Published online Oct 7, 2013. doi: 10.3748/wjg.v19.i37.6178
Revised: August 10, 2013
Accepted: August 16, 2013
Published online: October 7, 2013
Processing time: 109 Days and 23.3 Hours
AIM: To investigate cytokeratin 8 (CK8) overexpression during hepatitis C virus (HCV) infection and its pathogenesis, and the effect of ectopic CK8 expression on hepatoma cell lines.
METHODS: We successfully established an in vitro HCV cell culture system (HCVcc) to investigate the different expression profiles of CK8 in Huh-7-HCV and Huh-7.5-HCV cells. The expression of CK8 at the mRNA level was determined by real-time polymerase chain reaction (RT-PCR). The expression of CK8 at the protein level was evaluated by Western blotting. We then constructed a eukaryotic expression combination vector containing the coding sequence of human full length CK8 gene. CK8 cDNA was amplified by reverse transcription-PCR and inserted into pEGFP-C1 and the positive clone pEGFP-CK8 was obtained. After confirming the sequence, the recombinant plasmid was transfected into SMMC7721 cells with lipofectamine2000 and CK8 expression was detected using inverted fluorescence microscopy, RT-PCR and Western blotting. Besides, we identified biological function of CK8 on SMMC7721 cells, including cell proliferation, cell cycle and apoptosis detection.
RESULTS: RT-PCR showed that the expression level of CK8 in Huh-7-HCV and Huh-7.5-HCV cells was 2.88 and 2.95 times higher than in control cells. Western blot showed that CK8 expression in Huh-7-HCV and Huh-7.5-HCV cells was 2.53 and 3.26 times higher than that in control cells, respectively. We found that CK8 at mRNA and protein levels were both significantly increased in HCVcc. CK8 was up-regulated in SMMC7721 cells. CK8 expression at the mRNA level was significantly upregulated in SMMC7721/pEGFP-CK8 cells. CK8 expression in SMMC7721/ pEGFP-CK8 cells was 2.69 times higher than in SMMC7721 cells, and was 2.64 times higher than in SMMC7721/pEGFP-C1 cells. CK8 expression at the protein level in SMMC7721/pEGFP-CK8 cells was 2.46 times higher than in SMMC7721 cells, and was 2.29 times higher than in SMMC7721/pEGFP-C1 cells. Further analysis demonstrated that forced expression of CK8 slowed cell growth and induced apoptosis of SMMC7721 cells.
CONCLUSION: CK8 up-regulation might have a functional role in HCV infection and pathogenesis, and could be a promising target for the treatment of HCV infection.
Core tip: In this study, we observed that cytokeratin 8 (CK8) levels are elevated in hepatitis C virus (HCV) cell culture system and its ectopic expression decreased the proliferation and induced apoptosis of SMMC7721 cells. CK8 up-regulation might have a functional role in HCV infection and pathogenesis, and could be a promising target for the treatment of HCV infection.
- Citation: Sun MZ, Dang SS, Wang WJ, Jia XL, Zhai S, Zhang X, Li M, Li YP, Xun M. Cytokeratin 8 is increased in hepatitis C virus cells and its ectopic expression induces apoptosis of SMMC7721 cells. World J Gastroenterol 2013; 19(37): 6178-6187
- URL: https://www.wjgnet.com/1007-9327/full/v19/i37/6178.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i37.6178
Hepatitis C virus (HCV) infection is a significant global healthcare burden[1]. Current estimation suggests that a minimum of 3% of the world’s population is chronically infected, with a prevalence of up to 170 million people[2,3]. However, the mechanism of HCV infection is not fully understood. Recently, the development of HCV replicon technology has accelerated the understanding of the mechanism underlying HCV infection[4,5]. It has been reported that there were more than 100 abnormal expression proteins in HCV infected cells and hepatitis C patients[6-10]. Studies determining the changes in protein expression associated with HCV infection will help elucidate host/virus interactions, and provide further insight to HCV pathogenesis.
Cytokeratin 8 (CK8) is the major component of the intermediate filament cytoskeleton, belonging to the type-II keratin, and is primarily expressed in the epithelia of liver, intestine, and exocrine pancreas[11,12]. CK8 plays a crucial role in maintaining the structural integrity and the mechanical properties of cells[13]. Recent studies have suggested that CK8 is involved in several liver diseases. CK8 knock-out mice develop liver hemorrhage and are more susceptible to liver injury[14,15]. Some variants of CK8 are associated with disease severity and progression in patients with chronic liver diseases[16,17]. Thus, we hypothesized that CK8 contributed to cellular pathological processes and the infection and pathogenesis of HCV, leading to liver injury and chronic liver diseases.
In this study, we established an in vitro HCV cell culture system (HCVcc) and investigated whether HCV affects CK8 levels. Simultaneously, we established eukaryotic expression recombination vector containing the full length coding sequence of CK8, then transfected into hepatoma cells in vitro and investigated the biological and functional role of CK8 in hepatoma cells.
Huh-7 and Huh-7.5 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% heat-inactivated fetal bovine serum, 0.1 mmol/L nonessential amino acids and 1 × penicillin-streptomycin-glutamine. Plasmid pFL-J6/JFH, containing a chimeric full length HCV genome, was kindly provided by Professor Charles M Rice from Rockefeller University. Plasmid pFL-J6/JFH, containing a single Xba I restriction site and T7 RNA polymerase start site, is the chimera of HCV J6 strain (5’-NCR-NS2) and JFH strain (NS3-3’-NCR). Subsequently, plasmid pFL-J6/JFH encoding the full length HCV chimeric genome was transcribed to HCV RNA in vitro. HCVcc was established by electroporation of HCV RNA into Huh-7 and Huh-7.5 cells.
Huh7 and Huh-7.5 were used as negative controls of HCVcc. Huh-7-HCV and Huh-7.5-HCV cells were maintained under the same condition as Huh-7 and Huh-7.5 cells. Cells were cultured in an incubator at 37 °C supplemented with 5% CO2. During the cell culture, the supernatant of cell culture was collected at 24, 48, 72 and 96 h after electroporation in order to determine the HCV copies. Quantitative real-time polymerase chain reaction (qRT-PCR) was used to determine HCV copy number. At approximately 72 h after transfection, cells were washed three times with 1 × phosphate-buffered saline (PBS) and then harvested. In addition, indirect immuno-fluorescence was used to observe the expression of HCV core protein. Mouse monoclonal HCV core protein antibody (Novus Biologicals, United States) was used as the primary antibody, and goat anti-mouse conjugated with Fluorescein Isothiocyanate (FITC) was used as the secondary antibody. The harvested cells were fixed with 3% glutaraldehyde at 4 °C for 24 h, then washed twice by 0.1 mol/L arsenic acid dimethyl sodium buffer (pH 7.4) at 4 °C, fixed by 1% osmium tetroxide for 1 h, gradient acetone dehydration, embedded by Epon812, sliced by ultra-thin LKB-V slicer. H-7650 transmission electron microscope (HITACHI, Japan) was also used to observe the morphology of the viral particles and intracellular ultrastructure changes.
Total RNA was extracted from cells by TRIzol reagent (Invitrogen, United States) according to the manufacturer’s protocol. A two-step reverse transcription PCR was performed. The first-strand cDNA was synthesized from 1 μg of total RNA with AMV Reverse Transcriptaseb (TAKARA, Japan). To investigate the expression of CK8 at the mRNA level, the expression of CK8 and glyceraldehydes-3-phosphate dehydrogenase (GAPDH) genes was quantified by RT-PCR, and GAPDH was used as an internal control. A total of 20 ng cDNA was used as template in the reaction. All RT-PCR assays were performed in triplicate using SYBR green incorporation method with Bio-Rad IQ5 Multicolor RT-PCR Detection System (Bio-Rad, United States) based on the manufacture’s protocol. Table 1 shows the sequences of the primer sets for CK8 and GAPDH. Briefly, following a denaturation at 95 °C for 5 s, RT-PCR was carried out with 50 cycles at a melting temperature of 95 °C for 30 s, an annealing temperature of 65 °C for 30 s, and an extension temperature of 72 °C for 10 s. Data analysis was performed using the Sequence Detector System software. The relative quantification was calculated by the 2-ΔΔCt method with GAPDH as the housekeeping gene and the control cells as the baseline, and the results were expressed as fold-change.
| Name | Forward primer (5’-3’) | Reverse primer (5’-3’) |
| CK8 (172 bp) | AGCTGGAGTCTCGCCTGGAA | TGTGCCTTGACCTCAGCAATG |
| GAPDH (138 bp) | GCACCGTCAAGGCTGAGAAC | TGGTGAAGACGCCAGTGGA |
| CK8 (1465 bp) | ATGTCGACATGTCCATCAGGGTGAC | TAGGATCCCTTGGGCAGGACGTC |
Total proteins were prepared by RIPA cell lysate. Proteins of interest were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) with a 10% polyacrylamide gel, and 1 mg/mL protein was loaded onto a SDS-PAGE gel. Proteins were transferred to nitrocellulose membranes and then detected by Western blotting under the recommended conditions. Mouse anti-human CK8 IgG (Abcam, United States) was used as the primary antibody, goat anti-mouse IgG conjugated with horseradish peroxidase (HRP) was used as the secondary antibody, and GAPDH was used as the control. The antigen-antibody complex was detected by an enhanced chemiluminescence (ECL) kit following the manufacturer’s protocol. The experiments were repeated in triplicate. The chemiluminescent signal of each band was analyzed by gel image analysis system (Syngene, United States).
The BamHI and SalI restriction sites were introduced into the CK8 coding sequence (CDS) by high fidelity PCR (Thermo, United States). Sequences for the primers are listed in Table 1, with the amplified product being 1465 bp. The CK8 CDS was purified by gel extraction. CK8 CDS and pEGFP-C1 vector (TAKARA, Japan) were digested respectively by the restriction enzyme SalI and BamHI (TAKARA, Japan). The digestion products were examined on 1% agarose gel by electrophoresis. The ligation reaction (Ligation Kit, TAKARA, Japan) was carried out between both of the DNA fragments, followed by transformation into competent Escherichia coli DH5α cells at 37 °C overnight (12-16 h). Colony selection was performed by PCR, and the amplicons were examined on 1% agarose gel by electrophoresis. Plasmid extraction (E.Z.N.A.® Endo-Free Plasmid Midi Kit, Omega, United States) was carried out for positive colonies, and then sequenced and matched by Blast method.
SMMC7721 cells were seeded in 6-well plates in 4 mL of growth medium for 24 h prior to transfection. In each well, 0.8 × 105-4.0 × 105 adherent cells were seeded. Four microgram (4.0 μg) of DNA (pEGFP-CK8 vector or pEGFP-C1 vector) was diluted in 250 μL of serum-free DMEM. Lipofectamine2000 (Millipore, United States) was added (10 μL) to the diluted DNA and mixed immediately by pipetting. The mixture was incubated for 25 min at room temperature. The lipofectamine2000/DNA mixture (500 μL) was added dropwise to the four wells containing the pEGFP-CK8 plasmid, and another two wells to control cells containing the pEGFP-C1 plasmid. The plate was then gently rocked to achieve even distribution of the complexes and incubated at 37 °C in a 5% CO2 incubator.
The expression and distribution of CK8 was observed under an inverted fluorescence microscope (Nikon eclipse Ti, Japan) 24 h after transfection. Forty-eight hours after transfection, cellular RNA and total cellular proteins were determined by RT-PCR and Western blotting, respectively. Total RNA was extracted from SMMC7721, SMMC7721/ pEGFP-C1, and SMMC7721/pEGFP-CK8 cells by TRIzol reagent. Total proteins were prepared by RIPA cell lysate. Real time PCR assays (SYBR®Premix Ex Taq™ II, TAKARA, Japan) were performed in triplicate with Bio-Rad iQ5 Multicolor RT-PCR Detection System according to the manufacture’s protocol. Rabbit anti-human IgG (Santa, United States) was used as the primary antibody, goat anti-rabbit IgG conjugated with HRP was used as the secondary antibody and β-actin (Abcam, United States) was used as control. Cells were collected after 24, 48 and 72 h transfection to perform a proliferation assay by MTT reaction (MTT cell proliferation Assay kit, Trevigen, United States). Cells were also collected 48 h after transfection to detect apoptosis (Annexin V-FITC Apoptosis Detection Kit, Abcam, United States) using Flow Cytometry (guava easyCyte HT, Millipore, United States).
All experiments were performed in triplicate. Representative graphical data are presented as mean ± SD. Statistical analyses were performed using the SPSS 10.0 software package (SPSS Inc.). We used Student’s t test. P values below 0.05 were considered to be significant.
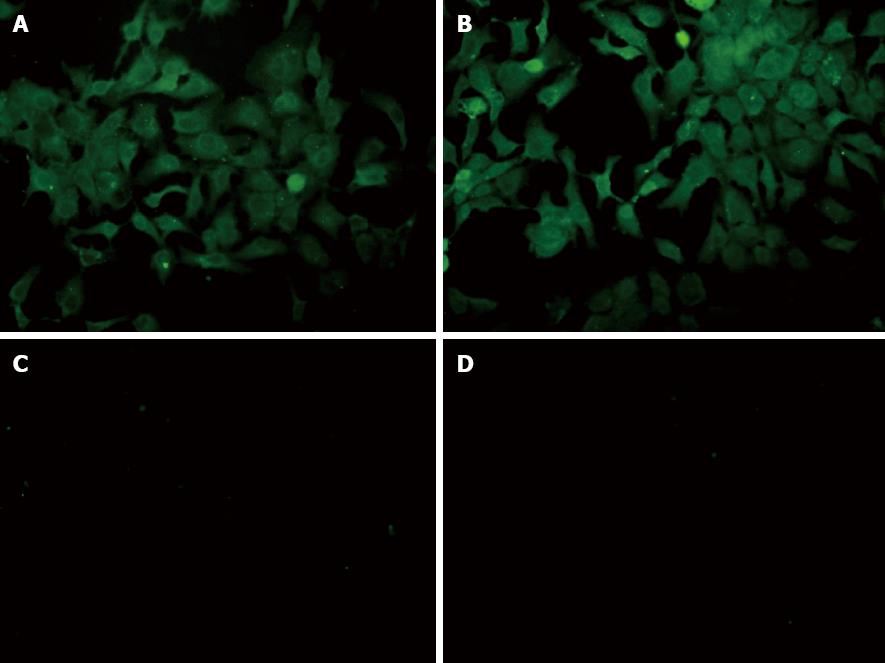
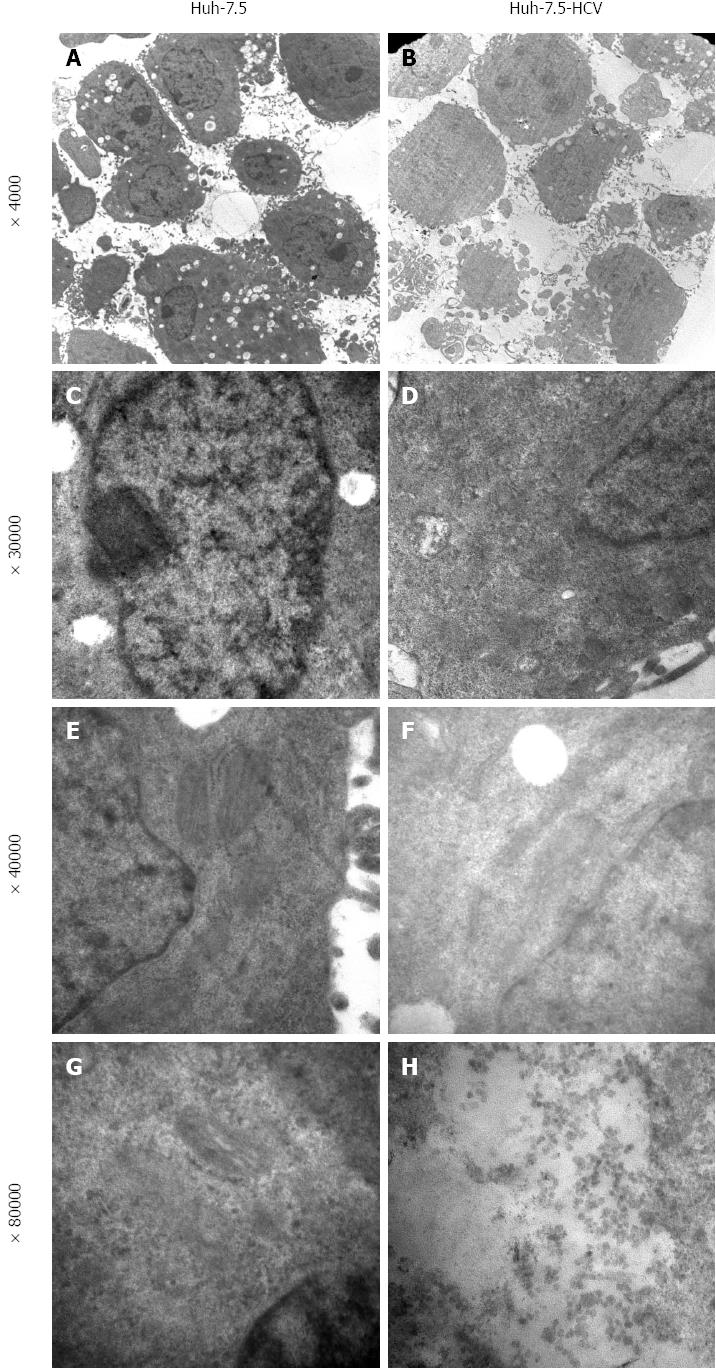
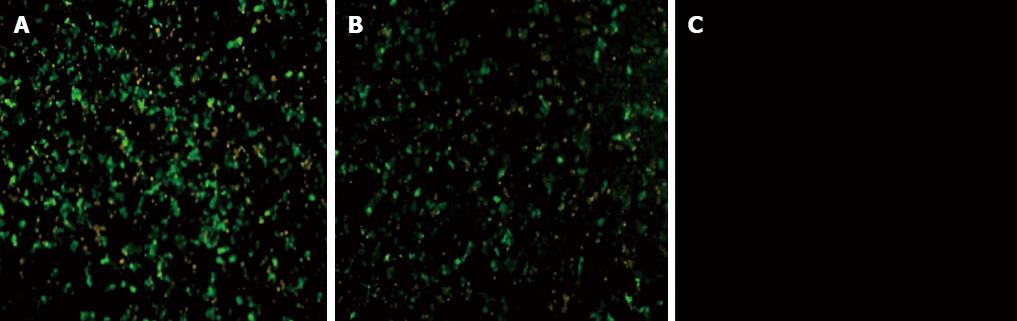
We determined HCV RNA copy number by performing qRT-PCR of viral supernatants obtained from HCV-transfected cells. High-level viral copies in the supernatant of transfected cells were observed at different time-points and reached its peak value at 48 h after transfection (Table 2). Indirect immuno-fluorescence also showed high expression of HCV core protein in the HCV-transfected cells. Huh-7-HCV and Huh-7.5 HCV cells were also labeled with GFP, further indicating that HCV core protein has been expressed in these cells compared to control cells (Figure 1). Transmission electron microscopy (TEM) revealed a large number of enveloped or unenveloped virus-like particles (VLPs) in HCVcc. Some characteristic structures of Flaviviridae virus infection were observed, including an increased number of endoplasmic reticulum, mitochondrial swelling, cristae disappearance, and cytoplasmic vacuolar structures. Also, a large number of HCV nucleocapsid-like particles of inclusion body were presented in HCVcc cells (Figure 2). Viral-like particles were not seen in the control cells. Moreover, hyperplasia, vacuolar membrane structure, and formation of inclusion bodies were not observed in the control cells.
| HCV RNA in supernatant of Huh-7-HCV cells | HCV RNA in supernatant of Huh-7.5-HCV cells | |
| 24 h | 5.73 × 105 | 9.48 × 105 |
| 48 h | 1.38 × 106 | 6.40 × 106 |
| 72 h | 3.00 × 104 | 9.29 × 104 |
| 96 h | 6.62 × 103 | 1.43 × 104 |
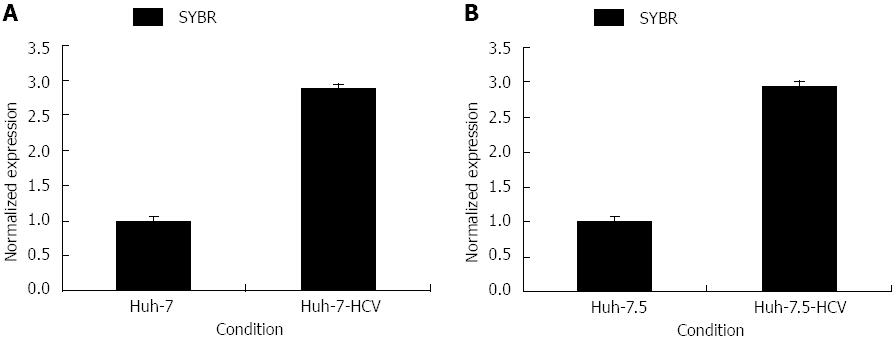
Extracted total cellular RNA was examined by electrophoresis on a 0.8% non-denaturing agarose gel. A 172 bp fragment of CK8 was successfully amplified by PCR without unspecific amplification. The melting and amplification curves of CK8 expression indicated that the primers were properly designed. CK8 expression in Huh-7-HCV cells was 2.88 times higher than that in Huh-7 cells, and CK8 expression in Huh-7.5-HCV cells was 2.95 times higher than that in Huh-7.5 cells (Figure 3). Therefore, CK8 was significantly highly expressed in HCVcc cells.
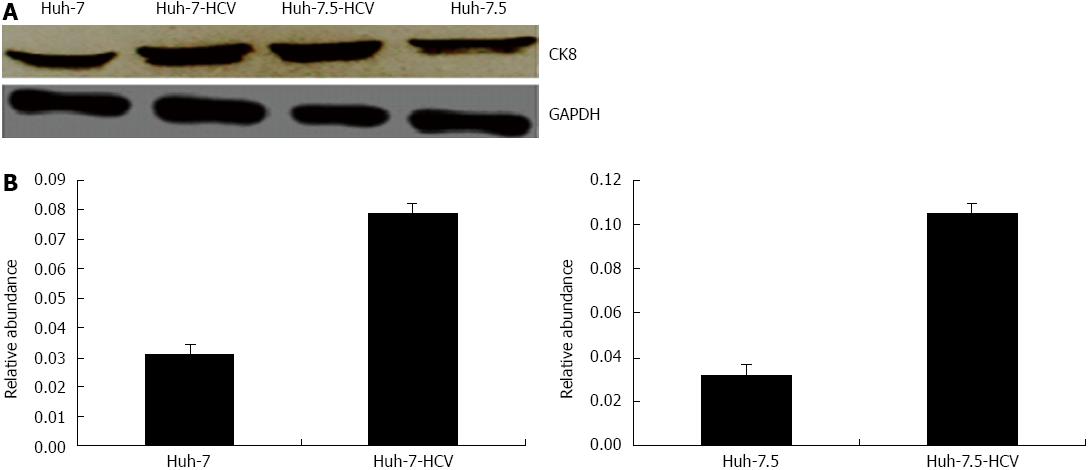
By Western blotting, we showed that the ratio of CK8/GAPDH was 0.079 ± 0.004 and 0.031 ± 0.003 in Huh-7-HCV cells and Huh-7 cells, respectively, which was 2.53 times higher. Furthermore, the ratio of CK8/GAPDH was 0.105 ± 0.004 in Huh-7.5-HCV cells, which was significantly higher than in Huh-7.5 cells (0.032 ± 0.002) and expression was 3.26 times higher (Figure 4). Therefore, we confirmed that HCVcc cells do have increased CK8 expression.
We next ectopically expressed CK8 in SMMC7721 cells. We confirmed the overexpression of CK8 in cells under inverted fluorescence microscope 24 h after transfection. Since the CK8 expression vector contains an EGFP marker, we observed that SMMC7721/pEGFP-C1 and SMMC7721/pEGFP-CK8 cells appeared bright green compared to control SMMC7721 cells (Figure 5). This data indicated that ectopic expression of CK8 was achieved in SMMC7721 cells.
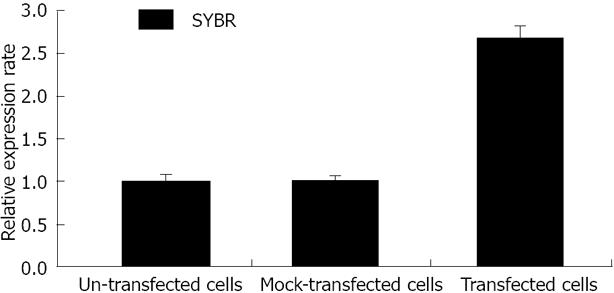
The 2-ΔΔCt value of CK8 mRNA levels in SMMC7721, SMMC7721/pEGFP-C1, and SMMC7721/pEGFP-CK8 cells are shown in Figure 6. Beta-actin was used as the housekeeping gene, while SMMC7721 cells were used for baseline detection. The results were expressed as fold-change. CK8 expression at the mRNA level was significantly upregulated in SMMC7721/pEGFP-CK8 cells. CK8 expression in SMMC7721/pEGFP-CK8 cells was 2.69 times higher than in SMMC7721 cells, and was 2.64 times higher than in SMMC7721/pEGFP-C1 cells.
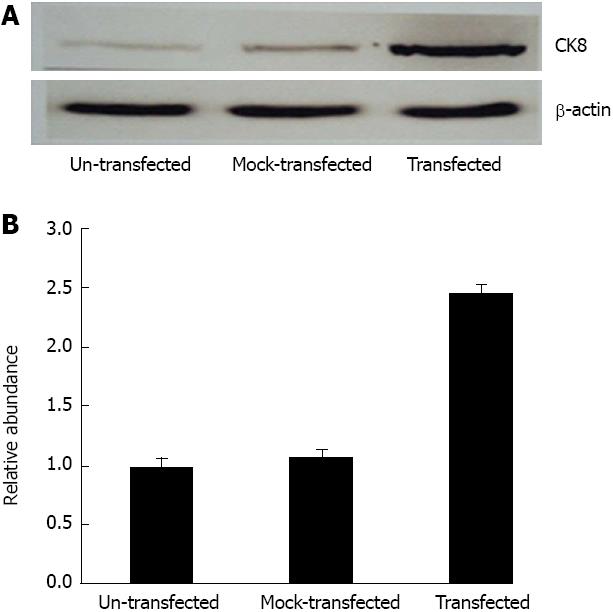
Using Western blotting, we compared the chemiluminescent signals of CK8 and β-actin in SMMC7721, SMMC7721/pEGFP-C1, and SMMC7721/pEGFP-CK8 cells. The ratio between CK8 and β-actin were reflective changes in CK8 expression. CK8 expression in SMMC7721/pEGFP-CK8 cells was 2.46 times higher than in SMMC7721 cells, and 2.29 times higher than in SMMC7721/pEGFP-C1 cells. This demonstrated that ectopic expression of CK8 was observed at the protein level in SMMC7721/pEGFP-CK8 cells (Figure 7). Therefore, we confirmed that CK8 expression was increased in SMMC7721 cells after transfection with pEGFP-CK8 vector.
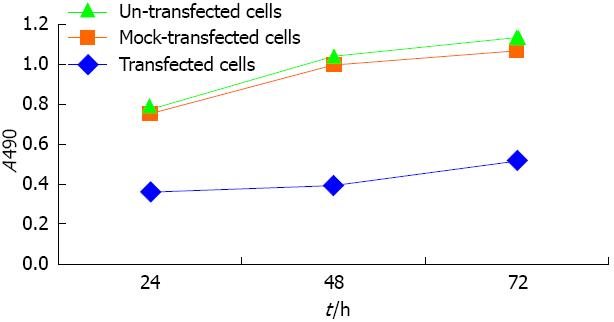
Using MTT detection, we determined the effects of ectopic CK8 expression on SMMC7721 cells 72 h after transfection. CK8 overexpression decreased the growth and proliferation of SMMC7721 cells compared to control cells and mock-transfected cells (Figure 8). This data indicated that ectopic CK8 expression decreased cell growth and proliferation of SMMC7721 cells.
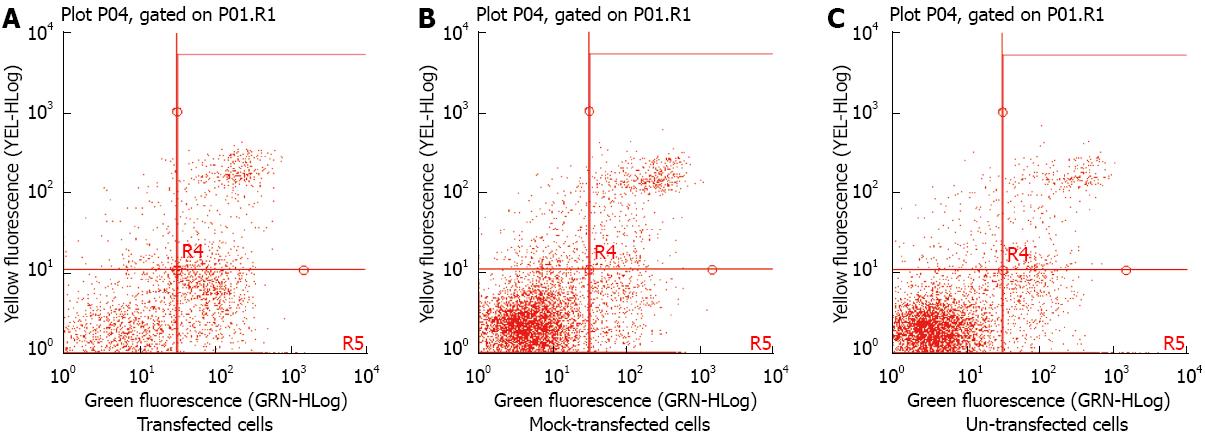
We determined the effects of ectopic CK8 expression on the apoptosis of SMMC7721 cells 48 h after transfection. Using flow cytometry, ectopic CK8 expression increased the apoptotic rate of SMMC7721 cells, compared to un-transfected and mock-transfected cells (Figure 9).
In this study, we established a full-length HCV genomic replication in Huh-7 and Huh-7.5 cells. Lohmann et al[18] reported that subgenomic HCV RNA replicons are capable of autonomously replicating in Huh7 cells. These di-cistronic replicons include the neomycin-resistant gene, making them selectable by G418, and most or all of the viral nonstructural genes[19,20]. This system provides a novel and powerful tool for the study of HCV replication mechanisms and for study of the interaction between host and viral factors involved in viral progression[21,22]. In our study, we transfected Huh-7 and Huh-7.5 cells to express HCV RNA and generated the HCVcc cell line. We used qRT-PCR, immunofluorescence, and TEM to detect HCV RNA, HCV core protein, and HCV particles, respectively. The results confirmed that HCV expression in Huh-7 and Huh-7.5 cells led to the production of HCV particles.
CK8 is a cytoskeletal intermediate filament protein that abundantly expresses in hepatocytes to maintain cell integrity, and prevent mechanical and non-mechanical cell injury[23,24]. Previous studies showed that CK8 was upregulated in HBV-infected liver tissues fromp21-HBx mice[25], and that its upregulation contributed to the development and progression of HCC-induced HBV. Tai DI found that CK8 was focally positive in a patient with a malignant liver patient infected with HCV[26]. Toivola et al[17] found that in chronic HCV infection, CK8 phosphorylation is a progression marker during HCV progression and regression. Furthermore, Strnad et al[27] found that a number of CK8 gene variants are increased in patients with chronic HCV infection. However, it is unclear about the relation between CK8 expression and HCVcc cells. We observed a concomitant increase in CK8 levels, which was confirmed by RT-PCR and Western blot analysis. CK8 mRNA expression in Huh-7-HCV and Huh-7.5-HCV cells was 2.88 and 2.95 times higher than in Huh-7 and Huh-7.5 cells, respectively. At the protein level, CK8 expression was 2.53 and 3.26 times higher in Huh-7-HCV and Huh-7.5-HCV cells, respectively, than Huh-7 and Huh-7.5 cells. This suggests that HCV up-regulates CK8 expression in HCVcc cells, and that CK8 expression is significantly associated with HCV.
CK8 plays a role in maintaining cellular structural integrity, signal transduction, and cellular differentiation[28-32]. Snider NT demonstrated that acetylation of CK8 was up-regulated in diabetic human livers[33]. We showed that HCV up-regulates CK8 expression in HCVcc cells. However, the biological function of ectopic CK8 in tumor cells is not fully elucidated. To further investigate the biological function of aberrant CK8 expression, we cloned the full length CDS of CK8 to establish the eukaryotic expression recombination vector pEGFP-CK8. To study the biological function of increased CK8 on cells independently, we chose another cell line called SMMC7721 cells in our laboratory. SMMC7721 cells were transfected by pEGFP-CK8 recombination vector, and under an inverted fluorescence microscope we observed the expression and distribution of GFP-tagged CK8. In addition, by RT-PCR and Western blot analysis, we found that CK8 mRNA levels in SMMC7721/pEGFP-CK8 cells was 2.69 and 2.64 times higher than in SMMC7721 cells and SMMC7721/pEGFP-C1 cells, respectively. At the protein level, CK8 expression in SMMC7721/pEGFP-CK8 cells was 2.46 and 2.29 times higher than in SMMC7721 and SMMC7721/pEGFP-C1 cells, respectively. These observations showed that CK8 gene was transcribed and expressed in SMMC7721 cells.
CK8 abnormal expression and mutations can lead to acute or sub-acute liver injury and promote tumor cells apoptosis[34,35]. The persistent expression of CK8 can induce tumor cell apoptosis through a number of transcription factors that regulate a large number of oncogenes[36]. In SMMC7721 transfected by pEGFP-CK8, we further observed the biological effects of increased CK8 on cells. We detected proliferation and apoptosis by MTT reaction and flow cytometry, respectively. We found that ectopic CK8 expression decreased cell growth and proliferation, and increased apoptosis of SMMC7721 cells. Therefore, we concluded that the abnormal expression of CK8 regulates cellular pathological injury. However, it is unclear what the mechanisms are by which CK8 affects cell cycle and apoptosis. In conclusion, these results suggest CK8 up-regulation might have a functional role during HCV infection and pathogenesis, and it could be a promising target for the treatment of HCV infection.
In summary, we successfully established and identified HCVcc and observed that CK8 is upregulated in HCVcc. Overexpression of CK8 in SMMC7721 cells inhibited cell proliferation and induced apoptosis. CK8 could be a potential target for the treatment of HCV infection. Future studies will (1) identify the interactions of CK8 with other proteins to mediate its effects; (2) assess how CK8 expression regulates a number of known oncogenes in HCV; and (3) determine how CK8 promotes apoptosis.
Currently, several proteins have been identified to be overexpressed during hepatitis C virus (HCV) infection and pathogenesis. Studies have suggested that cytokeratin 8 (CK8) is closely related to a number of liver diseases. CK8 knock-out mice develop liver hemorrhage and are more susceptible to liver injury. However, it remains unknown whether HCV affects CK8 levels in their established in vitro HCV cell culture system (HCVcc) and the biological and functional role of CK8 in hepatoma cells.
It has been reported that there are more than 100 abnormal proteins expressed in HCV-infected cells and hepatitis C patients. Studies determining the changes in protein expression associated with HCV infection will help to elucidate host/virus interactions, and provide further insight to HCV pathogenesis. CK8 plays a crucial role in maintaining the structural integrity and the mechanical properties of cells. Recent studies have suggested that CK8 is involved in several liver diseases. Much interest is shown to understand CK8 overexpression during HCV infection and to investigate the role of ectopic CK8 expression in hepatoma cell lines.
In this study, the authors transfected Huh-7 and Huh-7.5 cells to express HCV RNA and generated the HCVcc cell line. Previous studies showed that CK8 is upregulated in HBV-infected liver tissues from p21-HBx mice and in a patient with a malignant liver infected with HCV. However, it is unclear what the relation between CK8 expression and HCVcc cells is. The authors observed a concomitant increase in CK8 levels by real-time Polymerase chain reaction and Western blot analysis. The results show that HCV up-regulates CK8 expression in HCVcc cells. However, the biological function of ectopic CK8 in tumor cells is not fully elucidated. The authors found that ectopic CK8 expression decreased cell growth and proliferation, and increased apoptosis of SMMC7721 cells. Therefore, the authors concluded that the abnormal expression of CK8 regulates cellular pathological injury.
The results of this study suggest that CK8 up-regulation might have a functional role during HCV infection and pathogenesis, and it could be a promising target for the treatment of HCV infection.
This is a very well written manuscript. In this paper, the authors show the over-expression of CK8 in an in vitro HCV cell culture system. Large-scale proteome analyses of the in vitro HCV infection model have also been performed. Thus new hopes characterize the HCV field and new advances are reasonably expected. Here, CK8 is found up-regulated in Huh7 and Huh7.5 cells infected with chimeric full length HCV genome. The methodology is acceptable. The conclusions are interesting.
P- Reviewer AlricL A S- Editor Qi Y L- Editor Ma JY E- Editor Ma S
| 1. | Park J, Kang W, Ryu SW, Kim WI, Chang DY, Lee DH, Park do Y, Choi YH, Choi K, Shin EC. Hepatitis C virus infection enhances TNFα-induced cell death via suppression of NF-κB. Hepatology. 2012;56:831-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 2. | Mehrab-Mohseni M, Sendi H, Steuerwald N, Ghosh S, Schrum LW, Bonkovsky HL. Legalon-SIL downregulates HCV core and NS5A in human hepatocytes expressing full-length HCV. World J Gastroenterol. 2011;17:1694-1700. [PubMed] [DOI] [Full Text] |
| 3. | Wang YZ, Wang WB, Cao MM, Wang W, Zhao LJ, Xu G, Ren H, Qi ZT. Function of nonstructural 5A protein of genotype 2a in replication and infection of HCV with gene substitution. World J Gastroenterol. 2011;17:3398-3406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 4. | Banaudha K, Orenstein JM, Korolnek T, St Laurent GC, Wakita T, Kumar A. Primary hepatocyte culture supports hepatitis C virus replication: a model for infection-associated hepatocarcinogenesis. Hepatology. 2010;51:1922-1932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 5. | Boonstra A, van der Laan LJ, Vanwolleghem T, Janssen HL. Experimental models for hepatitis C viral infection. Hepatology. 2009;50:1646-1655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 69] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 6. | Schaller T, Appel N, Koutsoudakis G, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J Virol. 2007;81:4591-4603. [PubMed] |
| 7. | Sogawa K, Noda K, Umemura H, Seimiya M, Kuga T, Tomonaga T, Nishimura M, Kanai F, Imazeki F, Takizawa H. Serum fibrinogen alpha C-chain 5.9 kDa fragment as a biomarker for early detection of hepatic fibrosis related to hepatitis C virus. Proteomics Clin Appl. 2013;7:424-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 8. | Tripathi LP, Kambara H, Moriishi K, Morita E, Abe T, Mori Y, Chen YA, Matsuura Y, Mizuguchi K. Proteomic analysis of hepatitis C virus (HCV) core protein transfection and host regulator PA28γ knockout in HCV pathogenesis: a network-based study. J Proteome Res. 2012;11:3664-3679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Imakiire K, Uto H, Sato Y, Sasaki F, Mawatari S, Ido A, Shimoda K, Hayashi K, Stuver SO, Ito Y. Difference in serum complement component C4a levels between hepatitis C virus carriers with persistently normal alanine aminotransferase levels or chronic hepatitis C. Mol Med Rep. 2012;6:259-264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 10. | Qin S, Zhou Y, Lok AS, Tsodikov A, Yan X, Gray L, Yuan M, Moritz RL, Galas D, Omenn GS. SRM targeted proteomics in search for biomarkers of HCV-induced progression of fibrosis to cirrhosis in HALT-C patients. Proteomics. 2012;12:1244-1252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 11. | Fuchs E, Weber K. Intermediate filaments: structure, dynamics, function, and disease. Annu Rev Biochem. 1994;63:345-382. [PubMed] |
| 12. | Coulombe PA, Omary MB. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr Opin Cell Biol. 2002;14:110-122. [PubMed] |
| 13. | Busch T, Armacki M, Eiseler T, Joodi G, Temme C, Jansen J, von Wichert G, Omary MB, Spatz J, Seufferlein T. Keratin 8 phosphorylation regulates keratin reorganization and migration of epithelial tumor cells. J Cell Sci. 2012;125:2148-2159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 14. | Baribault H, Price J, Miyai K, Oshima RG. Mid-gestational lethality in mice lacking keratin 8. Genes Dev. 1993;7:1191-1202. [PubMed] |
| 15. | Loranger A, Duclos S, Grenier A, Price J, Wilson-Heiner M, Baribault H, Marceau N. Simple epithelium keratins are required for maintenance of hepatocyte integrity. Am J Pathol. 1997;151:1673-1683. [PubMed] |
| 16. | Zhong B, Strnad P, Selmi C, Invernizzi P, Tao GZ, Caleffi A, Chen M, Bianchi I, Podda M, Pietrangelo A. Keratin variants are overrepresented in primary biliary cirrhosis and associate with disease severity. Hepatology. 2009;50:546-554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 17. | Toivola DM, Ku NO, Resurreccion EZ, Nelson DR, Wright TL, Omary MB. Keratin 8 and 18 hyperphosphorylation is a marker of progression of human liver disease. Hepatology. 2004;40:459-466. [PubMed] |
| 18. | Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110-113. [PubMed] |
| 19. | Shahid I, Gull S, Ijaz B, Ahmad W, Ansar M, Asad S, Kausar H, Sarwar MT, Khan MK, Hassan S. Stable Huh-7 cell lines expressing non-structural proteins of genotype 1a of hepatitis C virus. J Virol Methods. 2013;189:65-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 20. | Butt S, Idrees M, Rehman IU, Ali L, Hussain A, Ali M, Ahmed N, Saleem S, Fayyaz M. Establishment of stable Huh-7 cell lines expressing various hepatitis C virus genotype 3a protein: an in-vitro testing system for novel anti-HCV drugs. Genet Vaccines Ther. 2011;9:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 21. | Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125:1808-1817. [PubMed] |
| 22. | Dang SS, Sun MZ, Yang E, Xun M, Ma L, Jia ZS, Wang WJ, Jia XL. Prohibitin is overexpressed in Huh-7-HCV and Huh-7.5-HCV cells harboring in vitro transcribed full-length hepatitis C virus RNA. Virol J. 2011;8:424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 23. | Sun Z, Wang HQ, Liu ZH, Chang L, Chen YF, Zhang YZ, Zhang WL, Gao Y, Wan ZY, Che L. Down-regulated CK8 expression in human intervertebral disc degeneration. Int J Med Sci. 2013;10:948-956. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 24. | Andratschke M, Luebbers CW, Johannson V, Schmitt B, Mack B, Zeidler R, Lang S, Wollenberg B, Gildehaus FJ. Biodistribution of 131I-labeled anti-CK8 monoclonal antibody in HNSCC in xenotransplanted SCID mice. Anticancer Res. 2011;31:3315-3321. [PubMed] |
| 25. | Sun Q, Wang Y, Zhang Y, Liu F, Cheng X, Hou N, Zhao X, Yang X. Expression profiling reveals dysregulation of cellular cytoskeletal genes in HBx-induced hepatocarcinogenesis. Cancer Biol Ther. 2007;6:668-674. [PubMed] |
| 26. | Tai DI, Shen YJ, Weng WH, Chen HW, Kang CC, Chen TC, Liao SK. New sarcomatoid cancer cell line SAR-HCV established from a hepatitis C virus-related liver tumour lesion. Anticancer Res. 2011;31:129-137. [PubMed] |
| 27. | Strnad P, Lienau TC, Tao GZ, Lazzeroni LC, Stickel F, Schuppan D, Omary MB. Keratin variants associate with progression of fibrosis during chronic hepatitis C infection. Hepatology. 2006;43:1354-1363. [PubMed] |
| 28. | Fortier AM, Asselin E, Cadrin M. Keratin 8 and 18 loss in epithelial cancer cells increases collective cell migration and cisplatin sensitivity through claudin1 up-regulation. J Biol Chem. 2013;288:11555-11571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 175] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 29. | Iyer SV, Dange PP, Alam H, Sawant SS, Ingle AD, Borges AM, Shirsat NV, Dalal SN, Vaidya MM. Understanding the role of keratins 8 and 18 in neoplastic potential of breast cancer derived cell lines. PLoS One. 2013;8:e53532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 30. | Wang Y, Zhu JF, Liu YY, Han GP. An analysis of cyclin D1, cytokeratin 5/6 and cytokeratin 8/18 expression in breast papillomas and papillary carcinomas. Diagn Pathol. 2013;8:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 31. | Mathew J, Loranger A, Gilbert S, Faure R, Marceau N. Keratin 8/18 regulation of glucose metabolism in normal versus cancerous hepatic cells through differential modulation of hexokinase status and insulin signaling. Exp Cell Res. 2013;319:474-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 32. | Usachov V, Nahon P, Lunova M, Ziol M, Rufat P, Sutton A, Beaugrand M, Strnad P. Keratin 8 variants are infrequent in patients with alcohol-related liver cirrhosis and do not associate with development of hepatocellular carcinoma. BMC Gastroenterol. 2012;12:147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 33. | Snider NT, Leonard JM, Kwan R, Griggs NW, Rui L, Omary MB. Glucose and SIRT2 reciprocally mediate the regulation of keratin 8 by lysine acetylation. J Cell Biol. 2013;200:241-247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 34. | Ku NO, Strnad P, Zhong BH, Tao GZ, Omary MB. Keratins let liver live: Mutations predispose to liver disease and crosslinking generates Mallory-Denk bodies. Hepatology. 2007;46:1639-1649. [PubMed] |
| 35. | Wang Y, He QY, Tsao SW, Cheung YH, Wong A, Chiu JF. Cytokeratin 8 silencing in human nasopharyngeal carcinoma cells leads to cisplatin sensitization. Cancer Lett. 2008;265:188-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 36. | Oshima RG, Baribault H, Caulín C. Oncogenic regulation and function of keratins 8 and 18. Cancer Metastasis Rev. 1996;15:445-471. [PubMed] |