Published online Dec 28, 2013. doi: 10.3748/wjg.v19.i48.9461
Revised: October 11, 2013
Accepted: November 2, 2013
Published online: December 28, 2013
Processing time: 134 Days and 0.5 Hours
AIM: To evaluate the association between the tumour necrosis factor alpha-308 (TNF-α-308) gene polymorphism and the risk of digestive system cancers.
METHODS: All eligible case-control studies published up to December 2012 were identified by searching PubMed, Web of Science, Embase and China National Knowledge Internet without language restrictions. The risk of digestive system cancers associated with the TNF-α-308 polymorphism was estimated for each study using odds ratio (OR) together with its 95%CI, respectively. Cochrane Collaboration RevMan 5.1 was used to perform the analysis. A χ2-test-based Q statistic test and an I2 test were performed to assess the between-study heterogeneity. When the Q test was significant (P < 0.05) or I2 > 50%, the random effects model was used, otherwise the fixed effects model was used.
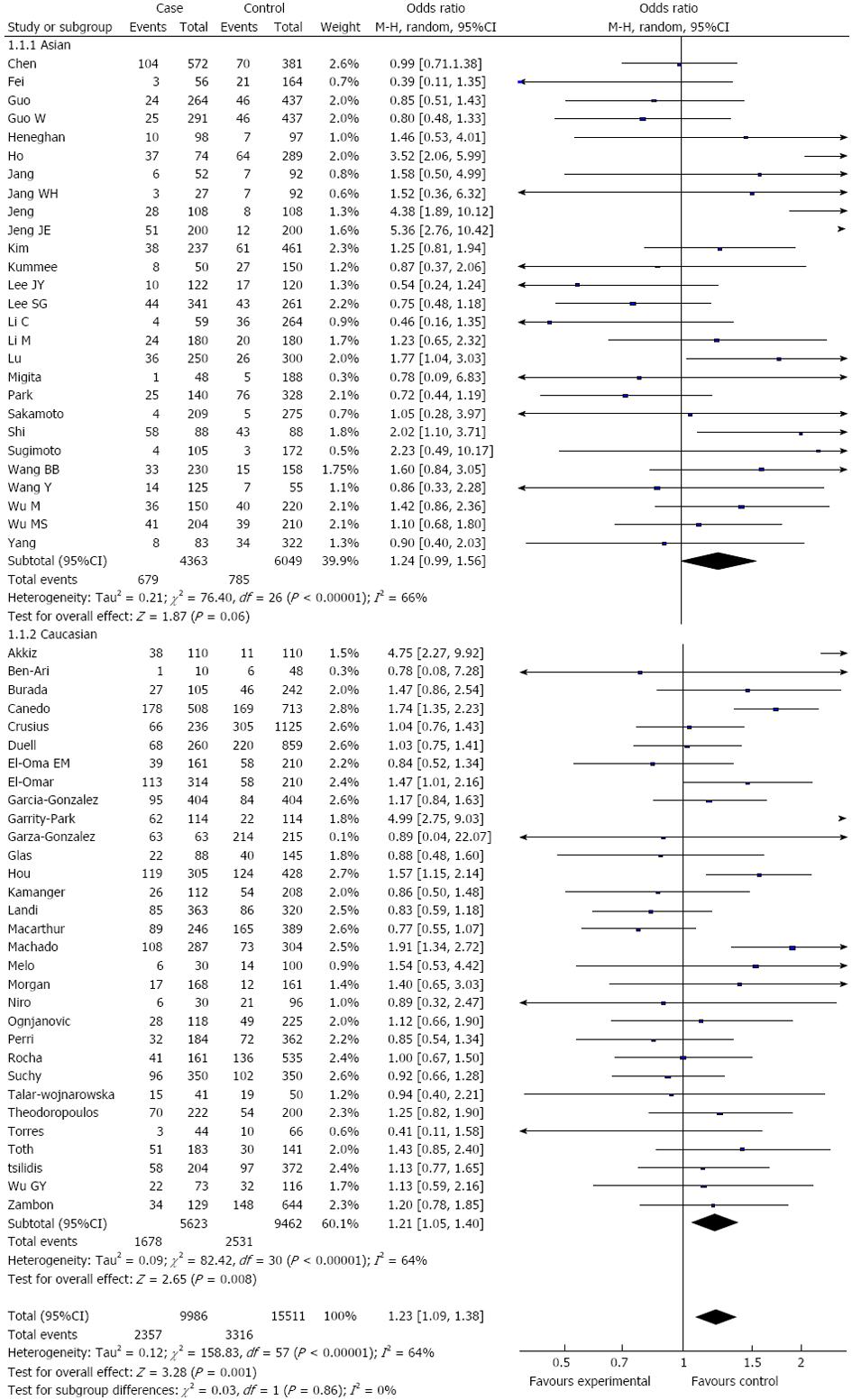
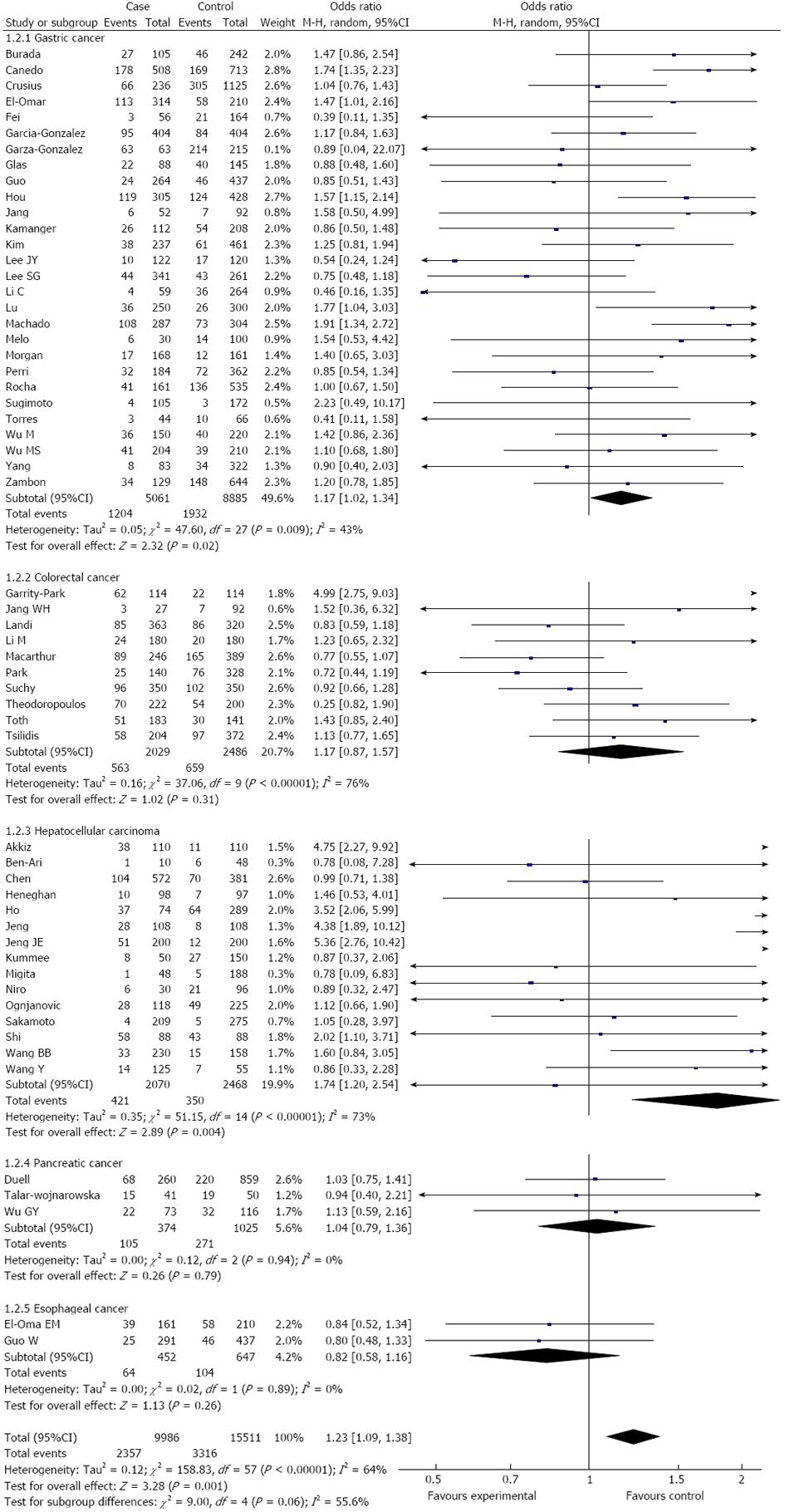
RESULTS: Fifty-eight studies from fifty-five publications with a total of 9986 cancer patients and 15511 healthy controls were included. Overall, a significant association was found between the TNF-α-308 polymorphism and the risk of digestive system cancers [dominant model: OR = 1.23, 95%CI: 1.09-1.39, (G/A) vs (G/G): OR = 1.15, 95%CI: 1.02-1.28, (A/A) vs (G/G): OR = 1.44, 95%CI: 1.19-1.73, recessive model: OR = 1.38, 95%CI: 1.15-1.66]. Furthermore, when the analysis was stratified by ethnicity, similar results were observed in both the Asian and Caucasian populations, except for the dominant model and heterozygote comparisons in the Asian population [dominant model: OR = 1.24, 95%CI: 0.99-1.56, (G/A) vs (G/G): OR = 1.09, 95%CI: 0.96-1.24]. When the cancer type subgroups were examined, similar results were detected in gastric and hepatocellular carcinomas; however, no significant association was observed among other digestive system cancers.
CONCLUSION: The TNF-α-308 gene polymorphism may be significantly associated with the risk of gastric and hepatocellular carcinomas, but not colorectal, pancreatic, or oesophageal cancer, in the Asian population.
Core tip: Genetic polymorphisms contribute to the risk of human malignant tumours. Many studies have reported the relationship between the tumour necrosis factor alpha-308 (TNF-α-308) gene polymorphism and risk of digestive system cancers. However, the results of these studies are inconsistent and contradictory. In this meta-analysis, our results suggest that the TNF-α-308 polymorphism is significantly associated with the risk of gastric and hepatocellular carcinomas in the Asian population (dominant model: 95%CI: 1.02-1.34, P < 0.05 and 95%CI: 1.20-2.54, P < 0.05, respectively). This finding indicates that certain polymorphisms and mutations at TNF-α-308 may increase susceptibility to digestive system cancers.
-
Citation: Guo XF, Wang J, Yu SJ, Song J, Ji MY, Cao Z, Zhang JX, Wang J, Dong WG.
TNF-α-308 polymorphism and risk of digestive system cancers: A meta-analysis. World J Gastroenterol 2013; 19(48): 9461-9471 - URL: https://www.wjgnet.com/1007-9327/full/v19/i48/9461.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i48.9461
Digestive system cancers are the most common malignant tumours worldwide, with 3.4 million new cases each year, and their mortality rates have increased gradually over the past decade[1,2]. Molecular epidemiology has confirmed that carcinogenesis is a complex, multifactorial and multistep event, in which the interaction of environmental triggers and genetic susceptibility may play an important role. However, the exact mechanism of carcinogenesis is still not fully understood.
Tumour necrosis factor-alpha (TNF-α), which is mainly produced by macrophages, is a multifunctional cytokine that plays an important role in the pathogenesis of inflammatory, autoimmune, and malignant diseases[3]. The TNF-α gene is located in the major histocompatibility complex class III region on the short arm of chromosome six. Several polymorphisms in the promoter region of the TNF-α gene have been identified and are implicated in the regulation of TNF-α transcription[4-5]. The TNF-α-308 polymorphism (rs1800629) is the most extensively studied polymorphism in digestive system cancers[6-9]. However, the results of the studies on TNF-α-308 have been inconclusive or inconsistent. Therefore, we conducted a meta-analysis to evaluate the association between the TNF-α-308 polymorphism and susceptibility to digestive system cancers.
A literature search was conducted using PubMed, Web of Science, Embase and CNKI for studies that were published up to December 2012 without language restrictions. The relevant studies were identified using the following terms: [“tumour necrosis factor alpha or TNF alpha or TNF-α”] AND [“genetic polymorphism or polymorphisms or variant”] AND [“digestive system cancer or gastric cancer or colorectal cancer or hepatocellular carcinoma or pancreatic cancer or oesophageal cancer”]. The search was restricted to humans. Additional studies were identified by a manual search of references of original or review articles on this topic. If more than one cancer type was reported in one study, the data for each type was extracted separately. If data or data subsets were published in more than one article, only the publication with the largest sample size was included.
Studies were included if they met the following criteria: (1) studies that evaluated the association between the TNF-α-308 polymorphism and digestive system cancer risk; (2) studies with a case-control study design; and (3) studies with detailed genotype frequencies for cases and controls or text that allowed for the calculation of these values. The major exclusion criteria were: (1) case-only studies, case reports, or review articles; (2) studies without raw data for the TNF-α-308G/A genotype; and (3) studies that compared the TNF-α-308G/A variants in precancerous lesions and other cancers.
Two investigators (Guo XF and Wang J) independently extracted the data and reached a consensus on each item. If the two investigators generated different results, they would check the data again and have a discussion to come to an agreement. If they could not reach an agreement, an expert (Dong WG) was invited to the discussion. The data extracted from the selected articles included the first author’s name, year of publication, country of origin, ethnicity, cancer type, genotyping methods, and number of cases and controls. The ethnicities were categorised as Asian or Caucasian. The cancer types were categorised as gastric, colorectal, hepatocellular, pancreatic, or oesophageal.
The meta-analysis was performed using the Cochrane Collaboration RevMan 5.1 software (Copenhagen, 2008). The association between the risk of digestive system cancers and the TNF-α-308 polymorphism was estimated for each study using the odds ratio (OR) and 95%CI. A χ2 test-based calculation of the Q statistic was performed to assess the between-study heterogeneity[10]. We also quantified the effect of heterogeneity with an I2 test. When the Q test was significant (P < 0.05) or I2 > 50%, indicating heterogeneity across studies, the random effects model was used[11]; otherwise, the fixed effects model was used[12]. Before estimating the relationship between the TNF-α-308 polymorphism and digestive system cancer risk, we tested whether the genotype frequencies of the controls were in Hardy-Weinberg equilibrium (HWE) using a χ2 test. We first estimated this relationship with the dominant model [G/A (GA) + A/A (AA) vs G/G (GG)] and the recessive model (AA vs GA + GG) and then with the co-dominant model (GA vs GG and AA vs GG). To evaluate the ethnicity-specific and cancer type-specific effects, we performed stratification analyses with respect to ethnicity and cancer type. Sensitivity analysis was performed to evaluate the stability of the results. Funnel plots were used to evaluate publication bias.
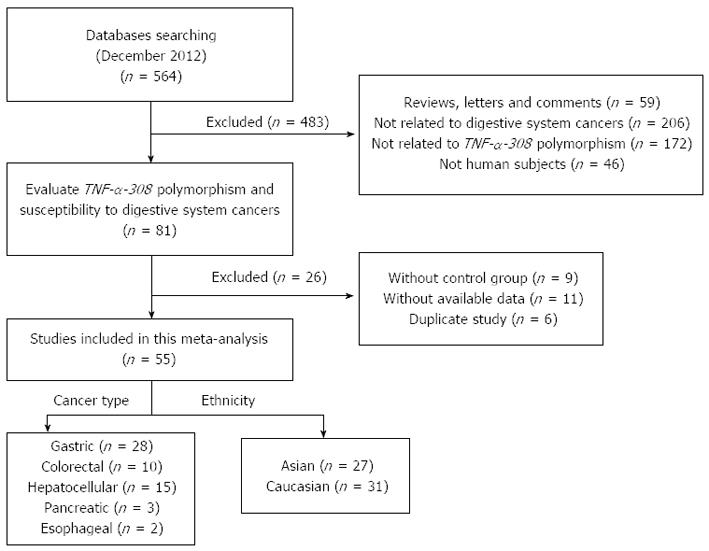
The search strategy retrieved 564 potentially relevant studies. According to the inclusion criteria, 55 studies with full-text were included in this meta-analysis and 509 studies were excluded. A flow chart of the study selection is shown in Figure 1. Because the studies of El-Omar et al[9], Guo et al[13] and Jang et al[14] each included separate analyses of two cancer types, we treated them separately in this meta-analysis[9,13,14]. Therefore, as shown in Table 1, there were 58 case-control studies from 55 publications on the TNF-α-308 polymorphism with a total of 9986 cancer cases and 15511 controls. Two ethnicities were addressed: 27 studies focused on Asian populations, and 31 studies focused on Caucasian populations. Five cancer types were addressed: 28 studies focused on gastric cancer[6-9,13-36], 10 studies on colorectal cancer[14,37-45], 15 studies on hepatocellular carcinoma[46-60], 3 studies on pancreatic cancer[61-63], and 2 studies on oesophageal cancer[9,13]. The genotype distribution in the controls was consistent with HWE for all of the selected studies, except for four studies on gastric cancer[7,13,33-34], one study on colorectal cancer[43], six studies on hepatocellular carcinoma[47,49,51-52,55-56], and one study on esophageal cancer[13].
| Ref. | Year | Country | Ethnicity | Cancer type | Genotyping method | Case | Control | P | ||||||
| Total | GG | GA | AA | Total | GG | GA | AA | |||||||
| Burada et al[6] | 2012 | Romania | Caucasian | Gastric | TaqMan | 105 | 78 | 26 | 1 | 242 | 196 | 44 | 2 | 0.78 |
| Canedo et al[7] | 2008 | Portugal | Caucasian | Gastric | TaqMan | 508 | 330 | 1781 | 713 | 544 | 1691 | NA | ||
| Crusius et al[8] | 2008 | Spain | Caucasian | Gastric | Real-time PCR | 236 | 170 | 64 | 2 | 1125 | 820 | 274 | 31 | 0.17 |
| El-Omar et al[9] | 2003 | United States | Caucasian | Gastric | TaqMan | 314 | 201 | 87 | 26 | 210 | 152 | 52 | 6 | 0.55 |
| Guo et al[13] | 2005 | China | Asian | Gastric | PCR-RFLP | 264 | 240 | 20 | 4 | 437 | 391 | 40 | 6 | < 0.01 |
| Jang et al[14] | 2001 | South Korea | Asian | Gastric | PCR-RFLP | 52 | 46 | 4 | 2 | 92 | 85 | 7 | 0 | 0.70 |
| Fei et al[15] | 2004 | China | Asian | Gastric | PCR | 56 | 53 | 3 | 0 | 164 | 143 | 20 | 1 | 0.74 |
| Garcia-Gonzalez et al[16] | 2007 | Spain | Caucasian | Gastric | TaqMan | 404 | 309 | 84 | 11 | 404 | 320 | 77 | 7 | 0.35 |
| Garza-Gonzalez et al[17] | 2005 | Mexico | Caucasian | Gastric | PCR-RFLP | 63 | 0 | 8 | 55 | 215 | 1 | 35 | 179 | 0.61 |
| Glas et al[18] | 2004 | Germany | Caucasian | Gastric | PCR-RFLP | 88 | 66 | 19 | 3 | 145 | 105 | 36 | 4 | 0.67 |
| Hou et al[19] | 2007 | Poland | Caucasian | Gastric | TaqMan | 305 | 186 | 98 | 21 | 428 | 304 | 109 | 15 | 0.19 |
| Kamangar et al[20] | 2006 | Finland | Caucasian | Gastric | TaqMan | 112 | 86 | 23 | 3 | 208 | 154 | 52 | 2 | 0.29 |
| Kim et al[21] | 2006 | South Korea | Asian | Gastric | PCR-RFLP | 237 | 199 | 34 | 4 | 461 | 400 | 59 | 2 | 0.91 |
| Lee et al[22] | 2004 | South Korea | Asian | Gastric | PCR | 341 | 297 | 43 | 1 | 261 | 218 | 42 | 1 | 0.49 |
| Lee et al[23] | 2005 | South Korea | Asian | Gastric | PCR-RFLP | 122 | 112 | 10 | 0 | 120 | 103 | 17 | 0 | 0.40 |
| Li et al[24] | 2005 | China | Asian | Gastric | PCR-RFLP | 59 | 55 | 4 | 0 | 264 | 228 | 34 | 2 | 0.56 |
| Lu et al[25] | 2005 | China | Asian | Gastric | PCR-DHPLC | 250 | 214 | 36 | 0 | 300 | 274 | 24 | 2 | 0.08 |
| Machado et al[26] | 2003 | Portugal | Caucasian | Gastric | PCR-SSCP | 287 | 179 | 105 | 3 | 304 | 231 | 69 | 4 | 0.65 |
| Melo et al[27] | 2009 | Brazil | Caucasian | Gastric | PCR-RFLP | 30 | 24 | 5 | 1 | 100 | 86 | 13 | 1 | 0.53 |
| Morgan et al[28] | 2006 | Honduras | Caucasian | Gastric | TaqMan | 168 | 151 | 17 | 0 | 161 | 149 | 12 | 0 | 0.62 |
| Perri et al[29] | 2005 | Italy | Caucasian | Gastric | PCR-RFLP | 184 | 152 | 30 | 2 | 362 | 290 | 65 | 7 | 0.15 |
| Rocha et al[30] | 2005 | Brazil | Caucasian | Gastric | PCR-RFLP | 161 | 120 | 37 | 4 | 535 | 399 | 123 | 13 | 0.34 |
| Sugimoto et al[31] | 2007 | Japan | Asian | Gastric | PCR-RFLP | 105 | 101 | 4 | 0 | 172 | 169 | 3 | 0 | 0.91 |
| Torres et al[32] | 2004 | Colombia | Caucasian | Gastric | PCR | 44 | 41 | 3 | 0 | 66 | 56 | 10 | 0 | 0.51 |
| Wu et al[33] | 2002 | China | Asian | Gastric | Direct sequencing | 150 | 114 | 27 | 9 | 220 | 180 | 27 | 13 | < 0.01 |
| Wu et al[34] | 2004 | China | Asian | Gastric | Direct sequencing | 204 | 163 | 29 | 12 | 210 | 171 | 26 | 13 | < 0.01 |
| Yang et al[35] | 2009 | South Korea | Asian | Gastric | SNaPshot | 83 | 75 | 8 | 0 | 322 | 288 | 34 | 0 | 0.32 |
| Zambon et al[36] | 2005 | Italy | Caucasian | Gastric | TaqMan | 129 | 95 | 31 | 3 | 644 | 496 | 138 | 10 | 0.91 |
| Garrity-Park et al[37] | 2008 | Ireland | Caucasian | Colorectal | PCR, sequencing | 114 | 52 | 49 | 13 | 114 | 92 | 20 | 2 | 0.46 |
| Jang et al[14] | 2001 | South Korea | Asian | Colorectal | PCR-RFLP | 27 | 24 | 3 | 0 | 92 | 85 | 7 | 0 | 0.70 |
| Landi et al[38] | 2003 | Spain | Caucasian | Colorectal | TaqMan | 363 | 278 | 80 | 5 | 320 | 234 | 76 | 10 | 0.22 |
| Li M et al[39] | 2011 | China | Asian | Colorectal | PCR-RFLP | 180 | 156 | 15 | 9 | 180 | 160 | 19 | 1 | 0.60 |
| Macarthur et al[40] | 2005 | Scotland | Caucasian | Colorectal | TaqMan | 246 | 157 | 74 | 15 | 389 | 224 | 145 | 20 | 0.58 |
| Park et al[41] | 1998 | South Korea | Asian | Colorectal | PCR-RFLP | 140 | 115 | 24 | 1 | 328 | 252 | 72 | 4 | 0.65 |
| Suchy et al[42] | 2008 | Poland | Caucasian | Colorectal | PCR-RFLP | 350 | 254 | 87 | 9 | 350 | 248 | 95 | 7 | 0.55 |
| Theodoropoulos et al[43] | 2006 | Greece | Caucasian | Colorectal | PCR-RFLP | 222 | 152 | 56 | 14 | 200 | 146 | 44 | 10 | 0.01 |
| Toth et al[44] | 2007 | Hungary | Caucasian | Colorectal | PCR-SSP | 183 | 132 | 48 | 3 | 141 | 111 | 30 | 0 | 0.16 |
| Tsilidis et al[45] | 2009 | United States | Caucasian | Colorectal | TaqMan | 204 | 146 | 55 | 3 | 372 | 275 | 90 | 7 | 0.91 |
| Akkiz et al[46] | 2009 | Turkey | Caucasian | Hepatocellular | PCR-RFLP | 110 | 72 | 35 | 3 | 110 | 99 | 11 | 0 | 0.58 |
| Ben-Ari et al[47] | 2003 | United States | Caucasian | Hepatocellular | PCR-SSP | 10 | 9 | 11 | 48 | 42 | 61 | NA | ||
| Chen et al[48] | 2005 | China | Asian | Hepatocellular | TaqMan | 572 | 468 | 95 | 9 | 381 | 311 | 67 | 3 | 0.77 |
| Heneghan et al[49] | 2003 | China | Asian | Hepatocellular | ASO-PCR | 98 | 88 | 10 | 0 | 97 | 90 | 6 | 1 | 0.03 |
| Ho et al[50] | 2004 | China | Asian | Hepatocellular | PCR-RFLP | 74 | 37 | 34 | 3 | 289 | 225 | 62 | 2 | 0.30 |
| Jeng et al[51] | 2007 | China | Asian | Hepatocellular | PCR-SSO | 108 | 80 | 281 | 108 | 100 | 81 | NA | ||
| Jeng JE et al[52] | 2009 | China | Asian | Hepatocellular | PCR-SSO | 200 | 149 | 511 | 200 | 188 | 121 | NA | ||
| Kummee et al[53] | 2007 | Thailand | Asian | Hepatocellular | PCR-RFLP | 50 | 42 | 8 | 0 | 150 | 123 | 26 | 1 | 0.77 |
| Migita et al[54] | 2005 | Japan | Asian | Hepatocellular | PCR-SSP | 48 | 47 | 1 | 0 | 188 | 183 | 5 | 0 | 0.85 |
| Niro et al[55] | 2005 | Italy | Caucasian | Hepatocellular | Direct sequencing | 30 | 24 | 61 | 96 | 75 | 211 | NA | ||
| Ognjanovic et al[56] | 2009 | United States | Caucasian | Hepatocellular | TaqMan | 118 | 90 | 281 | 225 | 176 | 491 | NA | ||
| Sakamoto et al[57] | 2008 | Japan | Asian | Hepatocellular | PCR-RFLP | 209 | 205 | 4 | 0 | 275 | 270 | 5 | 0 | 0.88 |
| Shi et al[58] | 2011 | China | Asian | Hepatocellular | PCR-RFLP | 88 | 30 | 43 | 15 | 88 | 45 | 35 | 8 | 0.75 |
| Wang et al[59] | 2003 | Japan | Asian | Hepatocellular | Direct sequencing | 125 | 111 | 13 | 1 | 55 | 48 | 6 | 1 | 0.16 |
| Wang et al[60] | 2010 | China | Asian | Hepatocellular | PCR-SSO | 230 | 197 | 30 | 3 | 158 | 143 | 15 | 0 | 0.53 |
| Duell et al[61] | 2006 | United States | Caucasian | Pancreatic | PCR-RFLP | 260 | 192 | 63 | 5 | 859 | 639 | 198 | 22 | 0.16 |
| Talor-wojnarowska et al[62] | 2009 | Poland | Caucasian | Pancreatic | PCR-RFLP | 41 | 26 | 12 | 3 | 50 | 31 | 17 | 2 | 0.86 |
| Wu GY et al[63] | 2010 | Germany | Caucasian | Pancreatic | PCR-RFLP | 73 | 51 | 20 | 2 | 116 | 84 | 30 | 2 | 0.72 |
| El-Omar et al[9] | 2003 | United States | Caucasian | Esophageal | TaqMan | 161 | 122 | 34 | 5 | 210 | 152 | 52 | 6 | 0.55 |
| Guo et al[13] | 2005 | China | Asian | Esophageal | PCR-RFLP | 291 | 266 | 21 | 4 | 437 | 391 | 40 | 6 | < 0.01 |
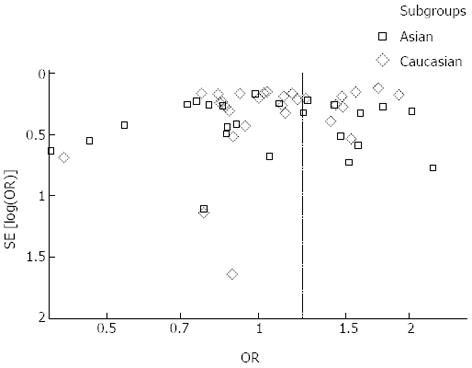
Overall, there was a significant difference in the TNF-α-308G/A genotype distribution between the digestive system cancer patients and the controls (dominant model: OR = 1.23, 95%CI: 1.09-1.39, P < 0.00001; GA vs GG: OR = 1.15, 95%CI: 1.02-1.28, P < 0.0001; AA vs GG: OR = 1.44, 95%CI: 1.19-1.73, P = 0.23; recessive model: OR = 1.38, 95%CI: 1.15-1.66, P = 0.50) (Table 2, Figure 2). In the analysis of the ethnic subgroups, similar results were observed in the Caucasian population; but in the Asian population, we found that there was no significant association between the TNF-α-308 polymorphism and the risk of digestive system cancers in the dominant model and heterozygote comparisons (GA + AA vs GG: OR = 1.24, 95%CI: 0.99-1.56, GA vs GG: OR = 1.09, 95%CI: 0.96-1.24) (Table 2, Figure 2). When stratified by cancer type, similar results were detected for gastric and hepatocellular carcinomas; however, no significant association was observed among the other digestive system cancer types (Table 2, Figure 3). Furthermore, we found that there was significant heterogeneity for the dominant model and heterozygote comparisons both overall and in the stratified analyses: I2 = 64% and 52% in the overall population, I2 = 66% and 45% (P = 0.008) in the Asian population, I2 = 64% and 58% in the Caucasian population, I2 = 76% and 70% in colorectal cancer, and I2 = 73% and 66% in hepatocellular carcinoma. In addition, there was evidence of heterogeneity in gastric cancer (dominant model: P = 0.009). Thus, the random effects model was employed in the OR calculations. Then, sensitivity analyses were conducted to determine whether modification of the inclusion criteria of the meta-analysis affected the final results. We examined the influence of these studies on the pooled OR by repeating the meta-analysis while excluding the study that was not in HWE. The estimated pooled OR did not show a significant change (Table 2), indicating that our results are statistically robust. The shapes of the funnel plots did not reveal any evidence of asymmetry, suggesting that there was no publication bias among the studies (Figure 4).
| Group | GA + AA vs GG | GA vs GG | AA vs GG | AA vs GA + GG | ||||||||
| n | OR (95%CI) | P1 | n | OR (95%CI) | P1 | n | OR (95%CI) | P1 | n | OR (95%CI) | P1 | |
| Overall | 58 | 1.23 (1.09, 1.38)2 | < 0.00001 | 52 | 1.14 (1.01, 1.28)2 | < 0.00001 | 44 | 1.43 (1.19, 1.73) | 0.26 | 44 | 1.38 (1.15, 1.66) | 0.55 |
| Studies with HWE | 46 | 1.18 (1.03, 1.34)2 | < 0.00001 | 46 | 1.14 (1.00, 1.29)2 | < 0.00001 | 38 | 1.54 (1.25, 1.90) | 0.15 | 38 | 1.48 (1.20, 1.81) | 0.40 |
| Cancer type | ||||||||||||
| Gastric | 28 | 1.23 (1.12, 1.34)2 | 0.009 | 27 | 1.15 (1.04, 1.27) | 0.07 | 22 | 1.38 (1.06, 1.80) | 0.63 | 22 | 1.33 (1.03, 1.72) | 0.67 |
| Colorectal | 10 | 1.17 (0.87, 1.57)2 | < 0.0001 | 10 | 1.10 (0.83, 1.45)2 | 0.0004 | 9 | 1.45 (0.76, 2.75)2 | 0.02 | 9 | 1.40 (0.99, 2.00) | 0.07 |
| Hepatocellular | 15 | 1.74 (1.20, 2.54)2 | < 0.00001 | 10 | 1.58 (1.05, 2.39)2 | 0.002 | 8 | 2.55 (1.38, 4.70) | 0.49 | 8 | 2.15 (1.19, 3.90) | 0.66 |
| Pancreatic | 3 | 1.04 (0.79, 1.36) | 0.94 | 3 | 1.04 (0.79, 1.38) | 0.88 | 3 | 0.99 (0.46, 2.14) | 0.63 | 3 | 0.99 (0.46, 2.13) | 0.60 |
| Esophageal | 2 | 0.82 (0.58, 1.16) | 0.89 | 2 | 0.80 (0.55, 1.15) | 0.89 | 2 | 1.01 (0.42, 2.43) | 0.95 | 2 | 1.05 (0.44, 2.51) | 0.92 |
| Ethnicity | ||||||||||||
| Asian | 27 | 1.24 (0.99, 1.56)2 | < 0.00001 | 25 | 1.07 (0.94, 1.22)2 | 0.008 | 19 | 1.55 (1.11, 2.17) | 0.43 | 19 | 1.47 (1.05, 2.06) | 0.60 |
| Caucasian | 31 | 1.21 (1.05, 1.40)2 | < 0.00001 | 27 | 1.17 (1.01, 1.35)2 | < 0.0001 | 25 | 1.38 (1.10, 1.74) | 0.18 | 25 | 1.34 (1.08, 1.67) | 0.39 |
TNF, an important pro-inflammatory cytokine, plays an important role in the regulation of cell differentiation, proliferation and death as well as in inflammation and the innate and adaptive immune response. TNF has also been implicated in a wide variety of human diseases. The presence of DNA sequence variations in the regulatory region might interfere with transcription of the TNF gene, influencing the circulating level of TNF and thus increasing susceptibility to human diseases, such as cancer[64]. The TNF enhancer polymorphism has been implicated in several diseases, and the TNF-α-308 polymorphism has been described as the most important TNF polymorphism in human disease susceptibility. The significance of these polymorphisms reflects their possible influence on the transcription of the TNF gene. However, the results of studies in this area are inconsistent. Canedo et al[7] found that the TNF-α-308G/A polymorphism increases the risk of gastric carcinoma. However, some studies have reported that no statistically significant association exists between the TNF-α-308G/A polymorphism and cancer risk[14,20].
The current meta-analysis, which included 58 case-control studies and 25497 subjects, was conducted to explore the association of the TNF-α-308 polymorphism with digestive system cancer risk. Overall, a significant association was identified between the TNF-α-308 polymorphism and the risk of digestive system cancers. When the analysis was stratified by ethnicity, we found a statistically significant association between this polymorphism and the risk of these cancers in the Caucasian population. However, no significant association was observed in the dominant model and heterozygote comparisons in the Asian population, which could be due to ethnic differences. When the analysis was stratified by cancer type, we found a significant association between this polymorphism and gastric and hepatocellular carcinoma risk under all four genetic models, but no significant association was observed among colorectal, pancreatic or oesophageal cancer.
Heterogeneity is a potential problem when interpreting the results of meta-analyses. In this meta-analysis, heterogeneity was found in the dominant model and heterozygote comparisons in both the overall and subgroup analyses; thus, the random effects model was used. Sensitivity analyses were also conducted by excluding the study that was not in HWE. With this exclusion, the estimated pooled OR did not change significantly, strengthening our confidence in our results. This finding suggests that the population selection and the study that was not in HWE were not sources of heterogeneity. Alternatively, lifestyle, environment and other unknown factors may be sources of heterogeneity. Moreover, no publication bias was shown, suggesting that our results are accurate.
Some limitations of this meta-analysis should be addressed. First, the number of published studies, especially for oesophageal and pancreatic cancers, was not sufficiently large for a comprehensive analysis, and some studies with small sample sizes may not have enough statistical power to prove authentic associations. Therefore, our analysis should be interpreted with caution, and more studies are needed. Second, our results were based on unadjusted estimates, and lack of information for the data analysis may cause serious confounding bias. Third, significant heterogeneity was found in some models, which may lead to failure to confirm marginal associations. In spite of these limitations, our meta-analysis had several advantages. First, a substantial number of cases and controls were pooled from different studies, which significantly increased the statistical power of the analysis. Second, the quality of the case-control studies included in the current meta-analysis was satisfactory and met our inclusion criteria. Third, we did not detect any publication bias, suggesting that the whole pooled result is unbiased.
In summary, this meta-analysis suggests that the TNF-α-308 polymorphism increases susceptibility to digestive system cancers in the Caucasian population. The TNF-α-308 AA genotype is closely related to the risk of digestive system cancers in people of Asian descent. The TNF-α-308 polymorphism may be significantly associated with the risk of gastric and hepatocellular carcinomas, but not colorectal, pancreatic, or oesophageal cancer. Future studies should use standardised unbiased genotyping methods, examine homogeneous cancer patients and well-matched controls, and include multiethnic groups.
We are very grateful to Mr. Hong Xia from the Key Laboratory of Hubei Province for Digestive System Disease for assistance in data collection.
Digestive system cancers are the most common malignant tumors worldwide. Tumor necrosis factor alpha-308 (TNF-α-308) polymorphism (rs1800629) is the most extensively studied polymorphism in digestive system cancers. However, the results are different or even inconsistent.
Molecular epidemiology has confirmed that carcinogenesis is a complex, multi-factorial, and multistep event, and genetic mutation play an important role in the process. Many studies have reported the association between the TNF-α-308 polymorphism and human malignant tumors, but no agreements have been reached till now.
This meta-analysis systemically assessed the association between TNF-α-308 polymorphism and risk of digestive system cancers. Results show that TNF-α-308 polymorphism may be significantly associated with the risk of gastric and hepatocellular carcinomas in Asians.
This study results indicate that TNF-α-308 polymorphism may be used as a detectable biomarker for gastric and hepatocellular carcinoma patients.
The authors present a meta-analysis study over the influence of a polymorphism of TNF-α on digestive system cancers. The manuscript is well written and interesting, especially because it is the first meta-analysis study on the subject.
P- Reviewers: Marcos R, Nagahara H, Swierczynski J S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Liu XM
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25534] [Article Influence: 1823.9] [Reference Citation Analysis (7)] |
| 2. | Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893-1907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1754] [Cited by in RCA: 1880] [Article Influence: 125.3] [Reference Citation Analysis (0)] |
| 3. | Bazzoni F, Beutler B. The tumor necrosis factor ligand and receptor families. N Engl J Med. 1996;334:1717-1725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 902] [Cited by in RCA: 857] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 4. | Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci USA. 1997;94:3195-3199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1608] [Cited by in RCA: 1698] [Article Influence: 60.6] [Reference Citation Analysis (0)] |
| 5. | Dunham I, Sargent CA, Trowsdale J, Campbell RD. Molecular mapping of the human major histocompatibility complex by pulsed-field gel electrophoresis. Proc Natl Acad Sci USA. 1987;84:7237-7241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 161] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 6. | Burada F, Angelescu C, Mitrut P, Ciurea T, Cruce M, Saftoiu A, Ioana M. Interleukin-4 receptor -3223T& amp; #8594; C polymorphism is associated with increased gastric adenocarcinoma risk. Can J Gastroenterol. 2012;26:532-536. [PubMed] |
| 7. | Canedo P, Durães C, Pereira F, Regalo G, Lunet N, Barros H, Carneiro F, Seruca R, Rocha J, Machado JC. Tumor necrosis factor alpha extended haplotypes and risk of gastric carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:2416-2420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 8. | Crusius JB, Canzian F, Capellá G, Peña AS, Pera G, Sala N, Agudo A, Rico F, Del Giudice G, Palli D. Cytokine gene polymorphisms and the risk of adenocarcinoma of the stomach in the European prospective investigation into cancer and nutrition (EPIC-EURGAST). Ann Oncol. 2008;19:1894-1902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 102] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 9. | El-Omar EM, Rabkin CS, Gammon MD, Vaughan TL, Risch HA, Schoenberg JB, Stanford JL, Mayne ST, Goedert J, Blot WJ. Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology. 2003;124:1193-1201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 672] [Cited by in RCA: 676] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 10. | Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1858] [Cited by in RCA: 1990] [Article Influence: 71.1] [Reference Citation Analysis (0)] |
| 11. | DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26739] [Cited by in RCA: 30332] [Article Influence: 777.7] [Reference Citation Analysis (0)] |
| 12. | MANTEL N, HAENSZEL W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719-748. [PubMed] |
| 13. | Guo W, Wang N, Li Y, Zhang JH. Polymorphisms in tumor necrosis factor genes and susceptibility to esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma in a population of high incidence region of North China. Chin Med J (Engl). 2005;118:1870-1878. [PubMed] |
| 14. | Jang WH, Yang YI, Yea SS, Lee YJ, Chun JH, Kim HI, Kim MS, Paik KH. The -238 tumor necrosis factor-alpha promoter polymorphism is associated with decreased susceptibility to cancers. Cancer Lett. 2001;166:41-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 127] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 15. | Fei BY, Xia B, Deng CS, Xia XQ, Xie M, Crusius JB, Pena AS. Association of tumor necrosis factor genetic polymorphism with chronic atrophic gastritis and gastric adenocarcinoma in Chinese Han population. World J Gastroenterol. 2004;10:1256-1261. [PubMed] |
| 16. | García-González MA, Lanas A, Quintero E, Nicolás D, Parra-Blanco A, Strunk M, Benito R, Angel Simón M, Santolaria S, Sopeña F. Gastric cancer susceptibility is not linked to pro-and anti-inflammatory cytokine gene polymorphisms in whites: a Nationwide Multicenter Study in Spain. Am J Gastroenterol. 2007;102:1878-1892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 105] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 17. | Garza-González E, Bosques-Padilla FJ, El-Omar E, Hold G, Tijerina-Menchaca R, Maldonado-Garza HJ, Pérez-Pérez GI. Role of the polymorphic IL-1B, IL-1RN and TNF-A genes in distal gastric cancer in Mexico. Int J Cancer. 2005;114:237-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 99] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 18. | Glas J, Török HP, Schneider A, Brünnler G, Kopp R, Albert ED, Stolte M, Folwaczny C. Allele 2 of the interleukin-1 receptor antagonist gene is associated with early gastric cancer. J Clin Oncol. 2004;22:4746-4752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 19. | Hou L, El-Omar EM, Chen J, Grillo P, Rabkin CS, Baccarelli A, Yeager M, Chanock SJ, Zatonski W, Sobin LH. Polymorphisms in Th1-type cell-mediated response genes and risk of gastric cancer. Carcinogenesis. 2007;28:118-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 81] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 20. | Kamangar F, Abnet CC, Hutchinson AA, Newschaffer CJ, Helzlsouer K, Shugart YY, Pietinen P, Dawsey SM, Albanes D, Virtamo J. Polymorphisms in inflammation-related genes and risk of gastric cancer (Finland). Cancer Causes Control. 2006;17:117-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 129] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 21. | Kim N, Cho SI, Yim JY, Kim JM, Lee DH, Park JH, Kim JS, Jung HC, Song IS. The effects of genetic polymorphisms of IL-1 and TNF-A on Helicobacter pylori-induced gastroduodenal diseases in Korea. Helicobacter. 2006;11:105-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 22. | Lee SG, Kim B, Yook JH, Oh ST, Lee I, Song K. TNF/LTA polymorphisms and risk for gastric cancer/duodenal ulcer in the Korean population. Cytokine. 2004;28:75-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 45] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 23. | Lee JY, Kim HY, Kim KH, Kim SM, Jang MK, Park JY, Lee JH, Kim JH, Yoo JY. Association of polymorphism of IL-10 and TNF-A genes with gastric cancer in Korea. Cancer Lett. 2005;225:207-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 82] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 24. | Li C, Xia B, Yang Y, Li J, Xia HH. TNF gene polymorphisms and Helicobacter Pylori infection in gastric carcinogenesis in Chinese population. Am J Gastroenterol. 2005;100:290-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 25. | Lu W, Pan K, Zhang L, Lin D, Miao X, You W. Genetic polymorphisms of interleukin (IL)-1B, IL-1RN, IL-8, IL-10 and tumor necrosis factor {alpha} and risk of gastric cancer in a Chinese population. Carcinogenesis. 2005;26:631-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 172] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 26. | Machado JC, Figueiredo C, Canedo P, Pharoah P, Carvalho R, Nabais S, Castro Alves C, Campos ML, Van Doorn LJ, Caldas C. A proinflammatory genetic profile increases the risk for chronic atrophic gastritis and gastric carcinoma. Gastroenterology. 2003;125:364-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 357] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 27. | Melo Barbosa HP, Martins LC, Dos Santos SE, Demachki S, Assumpção MB, Aragão CD, de Oliveira Corvelo TC. Interleukin-1 and TNF-alpha polymorphisms and Helicobacter pylori in a Brazilian Amazon population. World J Gastroenterol. 2009;15:1465-1471. [PubMed] |
| 28. | Morgan DR, Dominguez RL, Keku TO, Heidt PE, Martin CF, Galanko JA, Omofoye OA, Sandler RS. Gastric cancer and the high combination prevalence of host cytokine genotypes and Helicobacter pylori in Honduras. Clin Gastroenterol Hepatol. 2006;4:1103-1111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 29. | Perri F, Piepoli A, Bonvicini C, Gentile A, Quitadamo M, Di Candia M, Cotugno R, Cattaneo F, Zagari MR, Ricciardiello L. Cytokine gene polymorphisms in gastric cancer patients from two Italian areas at high and low cancer prevalence. Cytokine. 2005;30:293-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 30. | Rocha GA, Guerra JB, Rocha AM, Saraiva IE, da Silva DA, de Oliveira CA, Queiroz DM. IL1RN polymorphic gene and cagA-positive status independently increase the risk of noncardia gastric carcinoma. Int J Cancer. 2005;115:678-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 31. | Sugimoto M, Furuta T, Shirai N, Nakamura A, Xiao F, Kajimura M, Sugimura H, Hishida A. Different effects of polymorphisms of tumor necrosis factor-alpha and interleukin-1 beta on development of peptic ulcer and gastric cancer. J Gastroenterol Hepatol. 2007;22:51-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 99] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 32. | Torres MM, Acosta CP, Sicard DM, Groot de Restrepo H. [Genetic susceptibility and risk of gastric cancer in a human population of Cauca, Colombia]. Biomedica. 2004;24:153-162. [PubMed] |
| 33. | Wu MS, Huang SP, Chang YT, Shun CT, Chang MC, Lin MT, Wang HP, Lin JT. Tumor necrosis factor-alpha and interleukin-10 promoter polymorphisms in Epstein-Barr virus-associated gastric carcinoma. J Infect Dis. 2002;185:106-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 85] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 34. | Wu MS, Chen LT, Shun CT, Huang SP, Chiu HM, Wang HP, Lin MT, Cheng AL, Lin JT. Promoter polymorphisms of tumor necrosis factor-alpha are associated with risk of gastric mucosa-associated lymphoid tissue lymphoma. Int J Cancer. 2004;110:695-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 35. | Yang JJ, Ko KP, Cho LY, Shin A, Gwack J, Chang SH, Shin HR, Yoo KY, Kang D, Park SK. The role of TNF genetic variants and the interaction with cigarette smoking for gastric cancer risk: a nested case-control study. BMC Cancer. 2009;9:238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 36. | Zambon CF, Basso D, Navaglia F, Belluco C, Falda A, Fogar P, Greco E, Gallo N, Rugge M, Di Mario F. Pro- and anti-inflammatory cytokines gene polymorphisms and Helicobacter pylori infection: interactions influence outcome. Cytokine. 2005;29:141-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 162] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 37. | Garrity-Park MM, Loftus EV, Bryant SC, Sandborn WJ, Smyrk TC. Tumor necrosis factor-alpha polymorphisms in ulcerative colitis-associated colorectal cancer. Am J Gastroenterol. 2008;103:407-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 38. | Landi S, Moreno V, Gioia-Patricola L, Guino E, Navarro M, de Oca J, Capella G, Canzian F. Association of common polymorphisms in inflammatory genes interleukin (IL)6, IL8, tumor necrosis factor alpha, NFKB1, and peroxisome proliferator-activated receptor gamma with colorectal cancer. Cancer Res. 2003;63:3560-3566. [PubMed] |
| 39. | Li M, You Q, Wang X. Association between polymorphism of the tumor necrosis factor alpha-308 gene promoter and colon cancer in the Chinese population. Genet Test Mol Biomarkers. 2011;15:743-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 40. | Macarthur M, Sharp L, Hold GL, Little J, El-Omar EM. The role of cytokine gene polymorphisms in colorectal cancer and their interaction with aspirin use in the northeast of Scotland. Cancer Epidemiol Biomarkers Prev. 2005;14:1613-1618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 67] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 41. | Park KS, Mok JW, Rho SA, Kim JC. Analysis of TNFB and TNFA NcoI RFLP in colorectal cancer. Mol Cells. 1998;8:246-249. [PubMed] |
| 42. | Suchy J, Kłujszo-Grabowska E, Kładny J, Cybulski C, Wokołorczyk D, Szymańska-Pasternak J, Kurzawski G, Scott RJ, Lubiński J. Inflammatory response gene polymorphisms and their relationship with colorectal cancer risk. BMC Cancer. 2008;8:112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 43. | Theodoropoulos G, Papaconstantinou I, Felekouras E, Nikiteas N, Karakitsos P, Panoussopoulos D, Lazaris ACh, Patsouris E, Bramis J, Gazouli M. Relation between common polymorphisms in genes related to inflammatory response and colorectal cancer. World J Gastroenterol. 2006;12:5037-5043. [PubMed] |
| 44. | Tóth EK, Kocsis J, Madaras B, Bíró A, Pocsai Z, Fust G, Blaskó B, Karádi I, Adány R, Laki J. The 8.1 ancestral MHC haplotype is strongly associated with colorectal cancer risk. Int J Cancer. 2007;121:1744-1748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 45. | Tsilidis KK, Helzlsouer KJ, Smith MW, Grinberg V, Hoffman-Bolton J, Clipp SL, Visvanathan K, Platz EA. Association of common polymorphisms in IL10, and in other genes related to inflammatory response and obesity with colorectal cancer. Cancer Causes Control. 2009;20:1739-1751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 129] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 46. | Akkiz H, Bayram S, Bekar A, Ozdil B, Akgöllü E, Sümbül AT, Demiryürek H, Doran F. G-308A TNF-alpha polymorphism is associated with an increased risk of hepatocellular carcinoma in the Turkish population: case-control study. Cancer Epidemiol. 2009;33:261-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 47. | Ben-Ari Z, Mor E, Papo O, Kfir B, Sulkes J, Tambur AR, Tur-Kaspa R, Klein T. Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol. 2003;98:144-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 180] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 48. | Chen CC, Yang SY, Liu CJ, Lin CL, Liaw YF, Lin SM, Lee SD, Chen PJ, Chen CJ, Yu MW. Association of cytokine and DNA repair gene polymorphisms with hepatitis B-related hepatocellular carcinoma. Int J Epidemiol. 2005;34:1310-1318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 86] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 49. | Heneghan MA, Johnson PJ, Clare M, Ho S, Harrison PM, Donaldson PT. Frequency and nature of cytokine gene polymorphisms in hepatocellular carcinoma in Hong Kong Chinese. Int J Gastrointest Cancer. 2003;34:19-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 50. | Ho SY, Wang YJ, Chen HL, Chen CH, Chang CJ, Wang PJ, Chen HH, Guo HR. Increased risk of developing hepatocellular carcinoma associated with carriage of the TNF2 allele of the -308 tumor necrosis factor-alpha promoter gene. Cancer Causes Control. 2004;15:657-663. [PubMed] |
| 51. | Jeng JE, Tsai JF, Chuang LY, Ho MS, Lin ZY, Hsieh MY, Chen SC, Chuang WL, Wang LY, Yu ML. Tumor necrosis factor-alpha 308.2 polymorphism is associated with advanced hepatic fibrosis and higher risk for hepatocellular carcinoma. Neoplasia. 2007;9:987-992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 75] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 52. | Jeng JE, Tsai HR, Chuang LY, Tsai JF, Lin ZY, Hsieh MY, Chen SC, Chuang WL, Wang LY, Yu ML. Independent and additive interactive effects among tumor necrosis factor-alpha polymorphisms, substance use habits, and chronic hepatitis B and hepatitis C virus infection on risk for hepatocellular carcinoma. Medicine (Baltimore). 2009;88:349-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 53. | Kummee P, Tangkijvanich P, Poovorawan Y, Hirankarn N. Association of HLA-DRB1*13 and TNF-alpha gene polymorphisms with clearance of chronic hepatitis B infection and risk of hepatocellular carcinoma in Thai population. J Viral Hepat. 2007;14:841-848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 54. | Migita K, Miyazoe S, Maeda Y, Daikoku M, Abiru S, Ueki T, Yano K, Nagaoka S, Matsumoto T, Nakao K. Cytokine gene polymorphisms in Japanese patients with hepatitis B virus infection--association between TGF-beta1 polymorphisms and hepatocellular carcinoma. J Hepatol. 2005;42:505-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 120] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 55. | Niro GA, Fontana R, Gioffreda D, Valvano MR, Lacobellis A, Facciorusso D, Andriulli A. Tumor necrosis factor gene polymorphisms and clearance or progression of hepatitis B virus infection. Liver Int. 2005;25:1175-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 56. | Ognjanovic S, Yuan JM, Chaptman AK, Fan Y, Yu MC. Genetic polymorphisms in the cytokine genes and risk of hepatocellular carcinoma in low-risk non-Asians of USA. Carcinogenesis. 2009;30:758-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 54] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 57. | Sakamoto T, Higaki Y, Hara M, Ichiba M, Horita M, Mizuta T, Eguchi Y, Yasutake T, Ozaki I, Yamamoto K. Interaction between interleukin-1beta -31T/C gene polymorphism and drinking and smoking habits on the risk of hepatocellular carcinoma among Japanese. Cancer Lett. 2008;271:98-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 58. | Shi Z, Du C. Tumor necrosis factor alpha 308 G/A polymorphism and hepatocellular carcinoma risk in a Chinese population. Genet Test Mol Biomarkers. 2011;15:569-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 59. | Wang Y, Kato N, Hoshida Y, Yoshida H, Taniguchi H, Goto T, Moriyama M, Otsuka M, Shiina S, Shiratori Y. Interleukin-1beta gene polymorphisms associated with hepatocellular carcinoma in hepatitis C virus infection. Hepatology. 2003;37:65-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 137] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 60. | Wang B, Wang J, Zheng Y, Zhou S, Zheng J, Wang F, Ma X, Zeng Z. A study of TNF-alpha-238 and -308 polymorphisms with different outcomes of persistent hepatitis B virus infection in China. Pathology. 2010;42:674-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 61. | Duell EJ, Casella DP, Burk RD, Kelsey KT, Holly EA. Inflammation, genetic polymorphisms in proinflammatory genes TNF-A, RANTES, and CCR5, and risk of pancreatic adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2006;15:726-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 79] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 62. | Talar-Wojnarowska R, Gasiorowska A, Smolarz B, Romanowicz-Makowska H, Kulig A, Malecka-Panas E. Tumor necrosis factor alpha and interferon gamma genes polymorphisms and serum levels in pancreatic adenocarcinoma. Neoplasma. 2009;56:56-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 63. | Wu GY, Lu Q, Hasenberg T, Niedergethmann M, Post S, Sturm JW, Keese M. Association between EGF, TGF-{beta}1, TNF-{alpha} gene polymorphisms and cancer of the pancreatic head. Anticancer Res. 2010;30:5257-5261. [PubMed] |
| 64. | Qidwai T, Khan F. Tumour necrosis factor gene polymorphism and disease prevalence. Scand J Immunol. 2011;74:522-547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 116] [Article Influence: 8.3] [Reference Citation Analysis (0)] |