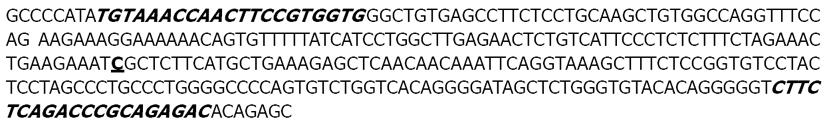Published online Jun 21, 2013. doi: 10.3748/wjg.v19.i23.3711
Revised: April 16, 2013
Accepted: May 16, 2013
Published online: June 21, 2013
Processing time: 96 Days and 18.2 Hours
In a recent study, Rafiei et al, reported a link between a C150T polymorphism in the human inducible nitric oxide gene and Helicobacter pylori infection as a risk factor for gastric cancer among an Iranian population. Subsequently, Abadi and Kusters published a letter to the editor questioning the validity of the study because of a supposed flaw in primer design. Here we respond to the claims of Abadi and Kusters and show that the results reported in the original article are valid.
Core tip: In a recent Letter to the Editor, Abadi and Kusters brought into question the validity of a study published by Rafiei et al. Herein we respond to the claims made by Abadi and Kusters, and show that the results reported in the article originally published by Rafiei et al, are valid.
-
Citation: Rafiei A, Gilbreath JJ, Merrell DS. Response to Abadi and Kusters,
World J Gastroenterol 19: 429-430. World J Gastroenterol 2013; 19(23): 3711-3712 - URL: https://www.wjgnet.com/1007-9327/full/v19/i23/3711.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i23.3711
In a recent letter to the editor, entitled “Association of inducible nitric oxide synthetase genotype and Helicobacter pylori infection gastric cancer risk may be due to faulty primer design”[1], Amin Talebi Bezmin Abadi and Johannes Kusters questioned the validity of the results published in our manuscript[2], the design of which was based on a previous study[3]. In their letter[1], the authors appraised our article[2] by two main points; variation in T allele frequency and primer design. In response to their first comment regarding the T allele frequency seen in our population, we stand by the conclusion that genetic polymorphisms depend on ethnic differences; the frequency of mutant genotypes varies in different human populations. Therefore, it is likely no surprise that the frequency of the T allele seen in our Caucasian population[2] differed from that of the East Asian population[3].
In regards to the second point made by Abadi et al[1], concerning the primer specificity, we note that there is a misprint in one primer sequence in our article[2]. The primer sequence published in our paper (5’-GTCTCTG-CGGGTCTGAAG-3’) differs from that of Shen et al[3] in that it is missing two base pairs in the 3’ end of the sequence. An erratum has been submitted to the journal to note the misprinted primer sequence [On page 4919[2], the primer sequence given for iNOS-R (5’-174 GTCTCTGCGGGTCTGAAG-3’) is missing two nucleotides and should appear as iNOS-R (5'-GTCTCTGCGGGTCTGAGAAG-3’)]. However, this 2 base misprint appears not to be the core issue since the correct primer sequence (5’-GTCTCTGCGGGTCTGAGAAG-3’) was previously published by Shen et al[3] and this publication was also called into question by Abadi and Kusters. We note that as shown in Figure 1, both of the primers used in the study are specific to exon 16 of the iNOS gene (Figure 1), which has been mapped to chromosome 17q11.2. Direct sequence analysis of exon 16 as submitted to GenBank by Shen et al[3] (http://www.ncbi.nlm.nih.gov/nuccore/x85772) shows that the primers used in our studies are completely conserved in this sequence. Furthermore, blast analysis of the inducible nitric oxidase synthase against the human database returns 3 hits to Homo sapiens chromosome 17. Analysis of the resulting alignments shows complete conservation of the forward primer in all 3 samples. Conservation of the reverse primer is not as high; the 5’ end of the primer shows several mismatches. However, the last 9 nucleotides found in the 3’ end of the primer are completely conserved across all 3 samples. Thus, there should be no overt obstacles to amplification of the gene in question. In conclusion, we stand by the results reported in our original study[2] and posit that the supposition by Abadi and Kusters that our study design is flawed is incorrect.
P- Reviewers Iera E, Naito Y S- Editor Gou SX L- Editor A E- Editor Zhang DN
| 1. | Abadi AT, Kusters JG. Association of inducible nitric oxide synthetase genotype and Helicobacter pylori infection gastric cancer risk may be due to faulty primer design. World J Gastroenterol. 2013;19:429-430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 1] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 2. | Rafiei A, Hosseini V, Janbabai G, Fazli B, Ajami A, Hosseini-Khah Z, Gilbreath J, Merrell DS. Inducible nitric oxide synthetase genotype and Helicobacter pylori infection affect gastric cancer risk. World J Gastroenterol. 2012;18:4917-4924. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 3. | Shen J, Wang RT, Wang LW, Xu YC, Wang XR. A novel genetic polymorphism of inducible nitric oxide synthase is associated with an increased risk of gastric cancer. World J Gastroenterol. 2004;10:3278-3283. [PubMed] |