaves in a very different way under oxidative stress condition in that translation is not only maintained, but is actually upregulated, suggesting a different or additional way of regulation under oxidative stress condition[14,174]. This is similar to the HIV and Nrf2 IRESs, in which translation is stimulated by oxidative stress[138-139,141]. Thus far two mechanisms have been proposed by which oxidative stress stimulates IRES translation, both of which involve ITAF, stressing the importance of ITAF in translational regulation during oxidative stress.
A positive regulatory mechanism in which oxidative stress stimulates IRES translation by increasing cytoplasmic level of ITAF, either by promoting its cytoplasmic shuttling or by one of the mechanisms mentioned above. An example can be seen in the Nrf2 IRES. H2O2 stimulated Nrf2 IRES translation by increasing shuttling of its ITAF, La, to the cytoplasm[141]. In this case, the H2O2-responsive element has been mapped to a region responsible for both basal and H2O2-induced IRES activity[139].
This is similar to the positive regulatory mechanism in which oxidative stress stimulates IRES translation by increasing cytoplasmic level of ITAF. However, in this case, the IRES activity is normally repressed by being locked into a weakly active conformation by a repressor protein. Oxidative stress induces eIF2α phosphorylation to shut down global protein synthesis including that of the repressor. As the repressor level drops, oxidative stress increases the cytoplasmic level of an activator ITAF, either by promoting its cytoplasmic shuttling or by one of the mechanisms mentioned above. The release of the repressor allows binding of the activator ITAF to induce a conformational change in the IRES to activate translation. In this case, the H2O2-responsive element has been mapped to a negatively regulating domain that inhibits basal IRES translation. An example can be found in the HIV IRES, although in this case the repressor and activator ITAFs have yet to be identified to support this derepression hypothesis[138].
As for HCV, we currently do not have evidence to suggest how H2O2 activates IRES translation. However, others have found that iron stimulates translation from the HCV IRES, via upregulation of eIF3 and La mRNAs[175,176]. Iron catalyzes the Fenton reaction in the conversion of H2O2 into the highly oxidizing and damaging .OH (Figure 4)[111]. Thus iron promotes oxidative stress and iron overload is frequent in HCV patients[105]. Although these studies did not show a direct correlation between oxidative stress and IRES translation, they provide an indication of how this might work and the similarity with the two proposed mechanisms in that they all involve an ITAF. Further work will be required to dissect the mechanisms of how H2O2 activates translation from the HCV IRES.
Still exactly how oxidative stress stimulates IRES translation is far from clear. Despite collectively known as IRES, each IRES is unique in terms of sequence, structure, use of eIF and ITAF, mechanism of translation and response to stress. Cellular IRESs are distinctly different from viral IRESs in that they are naturally capped, flatter and for most, depend on short motif rather than overall structure to function[38,39]. HIV-a retrovirus-has a capped mRNA which is translated by a cap-dependent mechanism under normal circumstances[177]. For HIV and some cellular genes, IRES-mediated translation serves as an alternative mechanism of translation under stress conditions[138]. In contrast, when IRES-mediated translation represents the main (sole) mechanism of translation in RNA viruses such as picornavirus and HCV the mRNAs are uncapped[38]. Thus it is anticipated that the mechanisms used to respond to oxidative stress would be as diverse as the IRES itself.
HCV genome exhibits a high degree of sequence variation, with > 30% difference between genotypes and 20%-25% between sub-types[178]. Due to structural constraint, the 5’ UTR (which contains the IRES element) is the most conserved region, but substitutions along the IRES region are common amongst genotypes, sub-types and even “quasispecies”/intra-host variants[93,179]. Substitutions have been mapped to the stem, loop and unpaired regions[36,180,181]. Most of the substitutions in the stem regions are co-variants thus preserving the structural integrity of the IRES element. A minority of substitutions in the stem regions results in loss of base-pairing and alteration in IRES structure and hence function. Substitutions mapped to the loop or unpaired regions are important as well, as they may contain binding sites for the ribosomal subunit, eIF3 and ITAFs[36,182,183]. Therefore, despite being a highly conserved region, slight alteration in the IRES sequence can have a profound effect on basal IRES translation and responsiveness to stresses. The efficiency of genotypic IRESs has been compared in various studies. IRESs from some genotypes or sub-types were more efficient in mediating basal translation (i.e., under non-stressed condition), however, the results are not consistent across studies, most probably due to the use of different IRES regions in their studies and the existence of intra-genotypic variation in the IRES sequences used in different studies[184-186]. Indeed, substitutions are commonly found in closely related IRES sequences isolated from a single patient and some of these substitutions impacted a substantial change in the translational efficiency, highlighting the fact that HCV exists as a swarm of variants each with slightly different IRES sequence and structure hence efficiency in basal translation and responsiveness to stresses/IFN[187]. It is well known that genotype is a determining factor in patients’ response to IFN treatment. Comparison of IRESs from six genotypes did not reveal any differences in their translational responsiveness to IFN, however, IRESs isolated from sustained responders of genotype 3a patients had lower translation efficiencies than that from non-responders and were more prone to IFN inhibition[188,189]. Other studies also identified marked differences in the distribution of substitutions between sustained responder and non-responder IRESs and between pre-treatment and post-treatment IRESs in non-responders, regardless of genotypes[190-193]. These results further emphasize the significance of intra-host IRES variants in determining stress/IFN responsiveness.
Variation in IRES sequence can also have an effect on virus replication via two mechanisms. First, as many of the translated proteins are required for virus replication, a change in the translation efficiency can alter the availability of proteins involved in virus replication. Second, the antisense IRES contains the promoter for the plus strand synthesis in virus replication, thus variation in IRES sequence can affect the rate of replication[194,195]. It is therefore interesting to see whether intra-host variation in IRES sequence will also result in a swarm of variants with different degrees of replication efficiencies under oxidative stress condition. This may also explain why opposing results were obtained regarding the effects of ROS on HCV replication[17,127-137].
Therefore, studies with viruses with high sequence variability such as HIV and HCV have been complicated by the existence of a population of intra-host variants in each patient. The collective response of this population of intra-host variants will ultimately determine the response to oxidative stress and outcome of infection (persistence).
HCV establishes a chronic infection[1]. To survive in a harsh environment the virus needs to deploy a number of machinery to evade the host anti-viral responses, one of which is oxidative stress[113]. H2O2 induces phosphorylation of eIF2α resulting in inhibition of global (including that of viral) protein synthesis and constitutes an important defence against virus infection[14]. Possession of an IRES element enables some viral and cellular genes to continue protein synthesis when the majority of protein synthesis is inhibited by phosphorylation of eIF2α, by means of (1) a phospho-eIF2α-dependent mechanism[161-163]; (2) an eIF-less mechanism[166,167]; and (3) an eIF2α- independent mechanism[158-160,168,170-173]. At present, there is no evidence to suggest which mechanism is operating to enable translation from the HCV IRES to proceed when eIF2α is phosphorylated by H2O2[14]. Thus far studies on other IRESs have led to the proposal of a positive regulatory and a derepression mechanism, both involving H2O2-responsive ITAF, such as the La autoantigen, implicating a pivotal role of ITAF in H2O2-regulated IRES translation[138-139,141].
HCV IRES appears to belong to a class of IRESs that is translationally upregulated by H2O2[14,138-142]. This category of IRESs includes a number of cellular IRESs that orchestrate the anti-oxidants response and IRESs from viruses that establish chronic infections in a highly oxidative environment. It is interesting to see whether viruses co-opted a cellular homeostatic IRES or it is an inherent property of the viral IRES in the facilitation of a persistent infection.
HCV exhibits a high degree of sequence variability[178]. One must therefore take into consideration the collective response of a swarm of intra-host variants, each with different IRES structure and function and hence different translation and replication efficiencies under oxidative stress condition and, as a functional unit, will ultimately determine how well the virus can survive a highly oxidative environment in the process leading to persistence.
Thanks must go to Paul MacCallum, Samantha Jack, Philip Egan and Benjamin McDermott, without their work, it will not be possible to write this review article. I would also like to thank the anonymous reviewers for their critical comments.
P- Reviewers: Enjoji M, Faintuch J, Vaughan G S- Editor: Gou SX L- Editor: A E- Editor: Zhang DN
| 1. | Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nat Rev Gastroenterol Hepatol. 2013;10:553-562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 704] [Cited by in RCA: 742] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 2. | Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837-849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 424] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 3. | Bowen DG, Walker CM. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature. 2005;436:946-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 570] [Cited by in RCA: 578] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 4. | Gale M, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 455] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 5. | Horner SM, Gale M. Regulation of hepatic innate immunity by hepatitis C virus. Nat Med. 2013;19:879-888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 220] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 6. | Toro F, Conesa A, Garcia A, Bianco NE, De Sanctis JB. Increased peroxide production by polymorphonuclear cells of chronic hepatitis C virus-infected patients. Clin Immunol Immunopathol. 1998;88:169-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 7. | Bureau C, Bernad J, Chaouche N, Orfila C, Béraud M, Gonindard C, Alric L, Vinel JP, Pipy B. Nonstructural 3 protein of hepatitis C virus triggers an oxidative burst in human monocytes via activation of NADPH oxidase. J Biol Chem. 2001;276:23077-23083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 122] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 8. | Gong G, Waris G, Tanveer R, Siddiqui A. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kappa B. Proc Natl Acad Sci USA. 2001;98:9599-9604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 485] [Cited by in RCA: 499] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 9. | Korenaga M, Wang T, Li Y, Showalter LA, Chan T, Sun J, Weinman SA. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J Biol Chem. 2005;280:37481-37488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 317] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 10. | Moriya K, Nakagawa K, Santa T, Shintani Y, Fujie H, Miyoshi H, Tsutsumi T, Miyazawa T, Ishibashi K, Horie T. Oxidative stress in the absence of inflammation in a mouse model for hepatitis C virus-associated hepatocarcinogenesis. Cancer Res. 2001;61:4365-4370. [PubMed] |
| 11. | Okuda M, Li K, Beard MR, Showalter LA, Scholle F, Lemon SM, Weinman SA. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology. 2002;122:366-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 690] [Cited by in RCA: 680] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 12. | Qadri I, Iwahashi M, Capasso JM, Hopken MW, Flores S, Schaack J, Simon FR. Induced oxidative stress and activated expression of manganese superoxide dismutase during hepatitis C virus replication: role of JNK, p38 MAPK and AP-1. Biochem J. 2004;378:919-928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 119] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 13. | Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476-1483. [PubMed] |
| 14. | MacCallum PR, Jack SC, Egan PA, McDermott BT, Elliott RM, Chan SW. Cap-dependent and hepatitis C virus internal ribosome entry site-mediated translation are modulated by phosphorylation of eIF2alpha under oxidative stress. J Gen Virol. 2006;87:3251-3262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 15. | Farias MS, Budni P, Ribeiro CM, Parisotto EB, Santos CE, Dias JF, Dalmarco EM, Fröde TS, Pedrosa RC, Wilhelm Filho D. Antioxidant supplementation attenuates oxidative stress in chronic hepatitis C patients. Gastroenterol Hepatol. 2012;35:386-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 16. | Gane EJ, Weilert F, Orr DW, Keogh GF, Gibson M, Lockhart MM, Frampton CM, Taylor KM, Smith RA, Murphy MP. The mitochondria-targeted anti-oxidant mitoquinone decreases liver damage in a phase II study of hepatitis C patients. Liver Int. 2010;30:1019-1026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 298] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 17. | Melhem A, Stern M, Shibolet O, Israeli E, Ackerman Z, Pappo O, Hemed N, Rowe M, Ohana H, Zabrecky G. Treatment of chronic hepatitis C virus infection via antioxidants: results of a phase I clinical trial. J Clin Gastroenterol. 2005;39:737-742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 93] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 18. | Simmonds P. The origin of hepatitis C virus. Curr Top Microbiol Immunol. 2013;369:1-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 19. | Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933-938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 602] [Cited by in RCA: 601] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 20. | Simmonds P, Alberti A, Alter HJ, Bonino F, Bradley DW, Brechot C, Brouwer JT, Chan SW, Chayama K, Chen DS. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology. 1994;19:1321-1324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 573] [Article Influence: 18.5] [Reference Citation Analysis (1)] |
| 21. | Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, Halfon P, Inchauspé G, Kuiken C, Maertens G. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962-973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1070] [Cited by in RCA: 1083] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 22. | Lauring AS, Andino R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010;6:e1001005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 698] [Cited by in RCA: 587] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 23. | Holmes EC. Does hepatitis C virus really form quasispecies? Infect Genet Evol. 2010;10:431-432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 24. | Chan SW, McOmish F, Holmes EC, Dow B, Peutherer JF, Follett E, Yap PL, Simmonds P. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. J Gen Virol. 1992;73:1131-1141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 273] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 25. | Simmonds P, McOmish F, Yap PL, Chan SW, Lin CK, Dusheiko G, Saeed AA, Holmes EC. Sequence variability in the 5’ non-coding region of hepatitis C virus: identification of a new virus type and restrictions on sequence diversity. J Gen Virol. 1993;74:661-668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 284] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 26. | Parsons J, Castaldi MP, Dutta S, Dibrov SM, Wyles DL, Hermann T. Conformational inhibition of the hepatitis C virus internal ribosome entry site RNA. Nat Chem Biol. 2009;5:823-825. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 131] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 27. | Prabhu R, Garry RF, Dash S. Small interfering RNA targeted to stem-loop II of the 5’ untranslated region effectively inhibits expression of six HCV genotypes. Virol J. 2006;3:100. [PubMed] |
| 28. | Dever TE. Translation initiation: adept at adapting. Trends Biochem Sci. 1999;24:398-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 112] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 29. | Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA. 2001;98:7029-7036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 572] [Cited by in RCA: 573] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 30. | Holcik M, Sonenberg N, Korneluk RG. Internal ribosome initiation of translation and the control of cell death. Trends Genet. 2000;16:469-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 177] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 31. | Jang SK. Internal initiation: IRES elements of picornaviruses and hepatitis c virus. Virus Res. 2006;119:2-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 51] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 32. | Vagner S, Galy B, Pyronnet S. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2001;2:893-898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 216] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 33. | Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292:513-529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 179] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 34. | Lyons AJ, Lytle JR, Gomez J, Robertson HD. Hepatitis C virus internal ribosome entry site RNA contains a tertiary structural element in a functional domain of stem-loop II. Nucleic Acids Res. 2001;29:2535-2541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 35. | Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 204] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 36. | Tang S, Collier AJ, Elliott RM. Alterations to both the primary and predicted secondary structure of stem-loop IIIc of the hepatitis C virus 1b 5’ untranslated region (5’UTR) lead to mutants severely defective in translation which cannot be complemented in trans by the wild-type 5’UTR sequence. J Virol. 1999;73:2359-2364. [PubMed] |
| 37. | Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem Soc Trans. 2005;33:1231-1241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 114] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 38. | Plank TD, Kieft JS. The structures of nonprotein-coding RNAs that drive internal ribosome entry site function. Wiley Interdiscip Rev RNA. 2012;3:195-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 39. | Baird SD, Lewis SM, Turcotte M, Holcik M. A search for structurally similar cellular internal ribosome entry sites. Nucleic Acids Res. 2007;35:4664-4677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 60] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 40. | Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci USA. 2000;97:1536-1541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 191] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 41. | Chappell SA, Edelman GM, Mauro VP. Biochemical and functional analysis of a 9-nt RNA sequence that affects translation efficiency in eukaryotic cells. Proc Natl Acad Sci USA. 2004;101:9590-9594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 42. | Xia X, Holcik M. Strong eukaryotic IRESs have weak secondary structure. PLoS One. 2009;4:e4136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 43. | Spriggs KA, Bushell M, Mitchell SA, Willis AE. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 2005;12:585-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 131] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 44. | Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028-2045. [PubMed] |
| 45. | Bonnal S, Boutonnet C, Prado-Lourenço L, Vagner S. IRESdb: the Internal Ribosome Entry Site database. Nucleic Acids Res. 2003;31:427-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 71] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 46. | Henis-Korenblit S, Shani G, Sines T, Marash L, Shohat G, Kimchi A. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc Natl Acad Sci USA. 2002;99:5400-5405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 123] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 47. | Anderson EC, Hunt SL, Jackson RJ. Internal initiation of translation from the human rhinovirus-2 internal ribosome entry site requires the binding of Unr to two distinct sites on the 5’ untranslated region. J Gen Virol. 2007;88:3043-3052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 48. | Damiano F, Rochira A, Tocci R, Alemanno S, Gnoni A, Siculella L. hnRNP A1 mediates the activation of the IRES-dependent SREBP-1a mRNA translation in response to endoplasmic reticulum stress. Biochem J. 2013;449:543-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 49. | Durie D, Lewis SM, Liwak U, Kisilewicz M, Gorospe M, Holcik M. RNA-binding protein HuR mediates cytoprotection through stimulation of XIAP translation. Oncogene. 2011;30:1460-1469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 74] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 50. | Evans JR, Mitchell SA, Spriggs KA, Ostrowski J, Bomsztyk K, Ostarek D, Willis AE. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22:8012-8020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 188] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 51. | Holcík M, Gordon BW, Korneluk RG. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol Cell Biol. 2003;23:280-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 132] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 52. | Verma B, Bhattacharyya S, Das S. Polypyrimidine tract-binding protein interacts with coxsackievirus B3 RNA and influences its translation. J Gen Virol. 2010;91:1245-1255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 53. | Lewis SM, Veyrier A, Hosszu Ungureanu N, Bonnal S, Vagner S, Holcik M. Subcellular relocalization of a trans-acting factor regulates XIAP IRES-dependent translation. Mol Biol Cell. 2007;18:1302-1311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 92] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 54. | Warnakulasuriyarachchi D, Cerquozzi S, Cheung HH, Holcík M. Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J Biol Chem. 2004;279:17148-17157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 114] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 55. | Back SH, Kim YK, Kim WJ, Cho S, Oh HR, Kim JE, Jang SK. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro). J Virol. 2002;76:2529-2542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 120] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 56. | Shiroki K, Isoyama T, Kuge S, Ishii T, Ohmi S, Hata S, Suzuki K, Takasaki Y, Nomoto A. Intracellular redistribution of truncated La protein produced by poliovirus 3Cpro-mediated cleavage. J Virol. 1999;73:2193-2200. [PubMed] |
| 57. | Sella O, Gerlitz G, Le SY, Elroy-Stein O. Differentiation-induced internal translation of c-sis mRNA: analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol Cell Biol. 1999;19:5429-5440. [PubMed] |
| 58. | Tinton SA, Schepens B, Bruynooghe Y, Beyaert R, Cornelis S. Regulation of the cell-cycle-dependent internal ribosome entry site of the PITSLRE protein kinase: roles of Unr (upstream of N-ras) protein and phosphorylated translation initiation factor eIF-2alpha. Biochem J. 2005;385:155-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 73] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 59. | Lewis SM, Holcik M. For IRES trans-acting factors, it is all about location. Oncogene. 2008;27:1033-1035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 71] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 60. | Cammas A, Pileur F, Bonnal S, Lewis SM, Lévêque N, Holcik M, Vagner S. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol Biol Cell. 2007;18:5048-5059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 121] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 61. | Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol. 2007;5:29-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 164] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 62. | Berry KE, Waghray S, Mortimer SA, Bai Y, Doudna JA. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure. 2011;19:1456-1466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 88] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 63. | Reynolds JE, Kaminski A, Carroll AR, Clarke BE, Rowlands DJ, Jackson RJ. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA. 1996;2:867-878. [PubMed] |
| 64. | Wang C, Le SY, Ali N, Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5’ noncoding region. RNA. 1995;1:526-537. [PubMed] |
| 65. | Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 574] [Cited by in RCA: 584] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 66. | Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959-1962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 383] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 67. | Fukushi S, Okada M, Stahl J, Kageyama T, Hoshino FB, Katayama K. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J Biol Chem. 2001;276:20824-20826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 89] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 68. | Hertz MI, Landry DM, Willis AE, Luo G, Thompson SR. Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol Cell Biol. 2013;33:1016-1026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 92] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 69. | Malygin AA, Shatsky IN, Karpova GG. Proteins of the human 40S ribosomal subunit involved in hepatitis C IRES binding as revealed from fluorescent labeling. Biochemistry (Mosc). 2013;78:53-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 70. | Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci USA. 2004;101:16990-16995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 141] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 71. | Ali N, Siddiqui A. Interaction of polypyrimidine tract-binding protein with the 5’ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J Virol. 1995;69:6367-6375. [PubMed] |
| 72. | Ali N, Siddiqui A. The La antigen binds 5’ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249-2254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 215] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 73. | Anwar A, Ali N, Tanveer R, Siddiqui A. Demonstration of functional requirement of polypyrimidine tract-binding protein by SELEX RNA during hepatitis C virus internal ribosome entry site-mediated translation initiation. J Biol Chem. 2000;275:34231-34235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 64] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 74. | Gosert R, Chang KH, Rijnbrand R, Yi M, Sangar DV, Lemon SM. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites In vivo. Mol Cell Biol. 2000;20:1583-1595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 102] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 75. | Hahm B, Kim YK, Kim JH, Kim TY, Jang SK. Heterogeneous nuclear ribonucleoprotein L interacts with the 3’ border of the internal ribosomal entry site of hepatitis C virus. J Virol. 1998;72:8782-8788. [PubMed] |
| 76. | Kim JH, Paek KY, Ha SH, Cho S, Choi K, Kim CS, Ryu SH, Jang SK. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol Cell Biol. 2004;24:7878-7890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 83] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 77. | Hwang B, Lim JH, Hahm B, Jang SK, Lee SW. hnRNP L is required for the translation mediated by HCV IRES. Biochem Biophys Res Commun. 2009;378:584-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 78. | Paek KY, Kim CS, Park SM, Kim JH, Jang SK. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J Virol. 2008;82:12082-12093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 69] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 79. | Park SM, Paek KY, Hong KY, Jang CJ, Cho S, Park JH, Kim JH, Jan E, Jang SK. Translation-competent 48S complex formation on HCV IRES requires the RNA-binding protein NSAP1. Nucleic Acids Res. 2011;39:7791-7802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 80. | Rivas-Aravena A, Ramdohr P, Vallejos M, Valiente-Echeverría F, Dormoy-Raclet V, Rodríguez F, Pino K, Holzmann C, Huidobro-Toro JP, Gallouzi IE. The Elav-like protein HuR exerts translational control of viral internal ribosome entry sites. Virology. 2009;392:178-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 53] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 81. | Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716-7729. [PubMed] |
| 82. | Costa-Mattioli M, Svitkin Y, Sonenberg N. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol Cell Biol. 2004;24:6861-6870. [PubMed] |
| 83. | Pudi R, Srinivasan P, Das S. La protein binding at the GCAC site near the initiator AUG facilitates the ribosomal assembly on the hepatitis C virus RNA to influence internal ribosome entry site-mediated translation. J Biol Chem. 2004;279:29879-29888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 84. | Petz M, Them N, Huber H, Beug H, Mikulits W. La enhances IRES-mediated translation of laminin B1 during malignant epithelial to mesenchymal transition. Nucleic Acids Res. 2012;40:290-302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 85. | Ray PS, Das S. La autoantigen is required for the internal ribosome entry site-mediated translation of Coxsackievirus B3 RNA. Nucleic Acids Res. 2002;30:4500-4508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 68] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 86. | Kim YK, Back SH, Rho J, Lee SH, Jang SK. La autoantigen enhances translation of BiP mRNA. Nucleic Acids Res. 2001;29:5009-5016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 76] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 87. | Kim YK, Jang SK. La protein is required for efficient translation driven by encephalomyocarditis virus internal ribosomal entry site. J Gen Virol. 1999;80:3159-3166. [PubMed] |
| 88. | Ali N, Pruijn GJ, Kenan DJ, Keene JD, Siddiqui A. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J Biol Chem. 2000;275:27531-27540. [PubMed] |
| 89. | Holcik M, Korneluk RG. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol Cell Biol. 2000;20:4648-4657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 182] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 90. | Bayfield MA, Yang R, Maraia RJ. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim Biophys Acta. 2010;1799:365-378. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 132] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 91. | Lindop R, Arentz G, Thurgood LA, Reed JH, Jackson MW, Gordon TP. Pathogenicity and proteomic signatures of autoantibodies to Ro and La. Immunol Cell Biol. 2012;90:304-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 92. | Pudi R, Abhiman S, Srinivasan N, Das S. Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by specific interaction of independent regions of human La autoantigen. J Biol Chem. 2003;278:12231-12240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 66] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 93. | Simmonds P. Genetic diversity and evolution of hepatitis C virus--15 years on. J Gen Virol. 2004;85:3173-3188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 622] [Cited by in RCA: 632] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 94. | Dasgupta A, Das S, Izumi R, Venkatesan A, Barat B. Targeting internal ribosome entry site (IRES)-mediated translation to block hepatitis C and other RNA viruses. FEMS Microbiol Lett. 2004;234:189-199. [PubMed] |
| 95. | Boni S, Lavergne JP, Boulant S, Cahour A. Hepatitis C virus core protein acts as a trans-modulating factor on internal translation initiation of the viral RNA. J Biol Chem. 2005;280:17737-17748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 96. | Kalliampakou KI, Kalamvoki M, Mavromara P. Hepatitis C virus (HCV) NS5A protein downregulates HCV IRES-dependent translation. J Gen Virol. 2005;86:1015-1025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 97. | She Y, Han T, Ye L, Wu Z. Hepatitis C virus NS2/3 protease regulates HCV IRES-dependent translation and NS5B RdRp activity. Arch Virol. 2009;154:1465-1473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 98. | She Y, Liao Q, Chen X, Ye L, Wu Z. Hepatitis C virus (HCV) NS2 protein up-regulates HCV IRES-dependent translation and down-regulates NS5B RdRp activity. Arch Virol. 2008;153:1991-1997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 99. | Bung C, Bochkaeva Z, Terenin I, Zinovkin R, Shatsky IN, Niepmann M. Influence of the hepatitis C virus 3’-untranslated region on IRES-dependent and cap-dependent translation initiation. FEBS Lett. 2010;584:837-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 100. | Song Y, Friebe P, Tzima E, Jünemann C, Bartenschlager R, Niepmann M. The hepatitis C virus RNA 3’-untranslated region strongly enhances translation directed by the internal ribosome entry site. J Virol. 2006;80:11579-11588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 108] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 101. | Romero-López C, Berzal-Herranz A. A long-range RNA-RNA interaction between the 5’ and 3’ ends of the HCV genome. RNA. 2009;15:1740-1752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 102] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 102. | Schwarz KB. Oxidative stress during viral infection: a review. Free Radic Biol Med. 1996;21:641-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 470] [Cited by in RCA: 451] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 103. | Fujita N, Sugimoto R, Ma N, Tanaka H, Iwasa M, Kobayashi Y, Kawanishi S, Watanabe S, Kaito M, Takei Y. Comparison of hepatic oxidative DNA damage in patients with chronic hepatitis B and C. J Viral Hepat. 2008;15:498-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 99] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 104. | Konishi M, Iwasa M, Araki J, Kobayashi Y, Katsuki A, Sumida Y, Nakagawa N, Kojima Y, Watanabe S, Adachi Y. Increased lipid peroxidation in patients with non-alcoholic fatty liver disease and chronic hepatitis C as measured by the plasma level of 8-isoprostane. J Gastroenterol Hepatol. 2006;21:1821-1825. [PubMed] |
| 105. | 53 Venturini D, Simão AN, Barbosa DS, Lavado EL, Narciso VE, Dichi I, Dichi JB. Increased oxidative stress, decreased total antioxidant capacity, and iron overload in untreated patients with chronic hepatitis C. Dig Dis Sci. 2010;55:1120-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 106. | Yoshida Y, Imai Y, Sawai Y, Saito Y, Cao J, Fukuda K, Niki E. Hydroxyoctadecadienoic acid as a potential biomarker for oxidative stress in patients with chronic hepatitis C. J Gastroenterol Hepatol. 2010;25:107-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 107. | De Maria N, Colantoni A, Fagiuoli S, Liu GJ, Rogers BK, Farinati F, Van Thiel DH, Floyd RA. Association between reactive oxygen species and disease activity in chronic hepatitis C. Free Radic Biol Med. 1996;21:291-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 165] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 108. | Paradis V, Kollinger M, Fabre M, Holstege A, Poynard T, Bedossa P. In situ detection of lipid peroxidation by-products in chronic liver diseases. Hepatology. 1997;26:135-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 159] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 109. | Yamashita T, Kaneko S, Hashimoto S, Sato T, Nagai S, Toyoda N, Suzuki T, Kobayashi K, Matsushima K. Serial analysis of gene expression in chronic hepatitis C and hepatocellular carcinoma. Biochem Biophys Res Commun. 2001;282:647-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 110. | Diamond DL, Jacobs JM, Paeper B, Proll SC, Gritsenko MA, Carithers RL, Larson AM, Yeh MM, Camp DG, Smith RD. Proteomic profiling of human liver biopsies: hepatitis C virus-induced fibrosis and mitochondrial dysfunction. Hepatology. 2007;46:649-657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 92] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 111. | Bindoli A, Rigobello MP. Principles in redox signaling: from chemistry to functional significance. Antioxid Redox Signal. 2013;18:1557-1593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 148] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 112. | Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Mol Cell. 2007;26:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1348] [Cited by in RCA: 1156] [Article Influence: 64.2] [Reference Citation Analysis (0)] |
| 113. | Nathan C, Cunningham-Bussel A. Beyond oxidative stress: an immunologist’s guide to reactive oxygen species. Nat Rev Immunol. 2013;13:349-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 910] [Cited by in RCA: 1059] [Article Influence: 88.3] [Reference Citation Analysis (0)] |
| 114. | Pal S, Polyak SJ, Bano N, Qiu WC, Carithers RL, Shuhart M, Gretch DR, Das A. Hepatitis C virus induces oxidative stress, DNA damage and modulates the DNA repair enzyme NEIL1. J Gastroenterol Hepatol. 2010;25:627-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 108] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 115. | Blackham S, Baillie A, Al-Hababi F, Remlinger K, You S, Hamatake R, McGarvey MJ. Gene expression profiling indicates the roles of host oxidative stress, apoptosis, lipid metabolism, and intracellular transport genes in the replication of hepatitis C virus. J Virol. 2010;84:5404-5414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 116] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 116. | Joyce MA, Walters KA, Lamb SE, Yeh MM, Zhu LF, Kneteman N, Doyle JS, Katze MG, Tyrrell DL. HCV induces oxidative and ER stress, and sensitizes infected cells to apoptosis in SCID/Alb-uPA mice. PLoS Pathog. 2009;5:e1000291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 126] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 117. | Boudreau HE, Emerson SU, Korzeniowska A, Jendrysik MA, Leto TL. Hepatitis C virus (HCV) proteins induce NADPH oxidase 4 expression in a transforming growth factor beta-dependent manner: a new contributor to HCV-induced oxidative stress. J Virol. 2009;83:12934-12946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 110] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 118. | de Mochel NS, Seronello S, Wang SH, Ito C, Zheng JX, Liang TJ, Lambeth JD, Choi J. Hepatocyte NAD(P)H oxidases as an endogenous source of reactive oxygen species during hepatitis C virus infection. Hepatology. 2010;52:47-59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 145] [Cited by in RCA: 148] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 119. | Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol. 2013;11:443-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 911] [Cited by in RCA: 1087] [Article Influence: 90.6] [Reference Citation Analysis (0)] |
| 120. | Storz G, Tartaglia LA, Ames BN. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science. 1990;248:189-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 653] [Cited by in RCA: 627] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 121. | McFadden G. Even viruses can learn to cope with stress. Science. 1998;279:40-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 122. | Shisler JL, Senkevich TG, Berry MJ, Moss B. Ultraviolet-induced cell death blocked by a selenoprotein from a human dermatotropic poxvirus. Science. 1998;279:102-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 118] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 123. | Stehbens WE. Oxidative stress in viral hepatitis and AIDS. Exp Mol Pathol. 2004;77:121-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 76] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 124. | Deshmane SL, Amini S, Sen S, Khalili K, Sawaya BE. Regulation of the HIV-1 promoter by HIF-1α and Vpr proteins. Virol J. 2011;8:477. [PubMed] |
| 125. | Pyo CW, Yang YL, Yoo NK, Choi SY. Reactive oxygen species activate HIV long terminal repeat via post-translational control of NF-kappaB. Biochem Biophys Res Commun. 2008;376:180-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 38] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 126. | Schreck R, Rieber P, Baeuerle PA. Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-kappa B transcription factor and HIV-1. EMBO J. 1991;10:2247-2258. [PubMed] |
| 127. | Choi J, Lee KJ, Zheng Y, Yamaga AK, Lai MM, Ou JH. Reactive oxygen species suppress hepatitis C virus RNA replication in human hepatoma cells. Hepatology. 2004;39:81-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 138] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 128. | Grønbaek K, Sonne J, Ring-Larsen H, Poulsen HE, Friis H, Bygum Krarup H. Viral load is a negative predictor of antioxidant levels in hepatitis C patients. Scand J Infect Dis. 2005;37:686-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 129. | Waris G, Turkson J, Hassanein T, Siddiqui A. Hepatitis C virus (HCV) constitutively activates STAT-3 via oxidative stress: role of STAT-3 in HCV replication. J Virol. 2005;79:1569-1580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 134] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 130. | Fillebeen C, Pantopoulos K. Iron inhibits replication of infectious hepatitis C virus in permissive Huh7.5.1 cells. J Hepatol. 2010;53:995-999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 131. | Kuroki M, Ariumi Y, Ikeda M, Dansako H, Wakita T, Kato N. Arsenic trioxide inhibits hepatitis C virus RNA replication through modulation of the glutathione redox system and oxidative stress. J Virol. 2009;83:2338-2348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 132. | Lehmann E, El-Tantawy WH, Ocker M, Bartenschlager R, Lohmann V, Hashemolhosseini S, Tiegs G, Sass G. The heme oxygenase 1 product biliverdin interferes with hepatitis C virus replication by increasing antiviral interferon response. Hepatology. 2010;51:398-404. [PubMed] |
| 133. | Rivas-Estilla AM, Bryan-Marrugo OL, Trujillo-Murillo K, Pérez-Ibave D, Charles-Niño C, Pedroza-Roldan C, Ríos-Ibarra C, Ramírez-Valles E, Ortiz-López R, Islas-Carbajal MC. Cu/Zn superoxide dismutase (SOD1) induction is implicated in the antioxidative and antiviral activity of acetylsalicylic acid in HCV-expressing cells. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1264-G1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 134. | McCartney EM, Semendric L, Helbig KJ, Hinze S, Jones B, Weinman SA, Beard MR. Alcohol metabolism increases the replication of hepatitis C virus and attenuates the antiviral action of interferon. J Infect Dis. 2008;198:1766-1775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 135. | Seronello S, Ito C, Wakita T, Choi J. Ethanol enhances hepatitis C virus replication through lipid metabolism and elevated NADH/NAD+. J Biol Chem. 2010;285:845-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 136. | Yano M, Ikeda M, Abe K, Kawai Y, Kuroki M, Mori K, Dansako H, Ariumi Y, Ohkoshi S, Aoyagi Y. Oxidative stress induces anti-hepatitis C virus status via the activation of extracellular signal-regulated kinase. Hepatology. 2009;50:678-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 137. | McCartney EM, Helbig KJ, Narayana SK, Eyre NS, Aloia AL, Beard MR. Signal transducer and activator of transcription 3 is a proviral host factor for hepatitis C virus. Hepatology. 2013;58:1558-1568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 138. | Gendron K, Ferbeyre G, Heveker N, Brakier-Gingras L. The activity of the HIV-1 IRES is stimulated by oxidative stress and controlled by a negative regulatory element. Nucleic Acids Res. 2011;39:902-912. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 139. | Li W, Thakor N, Xu EY, Huang Y, Chen C, Yu R, Holcik M, Kong AN. An internal ribosomal entry site mediates redox-sensitive translation of Nrf2. Nucleic Acids Res. 2010;38:778-788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 104] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 140. | Daba A, Koromilas AE, Pantopoulos K. Alternative ferritin mRNA translation via internal initiation. RNA. 2012;18:547-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 141. | Zhang J, Dinh TN, Kappeler K, Tsaprailis G, Chen QM. La autoantigen mediates oxidant induced de novo Nrf2 protein translation. Mol Cell Proteomics. 2012;11:M111.015032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 142. | Yeh SH, Yang WB, Gean PW, Hsu CY, Tseng JT, Su TP, Chang WC, Hung JJ. Translational and transcriptional control of Sp1 against ischaemia through a hydrogen peroxide-activated internal ribosomal entry site pathway. Nucleic Acids Res. 2011;39:5412-5423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 143. | Belsham GJ. Divergent picornavirus IRES elements. Virus Res. 2009;139:183-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 113] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 144. | Sweeney TR, Dhote V, Yu Y, Hellen CU. A distinct class of internal ribosomal entry site in members of the Kobuvirus and proposed Salivirus and Paraturdivirus genera of the Picornaviridae. J Virol. 2012;86:1468-1486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 145. | Hellen CU, de Breyne S. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J Virol. 2007;81:5850-5863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 115] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 146. | Bieleski L, Hindley C, Talbot SJ. A polypyrimidine tract facilitates the expression of Kaposi’s sarcoma-associated herpesvirus vFLIP through an internal ribosome entry site. J Gen Virol. 2004;85:615-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 147. | Proud CG. eIF2 and the control of cell physiology. Semin Cell Dev Biol. 2005;16:3-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 305] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 148. | Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2430] [Cited by in RCA: 2559] [Article Influence: 98.4] [Reference Citation Analysis (0)] |
| 149. | Sood R, Porter AC, Olsen DA, Cavener DR, Wek RC. A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2alpha. Genetics. 2000;154:787-801. [PubMed] |
| 150. | Williams BR. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112-6120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 627] [Cited by in RCA: 641] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 151. | Chen JJ, London IM. Regulation of protein synthesis by heme-regulated eIF-2 alpha kinase. Trends Biochem Sci. 1995;20:105-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 232] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 152. | Jiang HY, Wek RC. GCN2 phosphorylation of eIF2alpha activates NF-kappaB in response to UV irradiation. Biochem J. 2005;385:371-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 132] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 153. | Mouton-Liger F, Paquet C, Dumurgier J, Bouras C, Pradier L, Gray F, Hugon J. Oxidative stress increases BACE1 protein levels through activation of the PKR-eIF2α pathway. Biochim Biophys Acta. 2012;1822:885-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 136] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 154. | Pyo CW, Choi JH, Oh SM, Choi SY. Oxidative stress-induced cyclin D1 depletion and its role in cell cycle processing. Biochim Biophys Acta. 2013;1830:5316-5325. [PubMed] |
| 155. | Lu L, Han AP, Chen JJ. Translation initiation control by heme-regulated eukaryotic initiation factor 2alpha kinase in erythroid cells under cytoplasmic stresses. Mol Cell Biol. 2001;21:7971-7980. [PubMed] |
| 156. | McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem. 2005;280:16925-16933. [PubMed] |
| 157. | O’Loghlen A, Pérez-Morgado MI, Salinas M, Martín ME. Reversible inhibition of the protein phosphatase 1 by hydrogen peroxide. Potential regulation of eIF2 alpha phosphorylation in differentiated PC12 cells. Arch Biochem Biophys. 2003;417:194-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 158. | Kim JH, Park SM, Park JH, Keum SJ, Jang SK. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 2011;30:2454-2464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 97] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 159. | Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 2008;27:1060-1072. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 122] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 160. | Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol. 2008;15:836-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 150] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 161. | Fernandez J, Bode B, Koromilas A, Diehl JA, Krukovets I, Snider MD, Hatzoglou M. Translation mediated by the internal ribosome entry site of the cat-1 mRNA is regulated by glucose availability in a PERK kinase-dependent manner. J Biol Chem. 2002;277:11780-11787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 66] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 162. | Fernandez J, Yaman I, Sarnow P, Snider MD, Hatzoglou M. Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2alpha. J Biol Chem. 2002;277:19198-19205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 156] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 163. | Gerlitz G, Jagus R, Elroy-Stein O. Phosphorylation of initiation factor-2 alpha is required for activation of internal translation initiation during cell differentiation. Eur J Biochem. 2002;269:2810-2819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 85] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 164. | Choi SY, Scherer BJ, Schnier J, Davies MV, Kaufman RJ, Hershey JW. Stimulation of protein synthesis in COS cells transfected with variants of the alpha-subunit of initiation factor eIF-2. J Biol Chem. 1992;267:286-293. [PubMed] |
| 165. | Chan SW, Egan PA. Effects of hepatitis C virus envelope glycoprotein unfolded protein response activation on translation and transcription. Arch Virol. 2009;154:1631-1640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 166. | Jan E, Kinzy TG, Sarnow P. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc Natl Acad Sci USA. 2003;100:15410-15415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 112] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 167. | Lancaster AM, Jan E, Sarnow P. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA. 2006;12:894-902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 82] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 168. | Redondo N, Sanz MA, Welnowska E, Carrasco L. Translation without eIF2 promoted by poliovirus 2A protease. PLoS One. 2011;6:e25699. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 169. | Garaigorta U, Chisari FV. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2009;6:513-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 186] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 170. | Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem. 2010;285:26779-26787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 128] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 171. | Robert F, Kapp LD, Khan SN, Acker MG, Kolitz S, Kazemi S, Kaufman RJ, Merrick WC, Koromilas AE, Lorsch JR. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2.GTP.Met-tRNA(i)(Met) ternary complex availability. Mol Biol Cell. 2006;17:4632-4644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 106] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 172. | Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787-1801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 208] [Cited by in RCA: 182] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 173. | Thakor N, Holcik M. IRES-mediated translation of cellular messenger RNA operates in eIF2α- independent manner during stress. Nucleic Acids Res. 2012;40:541-552. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 174. | Jack SC, Chan SW. The role of PERK and GCN2 in basal and hydrogen peroxide-regulated translation from the hepatitis C virus internal ribosome entry site. Virus Genes. 2011;43:208-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 175. | Cho H, Lee HC, Jang SK, Kim YK. Iron increases translation initiation directed by internal ribosome entry site of hepatitis C virus. Virus Genes. 2008;37:154-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 176. | Wang Q, Liu Y, An D, Diao H, Xu W, He X, Sun R, Wei L, Li L. Regulation of hepatitis C virus translation initiation by iron: role of eIF3 and La protein. Virus Res. 2012;167:302-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 177. | de Breyne S, Soto-Rifo R, López-Lastra M, Ohlmann T. Translation initiation is driven by different mechanisms on the HIV-1 and HIV-2 genomic RNAs. Virus Res. 2013;171:366-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 178. | Kuiken C, Simmonds P. Nomenclature and numbering of the hepatitis C virus. Methods Mol Biol. 2009;510:33-53. [PubMed] |
| 179. | Soler M, Pellerin M, Malnou CE, Dhumeaux D, Kean KM, Pawlotsky JM. Quasispecies heterogeneity and constraints on the evolution of the 5’ noncoding region of hepatitis C virus (HCV): relationship with HCV resistance to interferon-alpha therapy. Virology. 2002;298:160-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 44] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 180. | Honda M, Beard MR, Ping LH, Lemon SM. A phylogenetically conserved stem-loop structure at the 5’ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J Virol. 1999;73:1165-1174. [PubMed] |
| 181. | Odreman-Macchioli FE, Tisminetzky SG, Zotti M, Baralle FE, Buratti E. Influence of correct secondary and tertiary RNA folding on the binding of cellular factors to the HCV IRES. Nucleic Acids Res. 2000;28:875-885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 51] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 182. | Barría MI, González A, Vera-Otarola J, León U, Vollrath V, Marsac D, Monasterio O, Pérez-Acle T, Soza A, López-Lastra M. Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role in cap-independent translation. Nucleic Acids Res. 2009;37:957-971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 183. | Zhang J, Yamada O, Ito T, Akiyama M, Hashimoto Y, Yoshida H, Makino R, Masago A, Uemura H, Araki H. A single nucleotide insertion in the 5’-untranslated region of hepatitis C virus leads to enhanced cap-independent translation. Virology. 1999;261:263-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 184. | Buratti E, Gerotto M, Pontisso P, Alberti A, Tisminetzky SG, Baralle FE. In vivo translational efficiency of different hepatitis C virus 5’-UTRs. FEBS Lett. 1997;411:275-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 185. | Honda M, Rijnbrand R, Abell G, Kim D, Lemon SM. Natural variation in translational activities of the 5’ nontranslated RNAs of hepatitis C virus genotypes 1a and 1b: evidence for a long-range RNA-RNA interaction outside of the internal ribosomal entry site. J Virol. 1999;73:4941-4951. [PubMed] |
| 186. | Collier AJ, Tang S, Elliott RM. Translation efficiencies of the 5’ untranslated region from representatives of the six major genotypes of hepatitis C virus using a novel bicistronic reporter assay system. J Gen Virol. 1998;79:2359-2366. [PubMed] |
| 187. | Laporte J, Malet I, Andrieu T, Thibault V, Toulme JJ, Wychowski C, Pawlotsky JM, Huraux JM, Agut H, Cahour A. Comparative analysis of translation efficiencies of hepatitis C virus 5’ untranslated regions among intraindividual quasispecies present in chronic infection: opposite behaviors depending on cell type. J Virol. 2000;74:10827-10833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 188. | Hazari S, Patil A, Joshi V, Sullivan DE, Fermin CD, Garry RF, Elliott RM, Dash S. Alpha interferon inhibits translation mediated by the internal ribosome entry site of six different hepatitis C virus genotypes. J Gen Virol. 2005;86:3047-3053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 189. | Yasmeen A, Hamid S, Granath FN, Lindström H, Elliott RM, Siddiqui AA, Persson MA. Correlation between translation efficiency and outcome of combination therapy in chronic hepatitis C genotype 3. J Viral Hepat. 2006;13:87-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 190. | El Awady MK, Azzazy HM, Fahmy AM, Shawky SM, Badreldin NG, Yossef SS, Omran MH, Zekri AR, Goueli SA. Positional effect of mutations in 5’UTR of hepatitis C virus 4a on patients’ response to therapy. World J Gastroenterol. 2009;15:1480-1486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 18] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (4)] |
| 191. | Ogata K, Kashiwagi T, Iwahashi J, Hara K, Honda H, Ide T, Kumashiro R, Kohara M, Sata M, Watanabe H. A mutational shift from domain III to II in the internal ribosome entry site of hepatitis C virus after interferon-ribavirin therapy. Arch Virol. 2008;153:1575-1579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 192. | Thelu MA, Drouet E, Hilleret MN, Zarski JP. Lack of clinical significance of variability in the internal ribosome entry site of hepatitis C virus. J Med Virol. 2004;72:396-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 193. | Thélu MA, Leroy V, Ramzan M, Dufeu-Duchesne T, Marche P, Zarski JP. IRES complexity before IFN-alpha treatment and evolution of the viral load at the early stage of treatment in peripheral blood mononuclear cells from chronic hepatitis C patients. J Med Virol. 2007;79:242-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 194. | Friebe P, Lohmann V, Krieger N, Bartenschlager R. Sequences in the 5’ nontranslated region of hepatitis C virus required for RNA replication. J Virol. 2001;75:12047-12057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 258] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 195. | Masante C, Mahias K, Lourenço S, Dumas E, Cahour A, Trimoulet P, Fleury H, Astier-Gin T, Ventura M. Seven nucleotide changes characteristic of the hepatitis C virus genotype 3 5’ untranslated region: correlation with reduced in vitro replication. J Gen Virol. 2008;89:212-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |