Published online Dec 14, 2013. doi: 10.3748/wjg.v19.i46.8619
Revised: September 24, 2013
Accepted: September 29, 2013
Published online: December 14, 2013
Processing time: 201 Days and 20.1 Hours
AIM: To investigate the role of P115 in the proliferation of gastric cancer cells and the mechanism involved.
METHODS: The RNA and protein level of P115 and macrophage migration inhibitory factor (MIF) in gastric cancer and normal gastric tissue/cells were measured and the effect of P115 on cell proliferation was assessed. The role of P115 in cell cycle checkpoints was investigated and the related proteins and signaling pathways, such as cyclin D1, Mcm2, p53, PCNA as well as the MAPK signaling pathway were determined. The interaction between P115 and MIF and the effect of P115 on MIF secretion were examined. The data were analyzed via one-way ANOVA comparisons between groups and P < 0.05 was considered significant.
RESULTS: P115 and MIF were both specifically expressed in gastric cancer tissues compared with normal gastric mucosa (both P < 0.01). The mRNA and protein levels of P115 and MIF in gastric cancer cell lines MKN-28 and BGC-823 were higher than in the human gastric epithelial cell line GES-1 (both P < 0.01). In MKN-28 and BGC-823 cell lines, P115 promoted cell proliferation and G0-G1 to S phase transition. In addition, several cell cycle-related regulators, including cyclin D1, Mcm2, PCNA, pERK1/2 and p53 were up-regulated by P115. Furthermore, the interaction between P115 and MIF was confirmed by co-immunoprecipitation assay. ELISA showed that P115 stimulated the secretion of MIF into the culture supernatant (P < 0.01) and the compensative expression of MIF in cells was observed by Western blotting.
CONCLUSION: P115 promotes proliferation of gastric cancer cells through an interaction with MIF.
Core tip: Gastric cancer is one of the most common cancers. P115 is a tether protein which plays a key role in cell proliferation through combination with binding partners, including migration inhibitory factor (MIF). The present study showed that P115 and MIF were specifically expressed in gastric cancer tissues and cells. P115 promoted cell proliferation and G0-G1 to S phase transition. Cell cycle regulators, including cyclin D1, Mcm2, PCNA, pERK1/2 and p53 were up-regulated by P115. P115 interacted with MIF and stimulated the secretion of MIF into the culture supernatant. In summary, P115 promotes proliferation of gastric cancer cells through an interaction with MIF.
- Citation: Li XJ, Luo Y, Yi YF. P115 promotes growth of gastric cancer through interaction with macrophage migration inhibitory factor. World J Gastroenterol 2013; 19(46): 8619-8629
- URL: https://www.wjgnet.com/1007-9327/full/v19/i46/8619.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i46.8619
Gastric cancer is one of the most common cancers worldwide with a significant impact on human health[1]. Despite significant developments in the diagnosis and treatment of gastric cancer, the prognosis remains poor. Extensive surgery combined with chemotherapy is the most common therapy choice in the early stages of gastric cancer[2], while additional treatment options, such as gene therapy are desperately needed. With significant advances in genomics and proteomics, the discovery of a novel oncogene for therapeutic intervention remains a future challenge.
Cancer growth is a highly complex process involving alterations in gene expression and the interaction of many proteins. Golgi-vesicular transport protein P115 is a tether protein that plays an important role in many signal pathways required for cell proliferation[3] and has been extensively studied[4-6]. Macrophage migration inhibitory factor (MIF) was one of the first cytokines to be described and extensively studied[7]. More recently, MIF has been reported to be overexpressed in a number of cancers, including esophageal squamous cell carcinoma[8], glioblastoma[9], neuroblastoma[10], colonic cancer[11] and colorectal cancer[12]. The ability of MIF to promote tumor progression has been demonstrated and MIF has been shown to be a potential target for anti-cancer therapy. Hudson et al[13] and Jung et al[14] reported that MIF antagonized the activity of p53, which led to cancer progression. It was shown that the binding partner of MIF was JAB1/CSN5[15] which is known to be involved in the differentiation and morphogenesis of cells[16]. Furthermore, it is well known that upon binding to one of its receptors-CD74, MIF can increase the phosphorylation of Akt, ERK, MAPK and Stat3 which are all necessary for tumor proliferation.
Recently, a yeast two-hybrid interaction was examined to identify the intracellular proteins which might bind to MIF and mediate its secretion, and it was shown that P115 was a binding partner of MIF[17]. Previous research in our laboratory also demonstrated the same result. The objective of the present study was to evaluate the expression, the function in cell proliferation and the biological mechanism of P115 in gastric cancer.
Human gastric cancer cell lines BGC-823 and MKN-28 were obtained from the American Type Culture Collection (Manassas, VA, United States). The human gastric epithelial cell line GES-1 was obtained from the cell bank of the Fourth Military Medical University. The cells were cultured in RPMI 1640 medium (Gibco, Maryland, United States) supplied with 10% FBS (Gibco, Maryland, United States), 100 units/mL penicillin and 100 μg/mL streptomycin at 37 °C in humidified 5% CO2.
Thirty gastric cancer and 30 normal gastric mucosa specimens were obtained from the Department of Pathology, the First Affiliated Hospital, Chongqing Medical University from September 2008 to November 2009. Normal gastric mucosa specimens were obtained from normal tissues adjacent to the cancer tissue, and were pathologically confirmed as non-cancerous. The procedure was approved by the Ethics Committee. Samples were incubated with anti-P115 and anti-MIF rabbit polyclonal antibody (Cell Signaling Technology, United States) at 4 °C overnight, and then incubated with biotinylated goat anti-rabbit antibody (Santa Cruz Biotechnology, TX, United States) at room temperature for 15 min. DAB substrate was then used in the chromogenic reaction.
The pCD-shRNA was reconstructed from pGPU6/GFP/Neo. Four shRNAs targeting P115 and shNC were designed as shown in Table 1. ShRNAs were ligated into the BamH I and Bbs I-digested pGPU6/GFP/Neo vector. The P115 expressing plasmid, pEGFP-N1-P115, was obtained from Jikai Company (Shanghai, China). Cells were seeded in 6-well plates and were transfected with 2 μg plasmids after reaching 70%-80% confluence using Lipofectamine 2000 (Invitrogen, Carlsbad, United States) following the manufacturer’s instructions.
| shRNA name | Target site | Sequences | |
| P115-shRNA1 | 1117 bp | S | 5'-CACCGCAGCTTTGTACTATCCTAATTTCAAGAGA |
| ATTAGGATAGTACAAAGCTGCTTTTTTG-3' | |||
| A | 5'-GATCCAAAAAAGCAGCTTTGTACTATCCTAAT | ||
| TCTCTTGAAATTAGGATAGTACAAAGCTGC-3' | |||
| P115-shRNA2 | 1318 bp | S | 5'-CACCGCGCTGTGCTGTTCTCTATTGTTCAAGAGA |
| CAATAGAGAACAGCACAGCGCTTTTTTG-3' | |||
| A | 5'-GATCCAAAAAAGCGCTGTGCTGTTCTCTATTG | ||
| TCTCTTGAACAATAGAGAACAGCACAGCGC-3' | |||
| P115-shRNA3 | 1578 bp | S | 5'-CACCGCAACCCTCCAGTTTCTTTACTTCAAGAGA |
| GTAAAGAAACTGGAGGGTTGCTTTTTTG-3' | |||
| A | 5'-GATCCAAAAAAGCAACCCTCCAGTTTCTTTAC | ||
| TCTCTTGAAGTAAAGAAACTGGAGGGTTGC-3' | |||
| P115-shRNA4 | 1777 bp | S | 5'-CACCGCAGTTGGTCCAAGGCTTATGTTCAAGAGACATAAGCCTTGGACCAACTGCTTTTTTG-3' |
| A | 5'-GATCCAAAAAAGCAGTTGGTCCAAGGCTTATG | ||
| TCTCTTGAACATAAGCCTTGGACCAACTGC-3' | |||
| NC-shRNA | - | S | 5'-CACCGTTCTCCGAACGTGTCACGTTTCAAGAGA |
| ACGTGACACGTTCGGAGAACTTTTTTG-3' | |||
| A | 5'- GATCCAAAAAAGTTCTCCGAACGTGTCACGT | ||
| TCTCTTGAAACGTGACACGTTCGGAGAAC-3' |
RT-PCR was carried out using the AccessQuickTM One-Step reverse transcription-polymerase chain reaction [RT-PCR kit (Promega Co., Madison, United States)] according to the manufacturer’s protocol. The oligonucleotide primers used were as follows: P115 sense: 5’-AACCTGGTGGCTGAACGGCAAG-3’, P115 antisense: 5’-AGAAGCTTCACACCAGGCCAGC-3’. MIF sense: 5’-CGGGTTCCTCTCCGAGCTCACC3’, MIF antisense: 5’-TGATGTAGACCCTGTCCGGGCTGA-3’. β-actin sense: 5’-GACCCAGATCATGTTTGAGACC-3’, β-actin antisense: 5’-GCCAGGATAGAGCCACCAAT-3’. Total RNA was reverse transcribed to synthesize cDNA at 45 °C for 45 min. PCR was performed in a single reaction volume of 25 μL. The schedule consisted of incubation for 5 min at 94 °C followed by 30 cycles of 94 °C for 30 s, 56 °C for 45 s and 72 °C for 1 min, then incubation for 10 min at 72 °C. The PCR products were subjected to 1.5% agarose gel electrophoresis. Quantitative real-time RT-PCR was performed using specific sense and antisense primers in a 25 μL reaction volume containing 12.5 μL of Absolute™ QPCR SYBR Green mix (Invitrogen), 0.25 pmol of each primer, and 0.5 μg of mRNA. Oligonucleotide primers were as follows: P115 sense: 5’-GGAGGGGAACAGTGATGGAG-3’, P115 antisense: 5’-CAAAGCTGCTGCAATAACCC-3’. β-actin sense: 5’-CGGGAAATCGTGCGTGAC-3’, β-actin antisense: 5’-TGGAAGGTGGACAGCGAGG-3’. The number of amplification cycles was 35, and the reaction were performed for 3 min at 50 °C, 20 s at 95 °C, and 30 s at 60 °C, with an initial step at 95 °C for 3 min.
Cells were lysed in 100 μL RIPA lysis buffer (50 mmol/L Tris-HCl, pH 7.5, 1% NP-40, 150 mmol/L NaCl, 1 mg/mL aprotinin, 1 mg/mL leupeptin, 1 mmol/L Na3VO4, 1 mmol/L NaF) at 4 °C for 30 min. Cell debris was removed by centrifugation at 12000 ×g for 20 min at 4 °C. Protein concentrations were determined by the Bradford assay. An equal amount of lysate (40 μg) was resolved by SDS-polyacrylamide gel electrophoresis and transferred to a PVDF membrane (Millipore, Bedford, United States). The membranes were blocked with 5% nonfat milk at room temperature for 1 h and then incubated for 2 h with primary antibodies. The membranes were then incubated for 1 h with an appropriate horseradish peroxidase-linked secondary antibody (Santa Cruz Biotechnology, TX, United States). Antibodies to P115, MIF, cyclin D1, Mcm2, PCNA, p53 and β-actin were obtained from Cell Signaling Technology (MA, United States). Electrochemiluminescence was performed according to the manufacturer’s instructions using a Bio-Rad imaging system. Quantity One software was used to quantify the density of bands.
Cells were seeded in 96-well plates at a density of 2000 cells/well and allowed to proliferate for 24 h, 48 h and 72 h. Cell proliferation ability was assessed by MTT assay. Briefly, MTT (5 mg/mL) was added to each well and the plate was incubated for a further 4 h before removal of the media. DMSO was then added to each well to solubilize the formazan crystals. The absorbance was read at a wavelength of 595 nm using a microtiter plate reader. All experiments were carried out in triplicate.
Flow cytometric analysis was performed as previously described[18]. Forty-eight hours after transfection, cells were harvested and fixed with 75% ethanol at -20 °C overnight. Cells were stained with propidium iodide (25 mg/mL) and RNaseA (200 mg/mL) at 37 °C for 30 min. The DNA content was analyzed using a FACScan flow cytometer (Beckman Coulter, Germany).
Cells were lysed in lysis buffer at 4 °C for 30 min. Cell debris was removed by centrifugation at 14000 ×g for 5 min at 4 °C. To remove non-specific binding, protein G sepharose beads containing mouse IgG were added to 200 μL protein and shaken slowly for 2 h at 4 °C. The sample was then centrifuged at 2500 ×g for 5 min at 4 °C and the supernatant was carefully removed for immunoprecipitation. 1 μg MIF antibody was incubated with the supernatant overnight and 42 μL protein G Sepharose beads were then added. The mixture was incubated for 3 h at 4 °C on a tube roller to precipitate protein complexes. The beads were obtained by centrifugation at 1000 ×g for 60 s and washed twice with PBS. Finally, 20 μL loading buffer was added for SDS-polyacrylamide gel electrophoresis to assess P115.
MIF level in the culture supernatant was determined by ELISA according to the manufacturer’s recommendations. A polyclonal anti-MIF antibody was used as the capture antibody, and absorbance was measured at 450 nm in a microplate reader. The concentration of MIF in each sample was obtained by comparing absorbance values against the standard curve using r-MIF. Each experiment was performed in triplicate.
The data were expressed as mean ± SD of three independent experiments. The data were analyzed via one-way ANOVA comparisons between different groups with significance value set at P < 0.05.
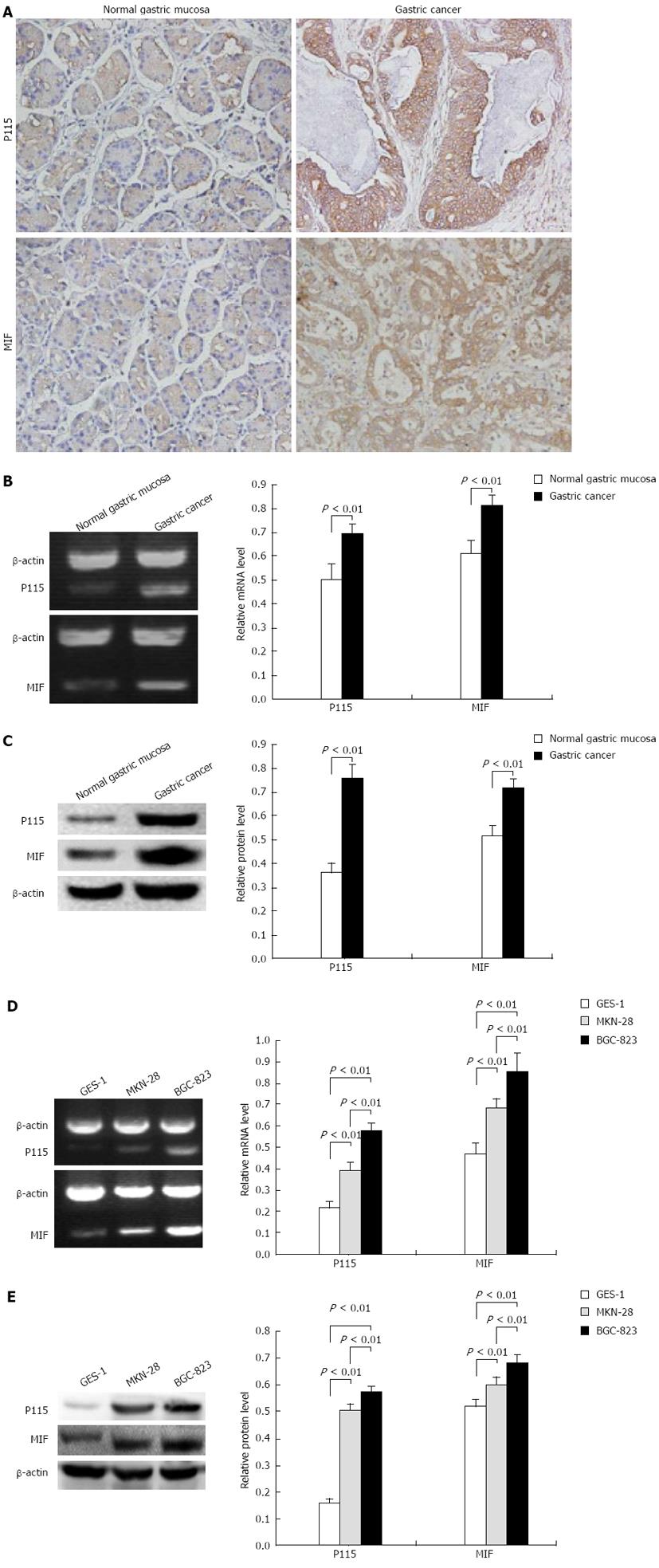
To examine whether P115 and MIF were specifically expressed in gastric cancer, the protein levels of P115 and MIF in human gastric tissue were first measured by immunohistochemistry (Figure 1A). It was shown that P115 was expressed in Golgi and cytoplasm near the nucleus, there were 12 positive samples (40.0%) in normal gastric mucosa and 22 positive samples (73.3%) in gastric cancer with 63.6% showing a strong positive rate (14 cases). MIF was expressed in cytoplasm and sparsely in membrane, there were 14 positive samples (46.7%) in normal gastric mucosa and 24 positive samples (80%) in gastric cancer with 66.7% showing a strong positive rate (16 cases).
Furthermore, the tissue homogenates of normal gastric mucosa and gastric cancer were lysed to measure P115 and MIF levels. Semi-quantitative RT-PCR analysis showed that P115 and MIF mRNA in gastric cancer (0. 694 ± 0. 046 and 0. 814 ± 0. 040, respectively, n = 3) were 1.377 and 1.326 times that in normal mucosa (0.504 ± 0.646 and 0.614 ± 0.054, respectively, n = 3) (Figure 1B, both P < 0.01). As shown in the semi-quantitative analysis of Western blotting results (Figure 1C), compared with normal gastric mucosa, the expression of P115 and MIF increased by 2.085- and 1.391-fold in gastric cancer (0.759 ± 0.058 vs 0.364 ± 0.037; 0.715 ± 0.040 vs 0.514 ± 0.044, respectively, n = 3; both P < 0.01).
P115 and MIF mRNA in different cell lines are shown in Figure 1D. P115 mRNA in the MKN-28 (0.391 ± 0.042, n = 3) and BGC-823 (0.513 ± 0. 038, n = 3) cell lines was 1.836- and 2.408-fold that in the normal gastric mucosa epithelial cell line GES-1 (0.213 ± 0.036, n = 3), respectively (both P < 0.01). Moreover, in the poorly differentiated cell line, BGC-823, it was 1.312 times that in MKN-28 cells (P < 0.01). Correspondingly, MIF mRNA in MKN-28 (0.683 ± 0.046, n = 3) and BGC-823 (0.895 ± 0.104, n = 3) cells was 1.453 and 1.904 times that in GES-1 (0.470 ± 0.052, n = 3) and BGC-823 cells was 1.310 times that in MKN-28 cells (all P < 0.01).
Similar to the results of RT-PCR, the protein levels of P115 and MIF were markedly higher in MKN-28 and BGC-823 cells than in GES-1 cells (Figure 1E), and were higher in BGC-823 cells. Semi-quantitative analysis showed that the expression of P115 in MKN-28 (0.507 ± 0. 020, n = 3) and BGC-823 (0.547 ± 0. 015, n = 3) cells was 3.229- and 3.484-fold that in GES-1 cells (0.157 ± 0.010, n = 3), and in BGC-823 it was 1.079-fold that in MKN-28 cells (all P < 0.01). The expression of MIF in MKN-28 (0.601 ± 0.017, n = 3) and BGC-823 (0.687 ± 0.015, n = 3) cells was 1.154- and 1.319-fold that in GES-1 cells (0.521 ± 0.020, n = 3), and in BGC-823 it was 1.143-fold that in MKN-28 cells (all P < 0.01).
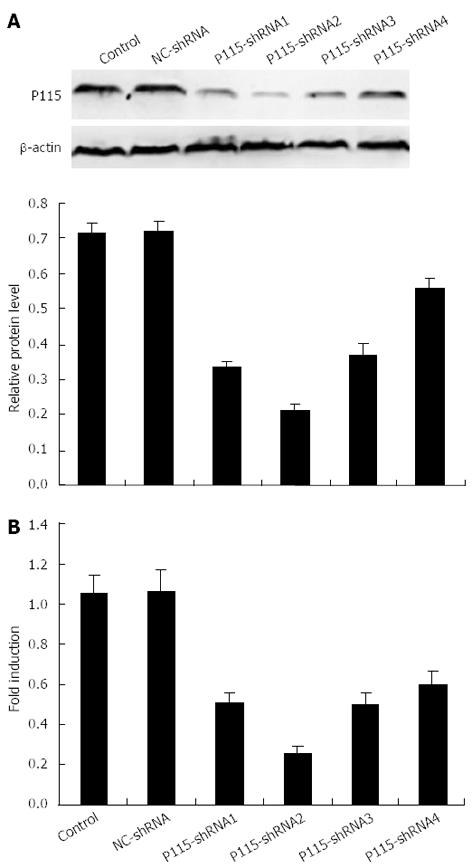
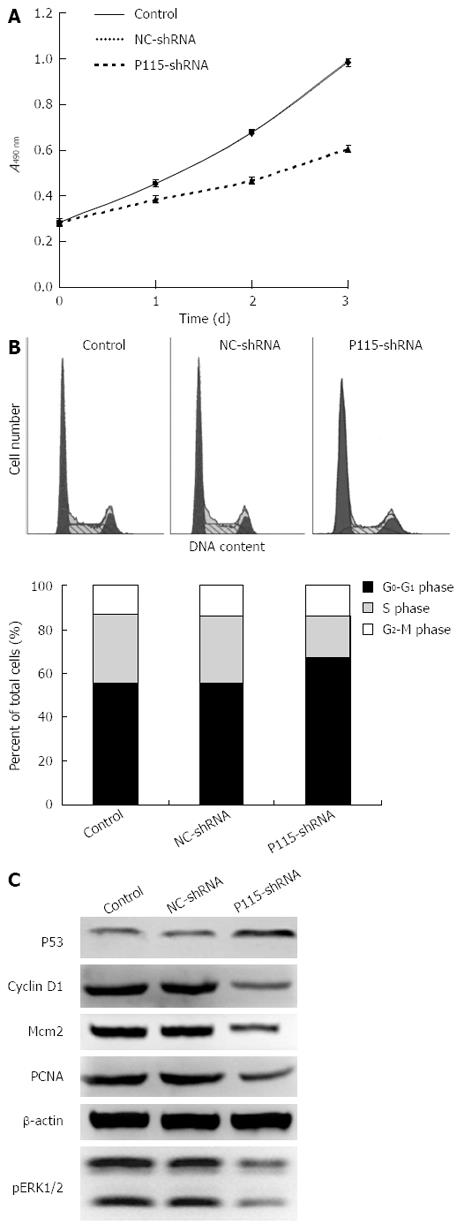
To explore the biological function of P115, P115-shRNA plasmids expressing siRNA were constructed. First, the silencing efficiencies of 4 different P115-shRNAs were tested using Western blotting (Figure 2A) and real-time PCR (Figure 2B), which showed that the level of P115 was down-regulated most by 2 μg P115-shRNA2 plasmids (protein: 0.259 ± 0.034, n = 3; mRNA: 0.211 ± 0.010, n = 3) after 36 h transfection in BGC-823 cells (expression of P115 was relatively high) compared with control cells (protein: 0.727 ± 0.018, n = 3; mRNA: 1.041 ± 0.086, n = 3) and NC-shRNA (protein: 0.735 ± 0.010, n = 3; mRNA: 1.054 ± 0.094, n = 3), with silencing efficacy up to 76.8%. Therefore, P115-shRNA2 was selected for subsequent study. The proliferation rate of BGC-823 cells was then determined by MTT assay 24 h, 48 h and 72 h after transfection, and showed that the growth rate of P115-shRNA treated BGC-823 cells was obviously decreased (Figure 3A) compared with NC-shRNA.
The role of P115 in the cell cycle checkpoints was investigated (Figure 3B). FACS analysis revealed that P115-shRNA resulted in an 11.3% and 11.18% increase in cell number at G0-G1 phase compared with control and NC-shRNA in BGC-823 cells.
As P115-shRNA caused cell cycle arrest, it was supposed that P115 could lead to a change in G0-G1 phase-related proteins and signaling pathways, such as cyclin D1, Mcm2, p53, PCNA as well as the MAPK signaling pathway. Therefore, the above proteins and phosphorylation of ERK1/2 were assessed. It was shown that cyclin D1, Mcm2, PCNA and pERK1/2 were significantly decreased by P115-shRNA in BGC-823 cells, which explained the effect of P115 on cell cycle phase. In addition, p53 was up-regulated by P115-shRNA (Figure 3C).
MIF in BGC-823 cells was extracted through protein precipitation with a multiple clone antibody and the protein complex was detected by Western blotting. As shown in Figure 4A, P115 was detected in the protein complex, indicating that there was an interaction between MIF and P115 protein.
Considering that MIF is a secretory protein, the level of MIF in the culture supernatant was assessed by ELISA. This assay showed that the secreted concentration of MIF in the culture supernatant in P115-shRNA treated BGC-823 cells was markedly reduced (1173.67 ± 63.47 pg/mL, n = 3) compared with control (1535.62 ± 77.25 pg/mL, n = 3) and NC-shRNA treated cells (1517.69 ± 102.51 pg/mL, n = 3), this difference was statistically significant (P < 0.01, Figure 4B). In addition, MIF mRNA and protein in cells were also detected. As shown in Figure 4C, P115-shRNA decreased the level of MIF mRNA in BGC-823 cells, and Western blotting showed the same trend as real-time PCR, in that P115-shRNA decreased the expression of MIF (Figure 4D).
The role of P115 in gastric cancer cells was assessed from another point of view. It was shown that after transfection with 2 μg pEGFP-N1-P115 for 24, 48 and 72 h, the proliferation rate of MKN-28 cells (expression of P115 was relatively low) was markedly increased (Figure 5A) and the transition of G0-G1 phase to S phase in MKN-28 cells was accelerated by 13.71% and 13.9%, respectively, compared with control and pEGFP-N1-NC (Figure 5B), suggesting that stimulation of cell growth by P115 was associated with the distribution of cell cycle phase. Correspondingly, cyclin D1, Mcm2, PCNA and pERK1/2 were significantly increased by pEGFP-N1-P115 in MKN-28 cells (Figure 5C).
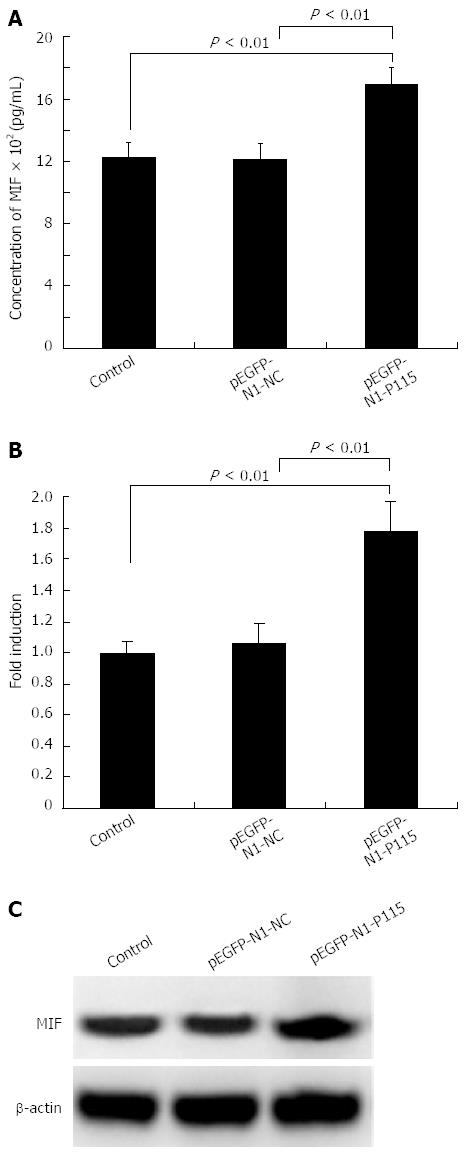
ELISA showed that the secreted concentration of MIF in the culture supernatant in pEGFP-N1-P115 treated MKN-28 cells was markedly increased (1696.38 ± 107.95 pg/mL, n = 3) compared with control (1227.64 ± 90.58 pg/mL, n = 3) and pEGFP-N1-NC treated cells (1208.63 ± 101.78 pg/mL, n = 3), and the difference was statistically significant (P < 0.01, Figure 6A). As shown in Figure 6B, MIF mRNA in pEGFP-N1-P115 treated cells was increased and Western blotting also indicated that pEGFP-N1-P115 increased the expression of MIF (Figure 6C).
The complicated molecular mechanisms of carcinogenesis and the interaction of multiple oncogenes in gastric cancer challenge our ability to identify novel and rational molecular therapeutic targets. The present study demonstrates that P115 may be a potential tumor biomarker and therapeutic target which is overexpressed in human gastric cancer. Interaction with MIF may be involved in its molecular mechanism.
P115 has been demonstrated to be involved in intra-Golgi transport[6] and can bind to the Golgi-associated proteins, GM130[19] and giantin[20], which both play an important role in mitosis, that is, P115 is essential for biogenesis of the Golgi apparatus[21,22]. MIF is a secretary protein which plays an important upstream role in the regulation of diverse cellular responses[23-25]. The role of MIF has been emphasized by the finding that high expression of MIF is associated with the incidence or the severity of oncologic diseases[26-28]. The data from this study showed that overexpression of P115 significantly enhanced the secretion of MIF, which indicated that P115 might be one of the stimuli inducing MIF secretion through direct interaction. Merk et al[29] reported that MIF was co-secreted with P115, indicating that P115 had a specific role in MIF export, which is consistent with our results. MIF lacks a signal sequence and is secreted by an unconventional route for protein export. Stimuli induce the rapid release of MIF from preformed and cytoplasmic pools, which is followed by an upregulation of MIF mRNA expression and a replenishment of intracellular protein content[30,31]. Therefore, the protein and mRNA expression of MIF in cells was detected. As expected, when P115 was overexpressed or silenced, MIF protein and mRNA in cells were also enhanced or reduced compensatively.
The biological mechanism of MIF on tumor growth includes the induction of growth-related protein expression and inhibition of apoptosis-related protein expression[32]. Jung et al[14] demonstrated that MIF interacted with p53 in vivo and directly promoted tumorigenesis by inhibiting p53 accumulation. Our data demonstrated that P115 knockdown enhanced the expression of p53, which was considered a result of MIF reduction. It is known that p53 is a classic tumor suppressor gene that can promote cell cycle arrest and apoptosis in response to DNA damage. Absence or down-regulation of p53 can interfere with these important checkpoints for maintaining genetic stability and allows cells to survive and proliferate. This may explain our results where knockdown of P115 led to the inhibition of cell proliferation and apoptosis (results not shown).
To further explore the molecular mechanism of P115 influencing cell growth, key proteins involved in the G0-G1 phase relevant signaling pathway were determined. It was reported that the ERK1/2 pathway was necessary for transcriptional induction of cyclin D1 which promoted progression from G1 to S phase[33]. In addition, Mcm2 and PCNA are both important proteins for initiation of DNA synthesis[34]. As shown in our results, cyclin D1, Mcm2 and PCNA, as well as pERK1/2 were markedly reduced by P115-shRNA, which was consistent with G0-G1 arrest. Researchers have reported that recombinant MIF can activate the ERK-MAP kinase pathway, and subsequently increase cell proliferation rate in fibroblasts and a colon cancer cell line[35]. Therefore, it is concluded that MIF is the key factor in the biological function of P115 in cell proliferation.
In conclusion, our study demonstrates that P115 is overexpressed in gastric cancer tissue and cells. Knockdown of P115 blocks cell proliferation in vitro, and the mechanism involves P115 stimulating the secretion of MIF directly by interacting with MIF, subsequently, leading to progression of cell cycle through relevant proteins. Although additional functional studies are required, P115 as well as the interaction between P115 and MIF may be a potential therapeutic target for the treatment of gastric cancer.
Cancer growth is a highly complex process involving alterations in gene expression and the interaction of many proteins. Golgi-vesicular transport protein P115 is a tether protein that plays an important role in many signal pathways required for cell proliferation. Some studies have reported the function of P115 in intra-Golgi transport, but there are few studies on the role of P115 in cancer cell proliferation and on its biological mechanism.
The ability of migration inhibitory factor (MIF) to promote tumor progression has been demonstrated. Recently, a yeast two-hybrid interaction was examined to identify the intracellular proteins which might bind to MIF and mediate its secretion, and it was shown that P115 was a binding partner of MIF.
This study showed that over-expression of P115 could significantly enhance the secretion of MIF, which indicated that P115 might be one of the stimuli inducing MIF secretion through a direct interaction. Through interaction with MIF, P115 promoted progression from G1 to S phase and then influenced the growth of cancer cells. Correspondingly, cyclin D1, Mcm2 and PCNA, as well as pERK1/2 were markedly reduced by P115-shRNA, being consistent with G0-G1 arrest.
The study results suggest that P115 as well as the interaction of P115 and MIF may be a potential therapeutic target for treatment of gastric cancer.
Protein-protein interaction: the majority of proteins in living systems function due to interaction with each other in stable or dynamic protein complexes. P115 is a tether protein which has been demonstrated to be involved in intra-Golgi transport. MIF is a secretary protein which plays an important role in regulation of diverse cellular responses.
This is a good descriptive study in which authors explore the expression and role of two important proteins in gastric cancer biology. The results are significant and suggest that P115 promote cells proliferation through interaction with MIF and provide a potential therapeutic target for treatment of gastric cancer.
P- Reviewers: Lu J, Raffaniello RD S- Editor: Wen LL L- Editor: Webster JR E- Editor: Zhang DN
| 1. | Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137-2150. [PubMed] |
| 2. | Bickenbach K, Strong VE. Comparisons of Gastric Cancer Treatments: East vs. West. J Gastric Cancer. 2012;12:55-62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 123] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 3. | An Y, Chen CY, Moyer B, Rotkiewicz P, Elsliger MA, Godzik A, Wilson IA, Balch WE. Structural and functional analysis of the globular head domain of p115 provides insight into membrane tethering. J Mol Biol. 2009;391:26-41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 4. | Sztul E, Lupashin V. Role of tethering factors in secretory membrane traffic. Am J Physiol Cell Physiol. 2006;290:C11-C26. [PubMed] |
| 5. | Short B, Haas A, Barr FA. Golgins and GTPases, giving identity and structure to the Golgi apparatus. Biochim Biophys Acta. 2005;1744:383-395. [PubMed] |
| 6. | Waters MG, Clary DO, Rothman JE. A novel 115-kD peripheral membrane protein is required for intercisternal transport in the Golgi stack. J Cell Biol. 1992;118:1015-1026. [PubMed] |
| 7. | Babu SN, Chetal G, Kumar S. Macrophage migration inhibitory factor: a potential marker for cancer diagnosis and therapy. Asian Pac J Cancer Prev. 2012;13:1737-1744. [PubMed] |
| 8. | Ren Y, Law S, Huang X, Lee PY, Bacher M, Srivastava G, Wong J. Macrophage migration inhibitory factor stimulates angiogenic factor expression and correlates with differentiation and lymph node status in patients with esophageal squamous cell carcinoma. Ann Surg. 2005;242:55-63. [PubMed] |
| 9. | Shkolnik T, Livni E, Reshef R, Lachter J, Eidelman S. Comparison of two lymphokines (macrophage migration inhibition, leukocyte adherence inhibition factors) and carcinoembryonic antigen, in colorectal cancer and colonic premalignant lesions. Am J Gastroenterol. 1987;82:1275-1278. [PubMed] |
| 10. | Al-Abed Y, VanPatten S. MIF as a disease target: ISO-1 as a proof-of-concept therapeutic. Future Med Chem. 2011;3:45-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 81] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 11. | Ren Y, Chan HM, Fan J, Xie Y, Chen YX, Li W, Jiang GP, Liu Q, Meinhardt A, Tam PK. Inhibition of tumor growth and metastasis in vitro and in vivo by targeting macrophage migration inhibitory factor in human neuroblastoma. Oncogene. 2006;25:3501-3508. [PubMed] |
| 12. | Lee H, Rhee H, Kang HJ, Kim HS, Min BS, Kim NK, Kim H. Macrophage migration inhibitory factor may be used as an early diagnostic marker in colorectal carcinomas. Am J Clin Pathol. 2008;129:772-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 13. | Hudson JD, Shoaibi MA, Maestro R, Carnero A, Hannon GJ, Beach DH. A proinflammatory cytokine inhibits p53 tumor suppressor activity. J Exp Med. 1999;190:1375-1382. [PubMed] |
| 14. | Jung H, Seong HA, Ha H. Critical role of cysteine residue 81 of macrophage migration inhibitory factor (MIF) in MIF-induced inhibition of p53 activity. J Biol Chem. 2008;283:20383-20396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 75] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 15. | Kleemann R, Hausser A, Geiger G, Mischke R, Burger-Kentischer A, Flieger O, Johannes FJ, Roger T, Calandra T, Kapurniotu A. Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature. 2000;408:211-216. [PubMed] |
| 16. | Fingerle-Rowson G, Petrenko O. MIF coordinates the cell cycle with DNA damage checkpoints. Lessons from knockout mouse models. Cell Div. 2007;2:22. [PubMed] |
| 17. | Nelson DS, Alvarez C, Gao YS, García-Mata R, Fialkowski E, Sztul E. The membrane transport factor TAP/p115 cycles between the Golgi and earlier secretory compartments and contains distinct domains required for its localization and function. J Cell Biol. 1998;143:319-331. [PubMed] |
| 18. | Zhao M, He HW, Sun HX, Ren KH, Shao RG. Dual knockdown of N-ras and epiregulin synergistically suppressed the growth of human hepatoma cells. Biochem Biophys Res Commun. 2009;387:239-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Gmachl MJ, Wimmer C. Sequential involvement of p115, SNAREs, and Rab proteins in intra-Golgi protein transport. J Biol Chem. 2001;276:18178-18184. [PubMed] |
| 20. | Sönnichsen B, Lowe M, Levine T, Jämsä E, Dirac-Svejstrup B, Warren G. A role for giantin in docking COPI vesicles to Golgi membranes. J Cell Biol. 1998;140:1013-1021. [PubMed] |
| 21. | Alvarez C, Fujita H, Hubbard A, Sztul E. ER to Golgi transport: Requirement for p115 at a pre-Golgi VTC stage. J Cell Biol. 1999;147:1205-1222. [PubMed] |
| 22. | Kondylis V, Rabouille C. A novel role for dp115 in the organization of tER sites in Drosophila. J Cell Biol. 2003;162:185-198. [PubMed] |
| 23. | Calandra T, Bernhagen J, Mitchell RA, Bucala R. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J Exp Med. 1994;179:1895-1902. [PubMed] |
| 24. | Bacher M, Metz CN, Calandra T, Mayer K, Chesney J, Lohoff M, Gemsa D, Donnelly T, Bucala R. An essential regulatory role for macrophage migration inhibitory factor in T-cell activation. Proc Natl Acad Sci USA. 1996;93:7849-7854. [PubMed] |
| 25. | Miller EJ, Li J, Leng L, McDonald C, Atsumi T, Bucala R, Young LH. Macrophage migration inhibitory factor stimulates AMP-activated protein kinase in the ischaemic heart. Nature. 2008;451:578-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 322] [Cited by in RCA: 360] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 26. | Mizue Y, Ghani S, Leng L, McDonald C, Kong P, Baugh J, Lane SJ, Craft J, Nishihira J, Donnelly SC. Role for macrophage migration inhibitory factor in asthma. Proc Natl Acad Sci USA. 2005;102:14410-14415. [PubMed] |
| 27. | Radstake TR, Sweep FC, Welsing P, Franke B, Vermeulen SH, Geurts-Moespot A, Calandra T, Donn R, van Riel PL. Correlation of rheumatoid arthritis severity with the genetic functional variants and circulating levels of macrophage migration inhibitory factor. Arthritis Rheum. 2005;52:3020-3029. [PubMed] |
| 28. | Grigorenko EL, Han SS, Yrigollen CM, Leng L, Mizue Y, Anderson GM, Mulder EJ, de Bildt A, Minderaa RB, Volkmar FR. Macrophage migration inhibitory factor and autism spectrum disorders. Pediatrics. 2008;122:e438-e445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 50] [Reference Citation Analysis (0)] |
| 29. | Merk M, Baugh J, Zierow S, Leng L, Pal U, Lee SJ, Ebert AD, Mizue Y, Trent JO, Mitchell R. The Golgi-associated protein p115 mediates the secretion of macrophage migration inhibitory factor. J Immunol. 2009;182:6896-6906. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 104] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 30. | Bacher M, Meinhardt A, Lan HY, Mu W, Metz CN, Chesney JA, Calandra T, Gemsa D, Donnelly T, Atkins RC. Migration inhibitory factor expression in experimentally induced endotoxemia. Am J Pathol. 1997;150:235-246. [PubMed] |
| 31. | Fingerle-Rowson G, Koch P, Bikoff R, Lin X, Metz CN, Dhabhar FS, Meinhardt A, Bucala R. Regulation of macrophage migration inhibitory factor expression by glucocorticoids in vivo. Am J Pathol. 2003;162:47-56. [PubMed] |
| 32. | Santos LL, Morand EF. Macrophage migration inhibitory factor: a key cytokine in RA, SLE and atherosclerosis. Clin Chim Acta. 2009;399:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 82] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 33. | Knudsen ES, Buckmaster C, Chen TT, Feramisco JR, Wang JY. Inhibition of DNA synthesis by RB: effects on G1/S transition and S-phase progression. Genes Dev. 1998;12:2278-2292. [PubMed] |
| 34. | Strzalka W, Ziemienowicz A. Proliferating cell nuclear antigen (PCNA): a key factor in DNA replication and cell cycle regulation. Ann Bot. 2011;107:1127-1140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 416] [Cited by in RCA: 523] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 35. | Bach JP, Deuster O, Balzer-Geldsetzer M, Meyer B, Dodel R, Bacher M. The role of macrophage inhibitory factor in tumorigenesis and central nervous system tumors. Cancer. 2009;115:2031-2040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 61] [Article Influence: 3.8] [Reference Citation Analysis (0)] |