Published online Jan 14, 2013. doi: 10.3748/wjg.v19.i2.290
Revised: April 28, 2012
Accepted: May 5, 2012
Published online: January 14, 2013
AIM: To test whether the status of positive cytomegalovirus (CMV) DNA detection adds to the predictive value of IL28B and to further categorize C/T allele carriers.
METHODS: This study included 166 chronic hepatitis C (CHC) patients who received combined interferon and ribavirin therapy for 48 wk, 84 spontaneous hepatitis C virus (HCV) resolvers who were positive for IgG anti-HCV antibody and negative for HCV RNA, and 100 healthy subjects who were negative for both HCV antibodies and RNA as controls. Genomic DNA from peripheral blood was used for IL28B rs.12979860 single nucleotide polymorphism (SNP) and CMV DNA detection. A 139 bp fragment containing IL28B SNP was amplified in all subjects by polymerase chain reaction using a specifically designed primer. Then the IL28B rs.12979860 SNP was detected by restriction fragment length polymorphism (RFLP) genotyping. The presence of CMV DNA was tested by amplification of the gB1 gene using nested polymerase chain reaction. The role of CMV and IL28B rs.12979860 SNP genotypes in determining the response rate to combined interferon therapy and clinical status of patients were statistically analyzed.
RESULTS: Current data showed that 67% of patients carrying the IL28B 12979860 C/C allele had a sustained viral response (SVR) while the genotypes C/T and TT were associated with lower SVR rates, 50% and 48%, respectively. SVR rates for the C/C allele were lower than other HCV genotypes and/or other populations. Genotype CC was associated with the response to interferon (P = 0.025). Genotype C/C was reduced from 48% in controls to 14% in CHC patients suggesting its protective role against progression to chronicity. The majority of spontaneously cleared subjects (86%) were C/C, confirming its protective role. The C/T allele was present in 71% of CHC patients compared with 38% of controls, so the use of IL28B SNP genotyping only in these patients may be of little value as a predictor of response. CMV reactivation occurred in 40% of CHC patients. Co-infection with CMV seriously diminished the response to interferon (IFN) therapy, with SVR rates in C/C genotypes 87.5% in CMV-negative patients and 12.5% in CMV-positive patients (P < 0.0001). SVR rates among C/T carriers were reduced to < 50% in patients with positive CMV DNA while the non-response rate doubled. These data indicate that a supplemental assay for CMV viremia adds to the prognostic value of IL28B genotyping.
CONCLUSION: The results suggest that both genetic (i.e., spontaneous) and therapeutic (IFN-based therapy) arms are complementary in the battle against HCV. CMV DNA testing may be of value to better predict the response to IFN, particularly in IL28B C/T carriers.
- Citation: El Awady MK, Bader El Din NG, Tabll A, El Hosary Y, Abdel Aziz AO, El Khayat H, Salama M, Abdelhafez TH. IL28B polymorphism and cytomegalovirus predict response to treatment in Egyptian HCV type 4 patients. World J Gastroenterol 2013; 19(2): 290-298
- URL: https://www.wjgnet.com/1007-9327/full/v19/i2/290.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i2.290
The current standard-of-care for chronic hepatitis C virus (HCV) infection includes weekly injections of pegylated interferon-α (peg-IFN) combined with daily oral ribavirin[1,2] for 48 wk[3]. During the first 3 mo of therapy, HCV viral loads usually fall to undetectable levels in response to interferon-α. However, in non-responder (NR) patients, HCV viral loads persist at or near pretreatment levels. Among those patients with an initial response to treatment, known as early virological responders, up to 50% will relapse after treatment is discontinued and are known as relapsed responders, whereas the remainder will have a sustained virological response (SVR) as determined by the absence of detectable viremia 6 mo after treatment has stopped[4]. In Egypt, several reports stated that approximately 50% of patients infected with genotype 4, the most common among Egyptian HCV patients, achieve a SVR to this regimen. Genome-wide association studies have recently revealed that single nucleotide polymorphisms (SNPs) within or adjacent to IL28B (19q13) that codes for interferon-λ3, predict spontaneous resolution of HCV[5,6] and a likely SVR to IFN-based treatment in chronic hepatitis C (CHC) patients infected with genotypes other than type 4[4,5,7-9]. Therefore, IL28B SNPs may have strong predictive value for the outcome of IFN-based therapy in the difficult to treat HCV genotype 4 patients, with the hope that IL28B SNPs will improve future decision-making in the management of CHC in Egypt. Nevertheless, it was estimated that IL28B variations account for about 15% of the inter-individual variability of SVR, thus supporting the necessity for additional predictor(s) of the response to treatment. Recent predictors include vitamin D deficiency, IFN-c-inducible protein-10 serum levels, steatosis/insulin resistance[10-17], or IFN-stimulated genes (ISGs) such as OAS1[18] and MxA[19]. Therefore, biological models for the prediction of SVR should include a variety of parameters besides IL28B genotyping, as this would offer better accuracy compared with IL28B typing alone[13]. Recently two predictors for SVR in genotype 4 patients were reported; first, reactivation of CMV infection[20] and second, SNP at exon 7 splice acceptor site of 2’ 5’OAS gene[18]. The role of several genetic factors that influence spontaneous or treatment-induced HCV clearance, such as ISGs and genes encoding the natural killer cell receptor KIR2DL3 and its ligand, and human leukocyte antigen C group 1[21-23] should be re-analyzed parallel with the IL28 genotype. A major difficulty in decision-making for prediction of the response at the start of treatment is the high frequency of heterozygous carriers rs 12979860 C/T among chronic HCV patients, ranging from 45% to 70% of the CHC populations and provide vague predictive results. This means that the predictive value of rs 12979860 SNP will not be useful in almost half the patient population. The search for additional predictive factors for response is mandatory. Co-infection with other pathogens is, in some instances, an interfering factor against host genotype-based prediction. Several studies revealed comparable associations between IL28B variations and treatment-induced HCV clearance in patients co-infected with human immunodeficiency virus (HIV)[5,24-26]. For example, one study in HIV co-infected patients reported 2-3-fold higher SVR rates for HCV genotypes 1 and 4 according to rs12979860 C/C vs C/T, T/T, but no difference for HCV genotype 3 according to rs12979860 C/C vs C/T, T/T[26]. Recently, active CMV infection was shown by our laboratory[20] to dramatically interfere with SVR rates in genotype 4 CHC patients, probably by inhibiting the JAK-STAT cascade. We, therefore, hypothesize that the role of CMV in the IL28B association with SVR should be a strong candidate for further categorization of C/T genotype carriers.
The present study comprised 350 subjects, including 166 CHC patients, 84 spontaneous HCV resolvers and 100 healthy control subjects.
Chronic HCV patients: All 166 patients had mild liver disease F0-F3 according to the Metavir classification[27]. All subjects had undetectable HBV surface antigen (HBsAg), Anti-schistosoma antibodies (Abs), and autoimmune Abs. None of the patients had a history of long-term drug use, aflatoxins consumption or alcohol intake. All patients were infected with HCV genotype 4. Patients included 105 males and 61 females, with an age range of 19-57 years. None had uncontrolled type II diabetes mellitus, thyroid dysfunction or obesity. All patients received weekly injection of peg-IFN-α plus daily oral ribavirin treatment for 48 wk. Eighty six patients achieved a SVR, namely undetectable HCV RNA 24 wk after the end of the treatment response that was achieved after 48 wk, i.e., a total of 72 wk. The remaining 80 patients were NR who failed to achieve a SVR, i.e., either did not respond to therapy after 12 wk or achieved undetectable viremia but failed to maintain the state of no viremia till 48 wk (breakthrough) or post 48 wk (viral relapse).
Spontaneous HCV resolvers: Eighty four subjects who tested positive for IgG anti-HCV Abs (3rd generation) and negative for HCV RNA were enrolled in the study. Patients included 53 males and 31 females, with an age range of 18-55 years.
Controls: One hundred healthy subjects who tested negative for both IgG anti HCV Abs and HCV RNA served as controls for the IL28B SNP and CMV reactivation studies. Also, they were negative for HBsAg, anti-schistosoma Abs, autoimmune Abs and CMV DNA. Subjects included 50 males and 50 females, with an age range of 18-64 years. None of subjects had a history of liver insult, either viral, metabolic or drug exposure.
Extraction of peripheral blood DNA: Peripheral blood was withdrawn from all subjects into EDTA solution and DNA was extracted using genomic DNA extraction kit (Qiagen, Milan Italy). Plasma samples were separated before DNA extraction and utilized for testing HBsAg, anti schistosomiases Abs and autoimmune Abs (ANA, AMA, LKM1, ASMA). Purified genomic DNA was used for IL28B SNP analysis and for testing the presence of CMV DNA.
Detection of IL-28B rs12979860 C/T polymorphism: Genotyping for the IL-28B rs12979860 C/T polymorphism was performed according to Fabris et al[24] with minor modification by polymerase chain reaction based restriction fragment length polymorphism (PCR-RFLP). Using purified genomic DNA, a 139 base-pair (bp) product was obtained with the forward primer 5’-CCAGGGCCCCTAACCTCTGCA-3’ and the reverse primer 5’-GGGAGCGCGGAGTGCAATTCA-3’, newly designed with the aid of NCBI Primer-Blast Tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Amplification was carried out in a total volume of 10 μL containing 10 mmol/L Tris-HCl (pH 8.3), 50 mmol/L KCl, Tween-20 0.01%, 0.2 mmol/L deoxyribonucleotides, 2-4 pmol of each primer, 2.0 mmol/L MgCl2, 0.5 units hot-start Taq DNA polymerase (Righ Taq, Euroclone, Milan, Italy) and ~10 ng of genomic DNA. The thermal protocol for amplification included 35 cycles of denaturation (at 95 °C for 60 s), annealing (at 64 °C for 60 s), and polymerization (at 72 °C for 60 s). Ten μL of the amplicons were digested with 1 unit of the BstU-I restriction endonuclease (New England Biolabs, Hitchin, UK) in a total volume of 20 μL at 37 °C for 4-6 h or overnight. The fragments were resolved by electrophoresis in 4% agarose gel after staining with ethidium bromide. A band of 139 bp indicated the TT genotype, 109 bp indicated the CC genotype; 139 + 109 bp indicated the CT genotype.
PCR amplification of the CMV gB gene: Primers for the first-round PCR were derived from the gB region of CMV, and PCR protocols were followed as described previously[28,29] The amplification mixture contained 10 pmol of each primer (CMV1 and CMV2), 0.2 mmol/L from each dNTP (Promega, Madison, WI, United States), 0.1 U Taq polymerase (Promega, Madison, WI, United States), 1 µL Taq buffer, 5 µL template, and distilled water to a final volume of 20 µL. A negative control tube (in which water replaced the DNA sample) was incorporated into each run. A sample from the first PCR product was used as a template in a second-round PCR (nested). Briefly, 1 µL of first round product was added to the reaction mixture, which is the same as the first-round reaction mix except for use of CMV3 and CMV4 as nested primers. The thermal cycling protocol was as follows: 1 min at 94 °C, 1 min at 55 °C and 1 min at 72 °C for 30 cycles. Then, the nested PCR products were analyzed with 3% agarose gel electrophoresis and stained with ethidium bromide. A result was considered positive when a clear 100 bp product was visible on the gel.
Primers used for the first and the second-round PCR were CMV1: 5’-GAGGACAACGAAATCCTGTTGGGCA-3’; CMV2: 5’-GTCGACGGTGGAGATACTG CTGAGG-3’; CMV3: 5’-ACCACCGCACTGAGGAATGTCAG-3’; and CMV4: 5’-TCAATCATGCGTTTGAAGAGGTA-3’.
Assessment of SVR status (real-time and RT-PCR of HCV RNA: As reported in our previous study[20,29]. Disappearance of viremia was confirmed by a variety of tests including RT-PCR using nested primers derived from the highly conserved HCV 5’UTR and real-time PCR. These tests were done 3, 6, 12 and 18 mo after the start of IFN therapy. HCV RNA was extracted using the BIOZOL-total RNA extraction reagent kit (Hangzhou Bioer Technology, Hangzhou, China). Nested RT-PCR performed in a 25 µL reaction mixture containing 20 units of avian myeloblastosis virus reverse transcriptase (Clontech, Mountain View, CA, United States), 200-400 ng of total cellular RNA as template, 40 units of RNAsin (Clontech), 0.2 µmol/L from each dNTP (QBIOGENE, Carlsbad, CA, United States), and 50 pmol of the reverse primer P2. The reaction was incubated at 42 °C for 60 min and denatured at 98 °C for 10 min. Amplification of the highly conserved 5’-UTR sequences was done using two rounds of PCR with two pairs of nested primers. First-round amplification was done in 50 µL of reaction containing 50 pmol from each of the P1 forward primer and P2 reverse primer, 0.2 mmol/L from each dNTP, 10 µL from RT reaction mixture as template and two units of Taq DNA polymerase (Finnzyme, Woburn, MA, United States) in a 1 × buffer supplied with the enzyme. The thermal cycling protocol was as follows: 1 min at 94 °C, 1 min at 55 °C and 1 min at 72 °C for 30 cycles. The second round of amplification was done similar to the first round, except for use of the nested reverse primer p4 and forward primer p3 at 50 pmol each. A fragment of 171 bp length was identified in positive samples. Primer sequences were as follows: P1: 5’-AACTACTGTCTTCACGCAGAA-3’; P2: 5’-GGTGCACGGTCTACGAGACCTC-3’; P3: 5’-GTGCAGCCTCCAGGACCC-3’; and P4: 5’-ACTCGGCTAGCAGTCTCGCG-3’.
HCV quantitation was performed using the Fluorescence Quantitative Detection kit (Hangzhou Bioer Technology, Hangzhou, China). HCV RNA was reverse transcribed and a specific fragment was amplified with specific primers in a one-step RT-PCR reaction. The products were detected using specific Taqman-MGB Probes. The real-time RT-PCR reaction was done in 50 µL of reaction containing 25 µL RT-PCR mix, 2.5 µL Mn2+, 1.5 µL HCV probe mix, and 1 µL internal control. Then 20 µL of the corresponding HCV RNA template were added. For the standard, 20 µL of the standards control 1-4 were added. All PCR tubes were placed in the real-time PCR Detection Instrument. The thermal cycling protocol was as follows: one cycle (30 s at 90 °C, 20 min at 61 °C and 1 min at 95 °C) followed by 30 cycles and 50 cycles (15 s at 95 °C, 1 min at 60 °C).
The significance of the response to IFN therapy was investigated by multivariate logistic regression analysis. CMV significantly affected the HCV response rate in C/T heterozygous carriers. The frequency of rs 12979860 SNP was compared in the control group vs the CHC patient group. All statistical analyses were performed using SPSS 9.0 statistical software (SPSS Inc., Chicago, IL, United States). The odds ratio (OR) and 95% confidence interval (CI) were calculated to assess the relative risk confidence. A two tailed P-value 0.05 was considered statistically significant.
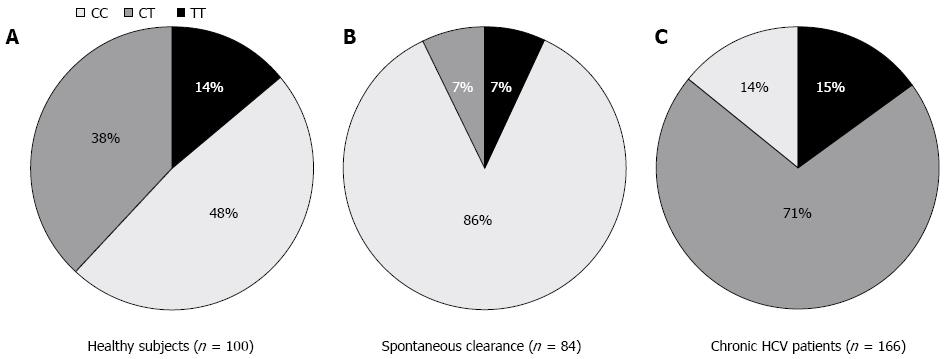
To test the hypothesis whether patients who spontaneously cleared HCV infection during or shortly following the acute phase bear the protective C/C allele more frequently than healthy subjects, we typed the IL28 B SNP (rs 12979860) in 84 spontaneous resolvers and 100 healthy subjects. The results of this study were previously described by our laboratory and are shown in Figure 1A and B, which clearly indicate the protective role of the C/C allele in patients who spontaneously cleared the virus, where C/C was present in 86% of spontaneous resolvers vs 48% of healthy subjects.
To examine the allele frequencies of the C/C genotype according to progression to chronic HCV infection, we compared the IL28B typing data derived from 166 chronic HCV patients with 84 subjects with spontaneous clearance of HCV. The results showed that the frequency of the protective C/C allele was markedly decreased from 86% in spontaneous resolvers to 48% in healthy controls to 14% in CHC patients regardless of their response to IFN therapy (Figure 1A-C). This decline was associated with a comparable increase in the heterozygous C/T from 7% in spontaneous resolvers to 38% in healthy subjects and to 71% in CHC patients.
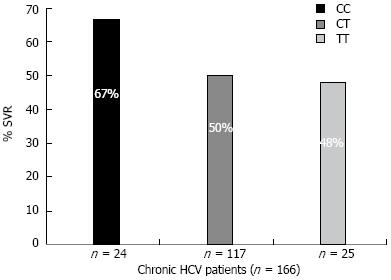
Among 166 CHC patients treated with combined IFN and ribavirin therapy, 79 patients did not achieve a SVR while 87 patients achieved a SVR. The effect of different IL28B genotypes and alleles on the HCV response to IFN treatment are shown in Table 1. The genotype CC was associated with response to IFN (P = 0.025). Of the 24 CHC patients bearing the C/C allele, 16 (67%) achieved a SVR, while the other two genotypes C/T and TT were associated with lower SVR rates: 50% and 48%, respectively (Figure 2).
| Non-responders (NR = 79) | Responders (SVR = 87) | Odds ratio | 95%CI | P value | |
| CC (n = 24) | 8 (33.3) | 16 (66.7) | 4.575 | 1.376-7.846 | 0.025 |
| CT (n = 117) | 58 (49.6) | 59 (50.4) | 0.858 | 0.538-1.449 | 0.589 |
| TT (n = 25) | 13 (52) | 12 (48) | 0.978 | 0.758-1.759 | 0.464 |
| C (n = 165) | 74 (44.8) | 91 (55.2) | 1.636 | 0.773-3.465 | 0.198 |
| T (n = 167) | 84 (50.3) | 83 (49.7) | 0.798 | 0.598-1.349 | 0.604 |
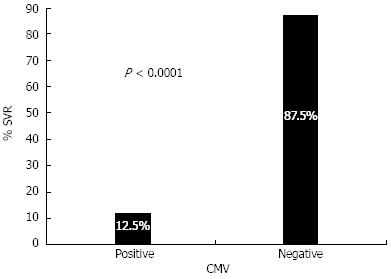
The PCR results of the CMV gB gene showed that 66/166 (40%) of chronic HCV patients have detectable CMV DNA. Since we have previously shown that coinfection with CMV seriously diminished the response to IFN therapy, it was tempting to examine whether this co-infection is associated with lower SVR rates in C/C holders. The data shown in Figure 3 clearly demonstrate that positive CMV DNA dramatically reduced the SVR rates in C/C genotypes as represented by a 12.5% SVR rate in CMV-positive patients compared with 87.5% in CMV-negative patients (P < 0.0001).
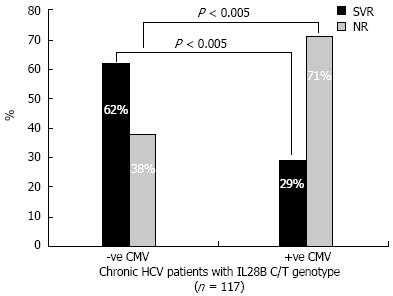
Since the C/T allele represents the majority of CHC patients (117/166) (71%), the use of IL28B SNP genotyping may be of little value as a predictor of response prior to starting the treatment regimen. Therefore, there is a great need to include another factor that was previously known to affect SVR rates. Table 2 shows the results of CMV testing in CT genotype patients. Specifically, CMV DNA was detected in 71% of NR patients and in 29% of SVR patients. In Figure 4, the SVR rates among C/T carriers was reduced to < 50% of its value in cases where patients had positive CMV DNA On the other hand, the NR rate doubled in cases of positive CMV DNA. These data indicate that a supplemental assay for CMV viremia adds to the prognostic value of IL28B genotyping.
| HCV response to treatment (n =117) | IL 28B genotype (CT) +ve CMV patients (n = 45) | IL 28B genotype (CT) -ve CMV patients (n = 72) |
| NR (n = 59) | 32 (71) | 27 (38) |
| SVR (n = 58) | 13 (29) | 45 (62) |
Several predictive factors have been reported recently for SVR rates for combination therapy in HCV infection. Viral factors, other than viral loads and genotypes were reported, including substitutions at core amino acids 70 and 91[30,31], IFN sensitivity determining region variations[32] and IFN and ribavirin response determining region variations[33]. SNPs within a number of host genes were found to be associated with different treatment outcomes to therapy. These include IFN-α pathway genes[34] and IFN-stimulated genes such as OAS[18] and MxA[19]. The widely studied IL28B SNP allele was reported to correlate with a favorable therapeutic response and was also associated with spontaneous clearance of HCV[6]. Thomas et al[6] reported that the allele related to HCV clearance, namely rs 12979860 C/C, was the major allele in the majority of Asian and European countries. Conversely, patients of African ancestry had lower C/C allele frequency and SVR rates. The current data indicate that the Egyptian population infected mainly with genotype 4 has an intermediate response between that of the populations of African and European ancestries, i.e., the C/C allele frequency in normal subjects was 48% vs > 50% in Europeans and ~35% in Africans while SVR rates in the currently studied CHC patients were 67% vs ~80% in Europeans and ~50% in Africans. One of the most interesting aspects of the current study is the dramatic decline in C/C frequency in CHC patients (from 48% in controls to 14% in genotype 4 infections), thus allowing speculation that protective IL28B variations provide a more substantial advantage in acute HCV genotype 4. In a similar study on a German cohort, the rs12979860 C/C genotype was found in 49% of uninfected subjects and declined in chronic infection to 42.7% of HCV genotype 2 and 3 patients and 33.9% of HCV genotype 1 patients[35]. Taking together the data from the German cohort and the data of the current study, we may speculate that HCV genotype 4 has the highest advantage of viral clearance during acute infection followed by genotype 1 and lastly genotypes 2 and 3.
Strong evidence from the current data on the significant protective effect of the IL28 B variant in acute HCV 4 is the high percentage of the C/C genotype among spontaneously cleared individuals, i.e., positive HCV Abs and negative HCV RNA compared with controls and CHC patients (86% vs 48 and 14%, respectively). The lack of a protective role of the C/C genotype in 14% of CHC patients suggests that, in CHC patients with the C/C genotype, other factors are involved in development of chronic infection. It is tempting to hypothesize that IL28B C/C in those 24 CHC patients was not sufficient to provide substantial tendency towards viral clearance and consequently explains the relatively lower figure for the C/C association with SVR rate in the present cohort compared with Asian and European studies. In a recent study from our laboratory co-infection with active CMV viremia induced severe failure to achieve acceptable rates of SVR[20]. Infection with CMV has evolved multiple mechanisms for disrupting the IFN-stimulated JAK/STAT signal transduction. It appears to inhibit IFN-α responsiveness by decreasing JAK1 protein, which is an essential component of IFN-α signaling[36]. It was reported that CMV blocks IFN-stimulated gene factor 3 (ISGF3)-dependent (MHC class I, 2’,5’-OAS, and MxA) and ISGF3-independent gene expression in infected cells. Moreover, the essential component of ISGF3, p48, is significantly decreased by CMV. Therefore decreased JAK1 and p48 protein would inhibit IFN-α stimulated signal transduction, transcription factor activation, and gene expression, thus it is likely to globally block IFN-stimulated responses in CMV-infected patients[37]. The dual analysis of both roles of IL28B SNP and CMV co-infection clearly showed that C/C carriers not infected with CMV have a 7-fold higher rate of SVR than those C/C patients co-infected with CMV. One of the major problems in decision-making on an individual patient basis is the overwhelming preponderance of the IL28B C/T genotype among the current CHC cohort as well as other cohorts[38,39]. The present results indicate that the IL28B SNP is not the only factor that influences the IFN-induced antiviral activities of host cells against viral infection. The present results showed a 2-fold increase in SVR rate in CMV-negative over CMV-positive C/T carriers and clearly highlight the significance of additional testing for CMV viremia besides IL28 genotyping to set a model for accurate prediction of response to combined therapy in C/T carriers.
In conclusion the current results allow us to speculate that both genetic (i.e., spontaneous) and therapeutic (IFN-based therapy) arms are complementary in the battle against HCV where poor spontaneous clearance is associated with higher IFN-mediated response as in genotypes 2 and 3. On the other hand, the difficult to treat genotypes 1 and 4 have clearly better chances for spontaneous resolution. Furthermore, CMV DNA testing may be of value at the moment to make better prognostication on response to IFN particularly in heterozygous carriers. Needless to say the search for supplemental predictive host factors will never stop at least in the near future.
The current recommended treatment for chronic hepatitis C virus (HCV) infection is the combination of pegylated α-interferon (IFN) and ribavirin given for 48 wk. Both viral and host factors may contribute to the phenomenon that some patients with chronic hepatitis C respond well to IFN therapy but others do not. The host genetic factors mediate the vigor of the immune response of the host through the control of inter-individual differences in the production of intracellular antiviral proteins or some cytokines. Different studies revealed that single nucleotide polymorphisms (SNPs) within or adjacent to IL28B (19q13) that codes for interferon-λ3, predict spontaneous resolution of HCV and likely SVR to IFN based treatment in chronic HCV patients infected with genotypes other than type 4. Since more than 93% of patients in Egypt are infected with HCV genotype 4, the authors investigated whether IL28B SNPs may be of predictive value for the outcome of IFN-based therapy in the difficult to treat HCV genotype 4 patients. Also the authors tested whether the status of positive cytomegalovirus (CMV) DNA detection added to the predictive value of IL28B and to further categorization of C/T allele carriers.
HCV is a global health problem that infects more than 170 million people around the world and more than 15% of the Egyptian population. The response rate to combined IFN therapy does not exceed 50% of patients infected with HCV genotype 4, the most prevalent subtype in Egypt. Timing and rules for this therapy without major complications are not very well defined, so that infected patients not only suffer from severe adverse effects but there is also a contribution to evolution of more resistant strains of the virus. A research focus is how to determine and understand which HCV patients are likely to develop persistent infection, progressive liver disease or do not respond significantly to IFN therapy. These criteria are of utmost importance in disease control programs and treatment strategies. Therefore, there is an urgent need for biological models containing a variety of prognostic parameters to predict the response to IFN treatment in HCV-infected patients and to help make better decisions on treatment strategy.
The use of IL28B SNP analysis in predicting the response to combined IFN therapy is informative only in cases of CC or TT genotypes. Since the heterozygous genotype CT represents more than 70% of chronic HCV patients, there is a need for supplemental markers for making a decision on whether the HCV patient is a potential responder to combined IFN therapy or not. The authors' data clearly improve the decision-making by detecting CMV reactivation in the CT genotype group. CMV reactivation has been shown to dramatically interfere with the response to combined IFN treatment. The data presented in this manuscript are indeed important for hepatologists regarding the anticipated response to standard combined therapy for HCV genotype 4 patients.
The data of this manuscript help hepatologists make better decisions on response rate to combined interferon therapy for HCV genotype 4 patients.
In this manuscript, the authors aimed to investigate whether the status of positive cytomegalovirus (CMV) DNA detection increases the predictive value of IL28B and to further categorize C/T allele carriers. The authors have found that CMV DNA testing might be valuable at the moment to make better decisions on response to IFN particularly in IL28B C/T carriers. Overall, this topic is very interesting.
P- Reviewer Hung CH S- Editor Lv S L- Editor Cant MR E- Editor Zhang DN
| 1. | Ferenci P. Current Treatment for Chronic Hepatitis C. Curr Treat Options Gastroenterol. 2004;7:491-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 2. | Hadziyannis SJ, Sette H, Morgan TR, Balan V, Diago M, Marcellin P, Ramadori G, Bodenheimer H, Bernstein D, Rizzetto M. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med. 2004;140:346-355. [PubMed] |
| 3. | Lagging M, Wejstål R, Uhnoo I, Gerdén B, Fischler B, Friman S, Josephson F, Karlström O, Sangfelt P, Schvarz R. Treatment of hepatitis C virus infection: updated Swedish Consensus recommendations. Scand J Infect Dis. 2009;41:389-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 4. | Knapp S, Yee LJ, Frodsham AJ, Hennig BJ, Hellier S, Zhang L, Wright M, Chiaramonte M, Graves M, Thomas HC. Polymorphisms in interferon-induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS-1 and PKR. Genes Immun. 2003;4:411-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 172] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 5. | Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338-145, 1338-145. [PubMed] |
| 6. | Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798-801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1706] [Cited by in RCA: 1686] [Article Influence: 105.4] [Reference Citation Analysis (0)] |
| 7. | Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2776] [Cited by in RCA: 2721] [Article Influence: 170.1] [Reference Citation Analysis (0)] |
| 8. | Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100-1104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1505] [Cited by in RCA: 1502] [Article Influence: 93.9] [Reference Citation Analysis (0)] |
| 9. | Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105-1109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1779] [Cited by in RCA: 1773] [Article Influence: 110.8] [Reference Citation Analysis (0)] |
| 10. | Zeuzem S, Berg T, Moeller B, Hinrichsen H, Mauss S, Wedemeyer H, Sarrazin C, Hueppe D, Zehnter E, Manns MP. Expert opinion on the treatment of patients with chronic hepatitis C. J Viral Hepat. 2009;16:75-90. [PubMed] |
| 11. | Berg T, Sarrazin C, Herrmann E, Hinrichsen H, Gerlach T, Zachoval R, Wiedenmann B, Hopf U, Zeuzem S. Prediction of treatment outcome in patients with chronic hepatitis C: significance of baseline parameters and viral dynamics during therapy. Hepatology. 2003;37:600-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 208] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 12. | Thompson AJ, Muir AJ, Sulkowski MS, Ge D, Fellay J, Shianna KV, Urban T, Afdhal NH, Jacobson IM, Esteban R. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120-9.e18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 534] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 13. | Bitetto D, Fattovich G, Fabris C, Ceriani E, Falleti E, Fornasiere E, Pasino M, Ieluzzi D, Cussigh A, Cmet S. Complementary role of vitamin D deficiency and the interleukin-28B rs12979860 C/T polymorphism in predicting antiviral response in chronic hepatitis C. Hepatology. 2011;53:1118-1126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 114] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 14. | Lange CM, Bojunga J, Ramos-Lopez E, von Wagner M, Hassler A, Vermehren J, Herrmann E, Badenhoop K, Zeuzem S, Sarrazin C. Vitamin D deficiency and a CYP27B1-1260 promoter polymorphism are associated with chronic hepatitis C and poor response to interferon-alfa based therapy. J Hepatol. 2011;54:887-893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 188] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 15. | Lange CM, von Wagner M, Bojunga J, Berg T, Farnik H, Hassler A, Sarrazin C, Herrmann E, Zeuzem S. Serum lipids in European chronic HCV genotype 1 patients during and after treatment with pegylated interferon-α-2a and ribavirin. Eur J Gastroenterol Hepatol. 2010;22:1303-1307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 16. | Petta S, Cammà C, Scazzone C, Tripodo C, Di Marco V, Bono A, Cabibi D, Licata G, Porcasi R, Marchesini G. Low vitamin D serum level is related to severe fibrosis and low responsiveness to interferon-based therapy in genotype 1 chronic hepatitis C. Hepatology. 2010;51:1158-1167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 325] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 17. | Lagging M, Askarieh G, Negro F, Bibert S, Söderholm J, Westin J, Lindh M, Romero A, Missale G, Ferrari C. Response prediction in chronic hepatitis C by assessment of IP-10 and IL28B-related single nucleotide polymorphisms. PLoS One. 2011;6:e17232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 127] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 18. | El Awady MK, Anany MA, Esmat G, Zayed N, Tabll AA, Helmy A, El Zayady AR, Abdalla MS, Sharada HM, El Raziky M. Single nucleotide polymorphism at exon 7 splice acceptor site of OAS1 gene determines response of hepatitis C virus patients to interferon therapy. J Gastroenterol Hepatol. 2011;26:843-850. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 19. | Hijikata M, Ohta Y, Mishiro S. Identification of a single nucleotide polymorphism in the MxA gene promoter (G/T at nt -88) correlated with the response of hepatitis C patients to interferon. Intervirology. 2000;43:124-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 98] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 20. | Bader el-Din NG, Abd el-Meguid M, Tabll AA, Anany MA, Esmat G, Zayed N, Helmy A, el-Zayady AR, Barakat A, el-Awady MK. Human cytomegalovirus infection inhibits response of chronic hepatitis-C-virus-infected patients to interferon-based therapy. J Gastroenterol Hepatol. 2011;26:55-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 21. | Askar M, Avery R, Corey R, Lopez R, Thomas D, Pidwell D, Eghtesad B, Miller C, Fung J, Zein NN. Lack of killer immunoglobulin-like receptor 2DS2 (KIR2DS2) and KIR2DL2 is associated with poor responses to therapy of recurrent hepatitis C virus in liver transplant recipients. Liver Transpl. 2009;15:1557-1563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 22. | Carneiro VL, Lemaire DC, Bendicho MT, Souza SL, Cavalcante LN, Angelo AL, Freire SM, Mendes CM, Santana N, Lyra LG. Natural killer cell receptor and HLA-C gene polymorphisms among patients with hepatitis C: a comparison between sustained virological responders and non-responders. Liver Int. 2010;30:567-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 23. | Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 916] [Cited by in RCA: 925] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 24. | Fabris C, Falleti E, Cussigh A, Bitetto D, Fontanini E, Bignulin S, Cmet S, Fornasiere E, Fumolo E, Fangazio S. IL-28B rs12979860 C/T allele distribution in patients with liver cirrhosis: role in the course of chronic viral hepatitis and the development of HCC. J Hepatol. 2011;54:716-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 146] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 25. | Nattermann J, Vogel M, Nischalke HD, Danta M, Mauss S, Stellbrink HJ, Baumgarten A, Mayr C, Bruno R, Tural C. Genetic variation in IL28B and treatment-induced clearance of hepatitis C virus in HIV-positive patients with acute and chronic hepatitis C. J Infect Dis. 2011;203:595-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 26. | Rallón NI, Naggie S, Benito JM, Medrano J, Restrepo C, Goldstein D, Shianna KV, Vispo E, Thompson A, McHutchison J. Association of a single nucleotide polymorphism near the interleukin-28B gene with response to hepatitis C therapy in HIV/hepatitis C virus-coinfected patients. AIDS. 2010;24:F23-F29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 163] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 27. | Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24:289-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2860] [Cited by in RCA: 3073] [Article Influence: 106.0] [Reference Citation Analysis (0)] |
| 28. | Jones RN, Neale ML, Beattie B, Westmoreland D, Fox JD. Development and application of a PCR-based method including an internal control for diagnosis of congenital cytomegalovirus infection. J Clin Microbiol. 2000;38:1-6. [PubMed] |
| 29. | Tabll A, Shoman S, Ghanem H, Nabil M, El Din NG, El Awady MK. Assessment of human cytomegalovirus co-infection in Egyptian chronic HCV patients. Virol J. 2011;8:343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 30. | Akuta N, Suzuki F, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Arase Y. Predictive factors of early and sustained responses to peginterferon plus ribavirin combination therapy in Japanese patients infected with hepatitis C virus genotype 1b: amino acid substitutions in the core region and low-density lipoprotein cholesterol levels. J Hepatol. 2007;46:403-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 218] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 31. | Akuta N, Suzuki F, Sezaki H, Suzuki Y, Hosaka T, Someya T, Kobayashi M, Saitoh S, Watahiki S, Sato J. Predictive factors of virological non-response to interferon-ribavirin combination therapy for patients infected with hepatitis C virus of genotype 1b and high viral load. J Med Virol. 2006;78:83-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 88] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 32. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 752] [Cited by in RCA: 733] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 33. | El-Shamy A, Nagano-Fujii M, Sasase N, Imoto S, Kim SR, Hotta H. Sequence variation in hepatitis C virus nonstructural protein 5A predicts clinical outcome of pegylated interferon/ribavirin combination therapy. Hepatology. 2008;48:38-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 110] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 34. | Welzel TM, Morgan TR, Bonkovsky HL, Naishadham D, Pfeiffer RM, Wright EC, Hutchinson AA, Crenshaw AT, Bashirova A, Carrington M. Variants in interferon-alpha pathway genes and response to pegylated interferon-Alpha2a plus ribavirin for treatment of chronic hepatitis C virus infection in the hepatitis C antiviral long-term treatment against cirrhosis trial. Hepatology. 2009;49:1847-1858. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 71] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 35. | Sarrazin C, Susser S, Doehring A, Lange CM, Müller T, Schlecker C, Herrmann E, Lötsch J, Berg T. Importance of IL28B gene polymorphisms in hepatitis C virus genotype 2 and 3 infected patients. J Hepatol. 2011;54:415-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 186] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 36. | Miller DM, Cebulla CM, Sedmak DD. Human cytomegalovirus inhibition of major histocompatibility complex transcription and interferon signal transduction. Curr Top Microbiol Immunol. 2002;269:153-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 37. | Miller DM, Zhang Y, Rahill BM, Waldman WJ, Sedmak DD. Human cytomegalovirus inhibits IFN-alpha-stimulated antiviral and immunoregulatory responses by blocking multiple levels of IFN-alpha signal transduction. J Immunol. 1999;162:6107-6113. [PubMed] |
| 38. | Scott J, Holte S, Urban T, Burgess C, Coppel E, Wang C, Corey L, McHutchison J, Goldstein D. IL28B genotype effects during early treatment with peginterferon and ribavirin in difficult-to-treat hepatitis C virus infection. J Infect Dis. 2011;204:419-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 39. | Lutz P, Wasmuth JC, Nischalke HD, Vidovic N, Grünhage F, Lammert F, Oldenburg J, Rockstroh JK, Sauerbruch T, Spengler U. Progression of liver fibrosis in HIV/HCV genotype 1 co-infected patients is related to the T allele of the rs12979860 polymorphism of the IL28B gene. Eur J Med Res. 2011;16:335-341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |