Published online Dec 21, 2013. doi: 10.3748/wjg.v19.i47.8902
Revised: October 30, 2013
Accepted: November 18, 2013
Published online: December 21, 2013
Processing time: 144 Days and 20.2 Hours
Many aspects of cellular physiology display circadian (approximately 24-h) rhythms. Dysfunction of the circadian clock molecular circuitry is associated with human health derangements, including neurodegeneration, increased risk of cancer, cardiovascular diseases and the metabolic syndrome. Viruses triggering hepatitis depend tightly on the host cell synthesis machinery for their own replication, survival and spreading. Recent evidences support a link between the circadian clock circuitry and viruses’ biological cycle within host cells. Currently, in vitro models for chronobiological studies of cells infected with viruses need to be implemented. The establishment of such in vitro models would be helpful to better understand the link between the clock gene machinery and viral replication/viral persistence in order to develop specifically targeted therapeutic regimens. Here we review the recent literature dealing with the interplay between hepatitis B and C viruses and clock genes.
Core tip: New antiviral strategies have been developed, including the interferon/ribavirin-free therapy, to control hepatitis viruses replication. Although, IFN-free regimens have generated excitement among scientists, for the reason that they are better tolerated, they are not still able to completely eradicate the viruses. Here we underline the circadian relationship between host cell and hosted hepatitis viruses, that has to be taken into account in order to optimize the timing of therapeutic regimens, not only to minimize the pharmacological agents’ toxicity but also to improve the efficacy of treatment modalities through optimized timing of therapeutic regimens, targeting in a better way virus replication.
- Citation: Vinciguerra M, Mazzoccoli G, Piccoli C, Tataranni T, Andriulli A, Pazienza V. Exploitation of host clock gene machinery by hepatitis viruses B and C. World J Gastroenterol 2013; 19(47): 8902-8909
- URL: https://www.wjgnet.com/1007-9327/full/v19/i47/8902.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i47.8902
Viruses are among the most important human carcinogens[1]. Numerous mechanisms have been described to be dysregulated by viruses, always focusing on the impairment of the most well known tumor suppressors and/or oncogene proteins and their signaling pathways[2,3].
It has been already established that alteration of the circadian clock molecular circuitry is involved in carcinogenesis. Circadian defects have also been associated with liver diseases, including hepatocellular carcinoma (HCC)[4,5], a condition in which viruses play a role in disease pathogenesis and progression.
Specifically, basic cell functions and processes, such as cell division, proliferation, growth, differentiation, autophagy, apoptosis and metabolism, show time-related fluctuations, and when the period of oscillation is about 24 h the rhythmicity is defined as circadian[6-10]. At the cellular level, circadian rhythmicity is driven by a molecular clockwork comprised of a translational-transcriptional feedback loop realized by a set of genes, called core clock genes, coding for proteins that in turn suppress gene expression in a cycle that completes itself each day. Clock genes are transcriptionally activated by the transcription factors circadian locomotor output cycle kaput (CLOCK) and aryl hydrocarbon receptor nuclear translocator-like (ARNTL). The latter two protein heterodimerize and bind to the E-box enhancer elements in the promoters of the Period (PER 1, 2 and 3) and Cryptochrome (CRY 1 and 2) genes. The PER and CRY mRNAs are translated into PER and CRY proteins that form a repression complex, which in turn translocates back into the nucleus, interacts directly with CLOCK and ARNTL blocking their activity[4,11-13].
Among the processes regulated by the clock gene machinery are pathways of cell metabolism and vesicle trafficking, suggesting the potential role for the circadian clock circuitry in the regulation of viral expression/replication[14]. A relationship between circadian dysfunction and tumorigenesis has also been found at both the cellular and the organismal levels, indicating that the circadian clock may impact on the development of cancer[15-17], a disease also influenced by viruses. Recently, scientific evidences support a functional connection between viral expression/replication and circadian dysfunction in the pathogenesis of liver diseases[14,18-20]. However, whether the circadian clock directly regulates viral cell cycle in mammalian cells, or whether viruses may play a role in the cycling of mammalian cell clocks is not yet totally clear.
The implication of viral expression/replication and circadian dysfunction in the pathogenesis of liver diseases suggests that a functional connection between these two processes may exist as it has been already showed[14,18-20]. Nevertheless, the relationship between circadian cycles and viral expression/replication is an intriguing area for future study and it has implications for multiple human diseases. The study of new causes which are able to influence the clock genes expression are under investigation as disruption of biological clocks is implicated in a variety of disorders including fatty liver disease, obesity and diabetes[21,22]. Exciting data reported the influence of hepatitis B and C viruses on the hepatic clock genes[18,19], demonstrating for the first time that these viruses are able to impair the inner molecular clockwork, presumably to better exploit the host-cell replication machinery. Hepatotropic viruses impair also liver functions, and this effect may be a cause or a consequence of the disruption of the inner cellular biological clock. At the present, the relationship between hepatitis viruses expression/replication and the circadian clock is poorly understood. Here we review the scientific reports addressing the interaction between hepatitis B and C viruses and the molecular clockwork.
The liver plays an important role in maintaining energy homeostasis within the organism. The major biochemical reactions occurring within the liver are involved in glucose breakdown/genesis, which is strictly linked to fatty acid metabolism (biosynthesis/beta oxidation). All these biochemical reactions and the metabolic networks must be finely coordinated in order to avoid unnecessary interference between the pathways[21]. To this end, reactions are separated locally and temporally. Hepatic metabolic functions show rhythmic fluctuations with 24-h periodicity[23], driven by molecular clockworks ticking through translational-transcriptional feedback loops and operated by a set of genes, called clock genes, encoding circadian proteins[4]. In the absence of environmental cues, specifically light:dark cycle, it has been demonstrated that rhythmic food intake influences the hepatic circadian oscillator[23,24]. Hence, the clock genes oscillations are not phase locked but are flexible to enable adjustment to the changing environments[23].
In the liver, gene expression profiling has shown that transcriptional processes display approximately 24-h rhythmicity and have a crucial role in metabolic processes. Energy and nutrient homeostasis at both cellular and organismal levels is guaranteed by nearly constant adjustments of metabolic gene expression, and the transcriptional networks that regulate glucose and lipid metabolism are sensitive to nutritional status, responding to diverse physiological signals[25]. The fractions of cyclic transcripts depending on systemic signals and local oscillators amount to approximately 14% and 86%, respectively. The systemically regulated liver genes include immediate early genes (IEG), which convey rhythmic signals to core clock genes of hepatocyte oscillators and thus are involved in the synchronization of liver clocks, and tissue specific output genes, directly participating in rhythmic liver physiology and metabolism. The IEG class contains several heat shock protein genes, known to be regulated by heat shock transcription factor 1 (HSF1) and target genes of serum response factor 1 (SRF1), and these immediate early transcription factors (IETFs) act as sensors of blood-borne signals, driving the synchronization of circadian clocks[26]. Metabolite sensing is linked to transcriptional responses in hepatocytes by nuclear receptors through switching between co-activator and co-repressor recruitment[27]. Nuclear hormone receptors comprise a unique class of transcriptional regulators that are capable of sensing the concentrations of metabolites, including lipids, oxysterols, heme, and bile acids[28]. An important role in the control of glucose, lipid, and mitochondrial oxidative metabolism is played by the expression of co-regulators, in particular the PGC-1α, which is highly responsive to nutritional status and other physiological signals[29]. The cross-talk between circadian rhythms and metabolism is operated also by the peroxisome proliferator-activated receptors (PPAR), in particular α and γ[30]. Both factors are already known to be dysregulated by hepatitis B and C viruses. PPARα regulates transcription of genes involved in lipid and glucose metabolism upon binding of endogenous free fatty acids[31]. PPARγ binds eicosanoids deriving from either omega-3 (ω-3) or omega-6 (ω-6) fatty acids and their oxidized counterparts, is rhythmically expressed, its expression is regulated by PER2 and in turn directly regulates ARNTL transcription[32]. The clock gene machinery drives the expression of a large array of enzymes involved in lipid metabolism, controls lipogenesis and regulates triglyceride packaging into chylomicrons (globules that transport dietary lipids) at the level of the intestine, whereas in the liver, clock disruption triggers lipid accumulation[33-35]. In liver ARNTL and CLOCK control gene expression of enzymes involved in glucose and lipid homeostasis, as well as in bile acid and apolipoprotein biosynthesis[36]. Diurnal oscillation characterizes a number of proteins involved in lipid metabolism [such as hepatic cytochrome P450 cholesterol 7 α-hydroxylase, 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase, or apolipoprotein AIV] show in both humans and rodents. REV-ERB α links the clock with the master pathway of hepatic lipid metabolism, is involved in bile acid synthesis and sterol regulatory element-binding protein (SREBP) signaling and SREBPs control both fatty acid and sterol biosynthesis through modulation of rate-limiting enzymes in these pathways[35]. Diurnal variations hallmark also glucose metabolism, and the rate-limiting enzymes for gluconeogenesis, glycolysis, glycogenesis and glycogenolysis show circadian variations of activity, determining the circadian rhythmicity of hepatic glucose production and glycogen content. The biological clock drives the circadian regulation of hepatic gluconeogenesis by CRY 1 and CRY2 via inhibition of cAMP signaling in response to G protein coupled receptor (GPCR) activation[37], and controls hepatic glycogen synthesis through transcriptional activation of glycogen synthase (GYS2) by CLOCK[38], and the disruption or mutation of the clock genes CLOCK and ARNTL results in disorders of glucose homeostasis[39,40].
Hepatitis B virus (HBV) belongs to the Hepadnaviridae family, which causes persistent liver infections[41]. With more than 2 billion people being infected worldwide and 400 million suffering from chronic hepatitis B, HBV infection is one of the most significant public health problems. Despite the advance of modern medicine in the development of new antiviral drugs, HBV infection remains a leading cause of liver cirrhosis and cancer[3].
HBV genome is a partial double-stranded DNA that replicates through the reverse transcription of pregenomic RNA[42]. The analysis of the entire sequence of HBV-DNA, constituted by a circular incomplete double-strand DNA molecule, of 3182 bp in length[43], reveals four Open Reading Frames (ORFs), overlapping each other, necessary for transcription and expression of HBV proteins. These ORF are named: ORF S, ORF C, ORF P and ORF X[44] and they encode for four proteins with specific structure and function[45]. HBV biology and life cycle were already described[46]. The X protein (encoded by ORF X), remains partially explored and its function needs to be established[47]. Cultured hepatocytes overexpressing the X-gene, reveal a crucial role of the X protein in trans-activating viral and cellular genes[48]. Moreover, some authors associated HBx protein with HCC due to its property of impairing cellular proliferation[49], although the X protein cannot induce infection by itself.
One study reported the ability of the HBx protein in modulating the clock genes in LO2 cells[19]. Cultured LO2 cells stably overexpressing the HBx protein displayed higher mRNA and protein levels of the CLOCK gene whilst ARNTL resulted to be decreased as compared to control cells. The authors suggest that the impairment of circadian rhythm of liver cells due to HBx expression may be one of the reasons leading to liver cancer development. It remains to elucidate how HBV impairs the clock gene machinery and to confirm the effect on liver cancer progression due to impairment of the cellular molecular clockwork by HBx.
Hepatitis C virus (HCV) is a hepatotropic virus belonging to the Flavivirus family. It is estimated that 170 million people worldwide are infected with HCV[50]. In the majorities of the cases, HCV infection leads to severe liver diseases and is considered one of the major risk factors for HCC development[51].
HCV genome consists in a positive-stranded RNA of approximately 9.6 kb, coding for a single polyprotein of about 3000 amino acids, processed co- and post-translationally by cellular and viral proteases cleaving it into three structural (core, E1 and E2), seven nonstructural (NS2, NS3, NS4A, NS4B NS5A and NS5B) mature proteins and an ion channel (p7)[52]. Despite the small sequence divergences HCV is classified into six major genotypes (further divided into different subtypes)[50]. Overwhelming lines of evidence have indicated that the pathogenicity of HCV and its effect on disease progression and treatment is genotype dependent[50].
We used two different in vitro models to investigate the relationship between HCV and clock genes, the OR6 cells harboring HCV replication and the Huh-7 cells expressing the HCV core proteins of genotype 1b or 3a. In both cases it was found that HCV down-regulated the expression of two crucial clock proteins CRY2 and PER2.
CRY2 protein is involved in NF-κB activation and pro-inflammatory processes[53], (see next section for discussion), while the role of PER2 on HCV replication is particularly interesting, as this circadian protein regulates the rhythms of IFNγ signaling, critical for innate and adaptive immunity against infection[54,55]. Exogenous overexpression of PER2 protein in OR6 cells hampered HCV-RNA replication, and consistently, PER2 overexpression influenced the HCV-dependent altered expression of Interferon stimulated genes (ISG) products (OAS1, Mx1, IRF9). PER2 potentiated the expression of OAS1 which activates RNase L resulting in viral RNA degradation and inhibition of viral replication[56].
Of note, when experiments were performed, cells were synchronized using serum shock procedure, a method previously reported to induce circadian gene expression in mammalian cultured cells[57], before RNA extractions at regular time points over 28 h period. This approach allows assessing differences in the time-related fluctuation of expression.
Hepatic injury in HCV infection is not only directly induced by viral cytopathic effects, but is principally related to host immune responses. Viral persistence is influenced by dynamic restriction of the host’s immune response, and the strength of immune response determines resultant acute viral clearance opposed to chronic persistence, leading to pathogenic mechanisms potentially responsible for HCC onset and progression during chronic hepatitis virus infection. Chronic immune-mediated liver cell injury triggers the development of HCC in the absence of viral transactivation, insertional mutagenesis, and genotoxic chemicals[58]. Circadian patterns of immune function have been maintained throughout evolution, are driven by the clock gene machinery, and the magnitude of immune response depends in part on the circadian timing of antigen challenge[59,60]. Alterations in the circadian regulation of the immune system may therefore lead to viral persistence or reactivation. The components of the immune system show time related variations with a period of 24 h. In particular, the levels of leukocyte populations in the blood of humans and rodents are characterized by circadian variations. Natural killer (NK) cells are critical for immune surveillance against viral infections and their function is under tight circadian control. NK cells bear no antigen receptor and therefore belong to the innate immune system, however they share several features with highly differentiated T lymphocytes, such as a high tissue migratory potential and the production of granzyme B and perforin, interferon (IFN)-γ, tumor necrosis factor (TNF)-α, and granular macrophage cell stimulating factor, allowing immediate cytotoxic effector defense in the periphery[61]. Circadian expression of negative and positive components of the molecular clock, as well as cytokines and cytolytic factors, are evident in NK cells, and perturbations of daily rhythms caused by external and internal stressors may compromise the first line of defense against infections[61,62]. In NK cells, expression of cytokines (IFN-γ and TNF-α) and cytolytic factors (granzyme B and perforin) are highly synchronized, peaking approximately during the middle of the active period in rats, and NK cell cytotoxic activity peaks at similar circadian phases. Similarly, NK cytotoxicity is maximal during periods of wakefulness in humans[60]. The clock genes drive circadian rhythmicity of NK cell function. Alterations of the molecular clockwork modify the harmonized expression of NK cell cytolytic factors. In particular, knock-down of Per2 or Arntl in rat-derived RNK16 NK cells changes in a diverse way the expression of genes encoding IFN-γ, TNF-α, granzyme B, and perforin[54]. Furthermore, knock-down of Per2 or Arntl changes protein levels of granzyme B and perforin, but not of IFN-γ and TNF-α[63,64]. In addition, distorted rhythms of granzyme B and perforin as well as altered rhythm and low levels of IFN-γ, together with changes in the rhythm of Arntl and Per2, were evidenced in Per2 mutant mice[62].
In the human blood, higher counts of total lymphocytes, T lymphocytes and B lymphocytes have been consistently observed in the night time, and when T lymphocyte subsets are considered, CD4+ (T helper) and CD8+ (cytotoxic) naive, central memory and effector memory T lymphocytes show peak numbers in the night, while CD4+ effector T cells show no rhythm and CD8+ effector T cells show a low amplitude rhythm with a peak in the day[65,66]. T and B lymphocytes are involved in the adaptive (i.e., antigen-specific) immune response, whereas granulocytes, monocytes and NK cells mainly belong to the innate (i.e., not antigen-specific) immune system. In rodents higher numbers of total leukocytes and of lymphocytes were reported in the day, while in humans higher levels in the counts of innate immune system cells were reported in the daytime or late day[67]. Hence, both nocturnal rodents and diurnal humans show higher lymphocyte counts during the rest period, and peaks of other cell types (granulocytes, neutrophils, monocytes) were found in the day in rats, while highest NK cell numbers were observed at the end of the night, i.e., at the beginning of the activity period.
Cellular immune rhythms are synchronized by the mammalian central pacemaker located in the suprachiasmatic nuclei (SCN) in the anterior hypothalamus via time dependent changes in the activity of the sympathetic nervous system (SNS), in the release of hormones (growth hormone, prolactin, melatonin, cortisol) and in behavior that is linked to the sleep-wake cycle[65,68,69]. The rest period is characterized by peak levels of pro-inflammatory hormones like growth hormone, prolactin (and melatonin in humans) and pro-inflammatory cytokines like interleukin (IL)-1 and TNF-α. Besides, T helper (h) 1 and Th2 responses are likewise highest during sleep[70]. During the active period the hypothalamus-pituitary-adrenal axis becomes activated and cortisol suppresses pro-inflammatory cytokine production, CD4+ T cell numbers and allergic reactions[71]. Disruption of this temporal organization of the immune system can lead to immunodeficiency and/or exceeding immune reactions (e.g., low grade systemic inflammation).
Oscillation across the day was observed also for the levels of cytokines and other effector molecules, in particular serum levels and in vitro production of IFN-γ, tumor necrosis factor TNF-α, IL-1, IL-2, IL-6 and IL-12 were all shown to present a rhythm in humans, with a peak generally observed at night or in the early morning[60]. Immune rhythms are influenced by hormone rhythms (e.g., cortisol, melatonin, norepinephrine), and in humans the rhythms of naive, central memory, and effector memory T cell counts are regulated by cortisol, whereas numbers of CD8+ effector T cells follow changes in endogenous epinephrine[65,72-74].
The presence of biological clocks in immune cells and lymphoid organs drives rhythms in the functions of cells within the immune system, but on the other hand immune responses and mediators influence behavioral and molecular circadian rhythms[54,62]. Whether circadian disruption of cellular-mediated immunity or neuroendocrine-immune interaction lead to viral reactivation is unclear.
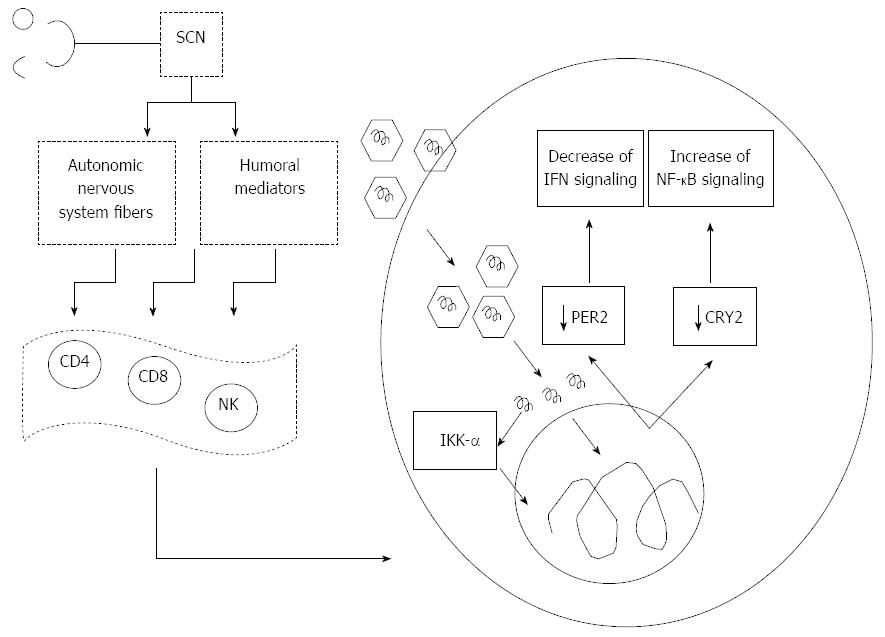
The cross-talk between the clock and innate immune functions is mediated among other circadian factors by CRY2, which transcriptionally regulates STAT3 and hampers activation of NF-κB signaling by negatively regulating the cAMP-PKA pathway[53]. Interestingly, we reported a severe down-regulation of CRY2 in OR6 cells replicating HCV genotype 1b[18], which could induce increase of cytokine production related to NF-κB signaling pathway[53]. This mechanism could enhance the effects deriving from direct activation of NF-κB by the HCV core protein, which may bind to the death domain of tumor necrosis factor receptor 1 (TNFR1) and to the cytoplasmic tail of lymphotoxin-beta receptor, with resistance to TNF-α-induced apoptosis, suggesting a mechanism by which HCV may evade the host’s immune surveillance leading to viral persistence and possibly to hepatocarcinogenesis[75]. On the other hand, HCV infection, and in particular core nonstructural protein (NS)4B and NS5B, reduce TNF-α-induced phosphorylation of IκB kinase (IKK, α, β and γ) and inhibitor of NF-κB (IκB), which are upstream regulators of NF-κB activation. HCV plays a role in immune-mediated liver injury in HCV infection also inhibiting nuclear translocation of NF-κB and expression of NF-κB-dependent anti-apoptotic proteins, such as B-cell lymphoma-extra large (Bcl-xL), X-linked inhibitor of apoptosis protein (XIAP), and the long form of cellular-FLICE inhibitory protein (c-FLIP)[76]. Furthermore, a crucial host factor for HCV is represented by IKK-α (Figure 1). HCV interacts with DEAD box polypeptide 3, X-linked (DDX3X) through its 3’ untranslated region, and activates IKK-α, which translocates to the nucleus and induces a CBP/p300-mediated transcriptional program involving sterol regulatory element-binding proteins (SREBPs). HCV infection in this way utilizes a NF-κB-independent and the kinase-mediated nuclear function of IKK-α: making use of this intrinsic innate pathway and taking control of lipogenic genes and lipid metabolism, enhances core-associated lipid droplet formation to facilitate viral assembly, which in turn may contribute to high chronicity rates and the pathological hallmark of steatosis in HCV infection[77].
Up to date only few studies reported the influence of viruses on the clock gene machinery. Further studies are required to investigate the relationship between viruses and the clock genes as they could lead to new therapeutic strategies for future treatment options. Performing cell synchronization may be useful to observe in vitro differences in time related patterns of expression[18]. Consequently, we recommend a better set-up of the experiments and cell synchronization before investigating the biological clock at the molecular level, considering that single cells in culture are asynchronous and this may conditionate the results.
As for the new therapeutic strategies that can be developed based on the circadian regulation of viral replication, circadian rhythm-based treatments (i.e., chronotherapies), have been employed against several different pathological conditions[78,79]. Standard therapy for HCV patients involves administration of IFN-α and ribavirin (a nucleoside analogue)[50,56]. Recently, an interferon/ribavirin-free therapy based on newly identified and efficacious protease inhibitors (telaprevir, boceprevir) promisingly entered into the clinic to treat HCV patients[80]. In light of these findings, if the new strategies to inhibit viral replication take in consideration the circadian relationship between host cell and hosted viruses, this could not only minimize the pharmacological agents’ toxicity but can also improve the efficacy of treatment modalities through optimized timing of therapeutic regimens, targeting in a better way virus replication. As already suggested, administration of nucleoside analogues to inhibit viral DNA replication can be matched to parallel the diurnal peaks[14] considering the circadian pattern of host cell proliferation and differentiation.
P- Reviewer: Herichova I S- Editor: Cui XM L- Editor: A E- Editor: Liu XM
| 1. | Butel JS. Viral carcinogenesis: revelation of molecular mechanisms and etiology of human disease. Carcinogenesis. 2000;21:405-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 217] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 2. | Bergonzini V, Salata C, Calistri A, Parolin C, Palù G. View and review on viral oncology research. Infect Agent Cancer. 2010;5:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 3. | Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 581] [Cited by in RCA: 631] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 4. | Demarque M, Schibler U. Shedding new light on circadian clocks. Elife. 2013;2:e00659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 5. | Filipski E, Subramanian P, Carrière J, Guettier C, Barbason H, Lévi F. Circadian disruption accelerates liver carcinogenesis in mice. Mutat Res. 2009;680:95-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 110] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 6. | Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 884] [Cited by in RCA: 844] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 7. | Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 868] [Cited by in RCA: 896] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 8. | Ma D, Panda S, Lin JD. Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J. 2011;30:4642-4651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 196] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 9. | Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, Wada T, Aoyagi T, Tezuka M. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci USA. 2005;102:12071-12076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 426] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 10. | Sachdeva UM, Thompson CB. Diurnal rhythms of autophagy: implications for cell biology and human disease. Autophagy. 2008;4:581-589. [PubMed] |
| 11. | Edery I. Circadian rhythms in a nutshell. Physiol Genomics. 2000;3:59-74. [PubMed] |
| 12. | Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693-705. [PubMed] |
| 13. | Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919-922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 575] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 14. | Shadan FF. A circadian model for viral persistence. Med Hypotheses. 2007;68:546-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 15. | Wood PA, Yang X, Hrushesky WJ. Clock genes and cancer. Integr Cancer Ther. 2009;8:303-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 68] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 16. | Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 17. | Hua H, Wang Y, Wan C, Liu Y, Zhu B, Wang X, Wang Z, Ding JM. Inhibition of tumorigenesis by intratumoral delivery of the circadian gene mPer2 in C57BL/6 mice. Cancer Gene Ther. 2007;14:815-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | Benegiamo G, Mazzoccoli G, Cappello F, Rappa F, Scibetta N, Oben J, Greco A, Williams R, Andriulli A, Vinciguerra M. Mutual antagonism between circadian protein period 2 and hepatitis C virus replication in hepatocytes. PLoS One. 2013;8:e60527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Yang S, Pan X, Xiong Z, Wei B, Yao H. The influence of hepatitis B virus X protein on the clock genes in liver cells and its significance. Chinese-German J Clin Oncol. 2011;10:N8. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 20. | Kalamvoki M, Roizman B. The histone acetyltransferase CLOCK is an essential component of the herpes simplex virus 1 transcriptome that includes TFIID, ICP4, ICP27, and ICP22. J Virol. 2011;85:9472-9477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 57] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 21. | Schmutz I, Albrecht U, Ripperger JA. The role of clock genes and rhythmicity in the liver. Mol Cell Endocrinol. 2012;349:38-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 22. | Yamajuku D, Inagaki T, Haruma T, Okubo S, Kataoka Y, Kobayashi S, Ikegami K, Laurent T, Kojima T, Noutomi K. Real-time monitoring in three-dimensional hepatocytes reveals that insulin acts as a synchronizer for liver clock. Sci Rep. 2012;2:439. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 92] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 23. | Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci USA. 2009;106:21453-21458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 569] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 24. | Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950-2961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1680] [Cited by in RCA: 1758] [Article Influence: 70.3] [Reference Citation Analysis (0)] |
| 25. | Li S, Lin JD. Molecular control of circadian metabolic rhythms. J Appl Physiol (1985). 2009;107:1959-1964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 26. | Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 454] [Cited by in RCA: 469] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 27. | Duez H, Staels B. The nuclear receptors Rev-erbs and RORs integrate circadian rhythms and metabolism. Diab Vasc Dis Res. 2008;5:82-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 97] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 28. | Teboul M, Gréchez-Cassiau A, Guillaumond F, Delaunay F. How nuclear receptors tell time. J Appl Physiol (1985). 2009;107:1965-1971. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 29. | Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615-622. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1169] [Cited by in RCA: 1115] [Article Influence: 58.7] [Reference Citation Analysis (0)] |
| 30. | Charoensuksai P, Xu W. PPARs in Rhythmic Metabolic Regulation and Implications in Health and Disease. PPAR Res. 2010;2010:pii: 243643. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 31. | Hayashida S, Kuramoto Y, Koyanagi S, Oishi K, Fujiki J, Matsunaga N, Ikeda E, Ohdo S, Shimeno H, Soeda S. Proxisome proliferator-activated receptor-α mediates high-fat, diet-enhanced daily oscillation of plasminogen activator inhibitor-1 activity in mice. Chronobiol Int. 2010;27:1735-1753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 32. | Maury E, Ramsey KM, Bass J. Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circ Res. 2010;106:447-462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 393] [Cited by in RCA: 356] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 33. | Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315-1319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 586] [Cited by in RCA: 554] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 34. | Cretenet G, Le Clech M, Gachon F. Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 2010;11:47-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 148] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 35. | Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Lo Sasso G, Moschetta A, Schibler U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 326] [Cited by in RCA: 366] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 36. | Gälman C, Angelin B, Rudling M. Bile acid synthesis in humans has a rapid diurnal variation that is asynchronous with cholesterol synthesis. Gastroenterology. 2005;129:1445-1453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 170] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 37. | Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152-1156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 471] [Cited by in RCA: 437] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 38. | Doi R, Oishi K, Ishida N. CLOCK regulates circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2. J Biol Chem. 2010;285:22114-22121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 116] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 39. | Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627-631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1256] [Cited by in RCA: 1144] [Article Influence: 76.3] [Reference Citation Analysis (0)] |
| 40. | Sadacca LA, Lamia KA, deLemos AS, Blum B, Weitz CJ. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia. 2011;54:120-124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 270] [Cited by in RCA: 243] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 41. | Ganem D, Schneider R. Hepadnaviridae: the viruses and their replication. Fields virology. Philadelphia: Lippincott-Raven 2001; 2923-2970. |
| 42. | Locarnini S. Molecular virology of hepatitis B virus. Semin Liver Dis. 2004;24 Suppl 1:3-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 136] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 43. | Kay A, Zoulim F. Hepatitis B virus genetic variability and evolution. Virus Res. 2007;127:164-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 209] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 44. | Tiollais P, Pourcel C, Dejean A. The hepatitis B virus. Nature. 1985;317:489-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 934] [Cited by in RCA: 927] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 45. | Delius H, Gough NM, Cameron CH, Murray K. Structure of the hepatitis B virus genome. J Virol. 1983;47:337-343. [PubMed] |
| 46. | Pazienza V, Niro GA, Fontana R, Vinciguerra M, Andriulli A. Advance in molecular diagnostic tools for hepatitis B virus detection. Clin Chem Lab Med. 2013;51:1707-1717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 47. | Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725-12734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 386] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 48. | Guo Y, Kang W, Lei X, Li Y, Xiang A, Liu Y, Zhao J, Zhang J, Yan Z. Hepatitis B viral core protein disrupts human host gene expression by binding to promoter regions. BMC Genomics. 2012;13:563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 49. | Madden CR, Slagle BL. Stimulation of cellular proliferation by hepatitis B virus X protein. Dis Markers. 2001;17:153-157. [PubMed] |
| 50. | Ripoli M, Pazienza V. Impact of HCV genetic differences on pathobiology of disease. Expert Rev Anti Infect Ther. 2011;9:747-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 51. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264-1273.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2183] [Cited by in RCA: 2491] [Article Influence: 191.6] [Reference Citation Analysis (1)] |
| 52. | Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385-1395. [PubMed] |
| 53. | Narasimamurthy R, Hatori M, Nayak SK, Liu F, Panda S, Verma IM. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc Natl Acad Sci USA. 2012;109:12662-12667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 330] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 54. | Arjona A, Sarkar DK. The circadian gene mPer2 regulates the daily rhythm of IFN-gamma. J Interferon Cytokine Res. 2006;26:645-649. [PubMed] |
| 55. | Miyazaki K, Wakabayashi M, Chikahisa S, Sei H, Ishida N. PER2 controls circadian periods through nuclear localization in the suprachiasmatic nucleus. Genes Cells. 2007;12:1225-1234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 17] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 56. | Jonas MM. Interferon-alpha for viral hepatitis. J Pediatr Gastroenterol Nutr. 1996;23:93-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 57. | Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929-937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1479] [Cited by in RCA: 1514] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 58. | Nakamoto Y, Guidotti LG, Kuhlen CV, Fowler P, Chisari FV. Immune pathogenesis of hepatocellular carcinoma. J Exp Med. 1998;188:341-350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 283] [Cited by in RCA: 258] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 59. | Mazzoccoli G, Sothern RB, Greco A, Pazienza V, Vinciguerra M, Liu S, Cai Y. Time-related dynamics of variation in core clock gene expression levels in tissues relevant to the immune system. Int J Immunopathol Pharmacol. 2011;24:869-879. [PubMed] |
| 60. | Cermakian N, Lange T, Golombek D, Sarkar D, Nakao A, Shibata S, Mazzoccoli G. Crosstalk between the circadian clock circuitry and the immune system. Chronobiol Int. 2013;30:870-888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 197] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 61. | Arjona A, Sarkar DK. Circadian oscillations of clock genes, cytolytic factors, and cytokines in rat NK cells. J Immunol. 2005;174:7618-7624. [PubMed] |
| 62. | Logan RW, Sarkar DK. Circadian nature of immune function. Mol Cell Endocrinol. 2012;349:82-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 135] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 63. | Arjona A, Sarkar DK. Are circadian rhythms the code of hypothalamic-immune communication? Insights from natural killer cells. Neurochem Res. 2008;33:708-718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 64. | Liu J, Malkani G, Shi X, Meyer M, Cunningham-Runddles S, Ma X, Sun ZS. The circadian clock Period 2 gene regulates gamma interferon production of NK cells in host response to lipopolysaccharide-induced endotoxic shock. Infect Immun. 2006;74:4750-4756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 179] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 65. | Dimitrov S, Benedict C, Heutling D, Westermann J, Born J, Lange T. Cortisol and epinephrine control opposing circadian rhythms in T cell subsets. Blood. 2009;113:5134-5143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 255] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 66. | Mazzoccoli G, De Cata A, Greco A, Carughi S, Giuliani F, Tarquini R. Circadian rhythmicity of lymphocyte subpopulations and relationship with neuro-endocrine system. J Biol Regul Homeost Agents. 2010;24:341-350. [PubMed] |
| 67. | Mazzoccoli G, Sothern RB, De Cata A, Giuliani F, Fontana A, Copetti M, Pellegrini F, Tarquini R. A timetable of 24-hour patterns for human lymphocyte subpopulations. J Biol Regul Homeost Agents. 2011;25:387-395. [PubMed] |
| 68. | Dimitrov S, Lange T, Born J. Selective mobilization of cytotoxic leukocytes by epinephrine. J Immunol. 2010;184:503-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 166] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 69. | Logan RW, Arjona A, Sarkar DK. Role of sympathetic nervous system in the entrainment of circadian natural-killer cell function. Brain Behav Immun. 2011;25:101-109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 70. | Lange T, Dimitrov S, Fehm HL, Westermann J, Born J. Shift of monocyte function toward cellular immunity during sleep. Arch Intern Med. 2006;166:1695-1700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 102] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 71. | Lange T, Dimitrov S, Born J. Effects of sleep and circadian rhythm on the human immune system. Ann N Y Acad Sci. 2010;1193:48-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 398] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 72. | Esquifino AI, Alvarez MP, Cano P, Chacon F, Reyes Toso CF, Cardinali DP. 24-hour pattern of circulating prolactin and growth hormone levels and submaxillary lymph node immune responses in growing male rats subjected to social isolation. Endocrine. 2004;25:41-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 73. | Esquifino AI, Chacon F, Cano P, Marcos A, Cutrera RA, Cardinali DP. Twenty-four-hour rhythms of mitogenic responses, lymphocyte subset populations and amino acid content in submaxillary lymph nodes of growing male rats subjected to calorie restriction. J Neuroimmunol. 2004;156:66-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 74. | Fauci AS. Mechanisms of corticosteroid action on lymphocyte subpopulations. I. Redistribution of circulating T and b lymphocytes to the bone marrow. Immunology. 1975;28:669-680. [PubMed] |
| 75. | Tai DI, Tsai SL, Chen YM, Chuang YL, Peng CY, Sheen IS, Yeh CT, Chang KS, Huang SN, Kuo GC. Activation of nuclear factor kappaB in hepatitis C virus infection: implications for pathogenesis and hepatocarcinogenesis. Hepatology. 2000;31:656-664. [PubMed] |
| 76. | Park J, Kang W, Ryu SW, Kim WI, Chang DY, Lee DH, Park do Y, Choi YH, Choi K, Shin EC. Hepatitis C virus infection enhances TNFα-induced cell death via suppression of NF-κB. Hepatology. 2012;56:831-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 77. | Li Q, Pène V, Krishnamurthy S, Cha H, Liang TJ. Hepatitis C virus infection activates an innate pathway involving IKK-α in lipogenesis and viral assembly. Nat Med. 2013;19:722-729. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 156] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 78. | Kang TH, Sancar A. Circadian regulation of DNA excision repair: implications for chrono-chemotherapy. Cell Cycle. 2009;8:1665-1667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 79. | Quera Salva MA, Hartley S, Barbot F, Alvarez JC, Lofaso F, Guilleminault C. Circadian rhythms, melatonin and depression. Curr Pharm Des. 2011;17:1459-1470. [PubMed] |
| 80. | Lange CM, Zeuzem S. Perspectives and challenges of interferon-free therapy for chronic hepatitis C. J Hepatol. 2013;58:583-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |