INTRODUCTION
Classification of polyps in children and subsequent attempts to diagnose hereditary polyposis syndromes begin with histologic sub-typing. The juvenile polyp was first described by Diamond[1] and is characterized histologically by an edematous lamina propria with inflammatory cells and cystically dilated glands which are lined by cuboidal to columnar epithelium[2]. While sporadic juvenile polyps are fairly common in the first decade of life and may be found in up to 2% of the pediatric population[3,4], juvenile polyposis syndrome (JPS) is a rare hereditary polyposis syndrome occurring in 1:100 000[5] and entails an increased risk of colorectal cancer and to a lesser degree gastric cancer. Juvenile polyposis syndrome is defined (Jass Criteria) by the presence of five or more juvenile polyps in the colorectum, any number of juvenile polyps proximal to the colorectum or any number of juvenile polyps with a positive family history of juvenile polyposis[3,4,6]. JPS typically presents in adolescence or adulthood. Treatment consists of surveillance endoscopy with polypectomy. Endoscopy is typically performed on a regular basis after polyps are found. Prophylactic surgery is indicated if polyp burden is unmanageable endoscopically, when juvenile polyps display dysplasia or in the case of severe gastrointestinal bleeding. A severe form of JPS called juvenile polyposis of infancy (JPI) has also been described and is characterized by its early manifestations of generalized polyposis, diarrhea, gastrointestinal bleeding and protein losing enteropathy in the first two years of life resulting in death in infancy in some patients[7].
Three genes have been associated with juvenile polyps. In 45%-60% of patients with typical JPS a mutation can be found in either the SMAD4 or BMPR1A genes[8-12]. SMAD4 is a tumor suppressor gene located on chromosome 18q21 and is associated with hereditary hemorrhagic telangiectasia in some individuals in addition to JPS. BMPR1A is located on chromosome 10q22-23[12] and encodes for a receptor important in the BMP/growth factor signaling pathways. Additionally, a third gene, PTEN, also located on chromosome 10q22-23, has been associated with juvenile polyps, in association with Cowden syndrome, a familial cancer syndrome, and the lesser known Bannayan-Riley-Ruvalcaba syndrome[13] associated with macrocephaly, developmental delay and some minor dysmorphia. These conditions, now grouped together as the “PTEN-hamartoma syndrome” (PHTS)[14], may present with juvenile or hamartomatous polyps. In addition to an increased risk for breast, thyroid, colorectal and endometrial cancers in adulthood, skin lesions such as lipomas, trichelommomas and papillomatous lesions, and penile macules are common findings.
Of considerable interest are a small group of patients who have been reported to have a chromosome 10q23 deletion involving both the BMPR1A and PTEN genes and also developed juvenile polyposis. Less than twenty patients have been described with these mutations[8,15-26]. Many of these patients were originally tested by chromosome analysis or more recently by chromosome microarray due to congenital anomalies, macrocephaly and/or developmental delay. Many of them also developed aggressive juvenile polyposis and in some cases required colectomy. The most common physical finding in these children was macrocephaly and the majority also had developmental delay. Other findings seen in multiple patients were atrial septal defect and/or ventricular septal defect, hemangioma, club foot, hypotonia and speckling of the penis.
Herein, we describe a patient who presented with a microdeletion of chromosome 10q23 which resulted in the deletion of both BMPR1A and PTEN genes and an additional microdeletion involving chromosome 1p31.3 of uncertain significance. His polyposis history is compared to that of others with similar 10q23 deletions and contrasted with those with mutations in either BMPR1A, PTEN or SMAD4 alone. We have developed an algorithm for genetic testing for patients presenting with juvenile polyposis since those with microdeletions are subject to significant health risks, including malignancy. We will also focus on the optimal long term gastrointestinal surveillance for these patients.
CASE REPORT
The patient was delivered at 37 wk gestation by Caesarean delivery due to macrocephaly and weighed 9 lbs, 12 oz. Head circumference at birth was 38.7 cm (90th percentile), and at 11 mo of age he was significantly macrocephalic (+4 SD). At a few days of age, a heart murmur was noted and echocardiogram revealed a large ventricular septal defect (VSD), atrial septal defects (ASD) and several smaller VSDs. Repair was performed at 10 d of age. His postoperative course was complicated by ectopic atrial tachycardia which required short term amiodarone therapy. Tracheostomy was performed at 11 mo of age due to multiple episodes of respiratory distress, multiple pneumonias and a diagnosis of tracheobronchomalacia. Other phenotypic characteristics in this child were hypospadias requiring repair, sagittal craniosynostosis requiring surgical correction, exotropia, midface hypoplasia with large cheeks, a prominent Cupid’s bow of the upper lip, and deep palmar creases. Additionally, his medical history included adenoidal hypertrophy necessitating adenoidectomy and fundoplication for medically refractory gastroesophageal reflux exacerbating respiratory compromise. Developmental delay was also present with delayed speech and gross motor delays. Endocrine issues included short stature (< 3rd percentile), obesity (body mass index, BMI > 97 kg/m2), growth hormone deficiency and primary hypothyroidism. The patient was treated with growth hormone and L-thyroxine and his height eventually reached the 10th percentile.
Due to the multiple anomalies found in this patient, genetic consultation had been obtained at 4 years of age. He was reported to have had a normal 46, XY karyotype on previous testing. Microarray comparative genomic hybridization (aCGH) analysis was performed (Agilent 244k platform) and two genomic deletions were found in this patient. One is a 1.03 Mb deletion within chromosome band 1p31.3 involving seven annotated genes and transcripts: CACHD1, RAVER2, JAK1, AK3L1, DNAJC6, LEPR, LEPROT [chr1:64870449-65897852 (hg18)]. The other one is a 5.75 Mb deletion of chromosome 10q23.1q23.31 involving 26 annotated genes and transcripts including BMPR1A and PTEN [chr10: 84311235-90064565 (hg18)]. Parental analyses of these two deletions showed normal results indicating these deletions are de novo in origin.
At 4 years of age the patient was seen in our Pediatric Gastroenterology Clinic for consultation due to the concern regarding polyposis with the 10q23 deletion and also for his symptoms of abdominal distension of uncertain etiology. At age 5 years he underwent esophagogastroduodenoscopy (EGD) and colonoscopy with significant findings of five small (4-5 mm) duodenal polyps and approximately 30 polyps in the colon, from rectum to cecum. Histopathology revealed juvenile polyps in all cases, without any adenomatous transformation. Growth hormone was stopped at this point due to a concern for increased polyp growth.
Four months later blood was noted in the stool and repeat endoscopy was performed. The polyp burden had increased to approximately 50 small polyps (4-6 mm) in the duodenum and 75-100 polyps in the colon. The majority of these colonic polyps were less than 6 mm, however there were five to six larger polyps 1-2 cm in size. Histology of all polyps was consistent with juvenile polyps.
A third endoscopy was performed six months later and again 50 polyps were noted in the duodenum, with several of them increased in size to 8 mm. Colonoscopy revealed 50-100 polyps from sigmoid to cecum (Figure 1). Approximately half of these polyps were now > 1.5 cm with several larger than 3 cm in diameter. Subsequently, as a result of the polyp burden which precluded endoscopic removal, the child was referred for laparoscopic subtotal colectomy with ileorectal anastamosis. The resected colon contained greater than 50 polyps, ranging in size from 0.6-3.1 cm in diameter. The polyps were juvenile in all cases and there was no dysplasia found. Post-operatively the patient struggled with frequent stooling and skin breakdown but is improved with use of fiber and loperamide.
Figure 1 Endoscopic view.
A: Polyps noted during colonoscopy, prior to colectomy; B: Endoscopic view of colonic polyps in the patient described.
DISCUSSION
The 10q23 deletion encompassing both PTEN and BMPR1A is rare, but conveys significant multisystem health problems and is known to have a variable phenotype with many individuals harboring juvenile polyps. Some individuals with this deletion fit the description of JPI with aggressive and early onset gastrointestinal polyposis. Our patient did not meet the criteria for JPI, as there was not diarrhea, bloody stools or hypoalbuminemia in the first two years of life. However, he did have extensive juvenile polyposis at a young age which led to colectomy. This aggressive gastrointestinal phenotype is not expected in generalized JPS, which is typically diagnosed in adolescence or adulthood. Additional clinical features in our patient included cardiac defects, macrocephaly, developmental delay, tracheobronchomalacia necessitating tracheostomy, medically refractory gastroesophageal reflux requiring fundoplication, thyroid and growth hormone deficiency and hypospadias. Whether these additional features represent the variability of the 10q23 deletion syndrome or whether they are associated with the additional 1p31.3 deletion is unknown at this time.
This is the first report of co-existing deletions of 10q23 and 1p31.3. The 1.03 Mb deletion of 1p31.3 has not been reported before. There are no known benign copy number variants in the region (http://projects.tcag.ca/variation/). Petti et al[27] reported a 15-year-old boy who carried a heterozygous 3.2 Mb deletion, which covers and extends beyond the deletion in our patient and had obesity, behavioral problems, mild intellectual impairment and facial dysmorphism. Vauthier et al[28] reported a three-year-old boy with an 80 kb homozygous deletion of 1p31.3 which included part of DNAJC6 and LEPR genes. This patient showed early onset obesity, mild dysmorphic features, intellectual disability, and epilepsy. Eight additional family members were heterozygous for the 80 kb 1p31.3 deletion. Seven of the eight were either overweight or obese and none had intellectual impairment. Our current patient has a heterozygous deletion of the DANJC6 and LEPR genes. Since age 2 years, his weight has tracked above the 75th percentile and height at or below the 10th percentile with a BMI at the 99th percentile for age which may reflect the effect of the deleted LEPR gene as high BMI is not typically associated with the 10q23 deletion phenotype. Heterozygous loss of the DNAJC6 gene in the current patient is of unknown significance. A literature review did not reveal any significant clinical associations of other genes deleted in the region within 1p31.3. Therefore, it is unclear how the 1p31.3 deletion may have impacted the phenotype of 10q23 deletion in our patient other than contributing to his elevated BMI.
This patient’s most significant medical issues, including polyposis and subsequent colectomy, pertain to his chromosome 10q23 deletion and its disruption of the function of BMPR1A and PTEN. PTEN is an important tumor suppressor gene. Mutations (including sequence changes or deletions) of the PTEN gene are associated with PHTS as previously described. Both hamartomatous and juvenile polyps are seen in PHTS. Sequence changes or partial deletions of the BMPR1A gene result in loss-of-function of that gene and typically result in juvenile polyposis syndrome. Interestingly, among patients reported to have a deletion of chromosome 10q23 which includes BMPR1A but does not include PTEN[17,29], none have been reported to have polyposis thus far[30]. A combined and synergistic effect of the deletion of both BMPR1A and PTEN in 10q23 microdeletion may be involved in this aggressive polyposis. The functions of the PTEN protein include phosphatase activity down-regulating the PI3K/Akt pathway, which helps regulate cell growth, proliferation, and apoptosis[31]. The BMPR1A gene encodes a receptor for the BMP pathway binding proteins and this pathway inhibits cell proliferation, especially of the gastrointestinal tract[32,33]. Therefore, the deletion of both of these genes may lead to increased proliferation of gastrointestinal cells predisposing to polyps and potential gastrointestinal malignancies.
Gastrointestinal management of patients with 10q23 microdeletions is determined on an individual basis due to the variability in onset of polyps and severity of progression. In many patients with this deletion, including the patient described in this report, there is an accelerated rate of polyp development that occurs at a very early age and is more aggressive than that seen in PHTS or in BMPR1A-associated JPS. In fact, nearly half of the reported patients have been referred for colectomy[16-18,20,23,25,26] in childhood, with several requiring surgery before 2 years of age. When contemplating colectomy, the number of polyps, size of polyps, associated symptoms and level of concern for malignancy are all considered. Although there is an increased risk of colorectal cancer in adults with PHTS[34] and JPS[35], children are rarely diagnosed with gastrointestinal cancer and do not routinely undergo colectomy. However, in those with 10q23 microdeletions there are reports of early colorectal dysplasia and malignancy. Dysplastic polyps or colonic epithelial dysplasia were noted in the colon in three children[22,23,25] and the duodenum in one[20]. An additional patient developed rectal cancer at age 24 years[23] and is now deceased. These observations suggest children with 10q23 deletions require frequent endoscopic surveillance of not only the lower but also the upper gastrointestinal tracts. We propose yearly EGD and colonoscopy after diagnosis of these mutations. Some patients, such as the one described in this paper, may require more frequent endoscopy if polyps are rapidly increasing in size or number and all polyps cannot be removed during one endoscopy. Small bowel surveillance with capsule endoscopy should also be considered. As in our patient, this risk for early gastrointestinal malignancy should prompt consideration for colectomy when the polyp burden becomes too great to manage through serial polypectomy or when dysplasia develops. Additionally, post-colectomy endoscopic surveillance is also warranted by the presence of upper intestinal polyps in a majority of reported cases, the high recurrence rates of polyps in the remnant rectum and the pouch and the fact that even after colectomy there is continued risk for duodenal or rectal cancer.
Extra-intestinal workup should also be considered due to the frequent non-gastrointestinal manifestations. In the patients’ reported, common findings include cardiac (ASD, VSD), developmental delay, hypotonia, lipoma and hemangioma. Extraintestinal malignancies reported in 10q23 microdeletions include thyroid cancer[8] and mucinous cystadenoma of the ovaries[26]. These observations suggest neurodevelopmental assessment, close monitoring of growth parameters, careful dermatologic exam, thyroid exam and/or ultrasound and echocardiography should all be considered in these patients both at time of diagnosis and throughout life.
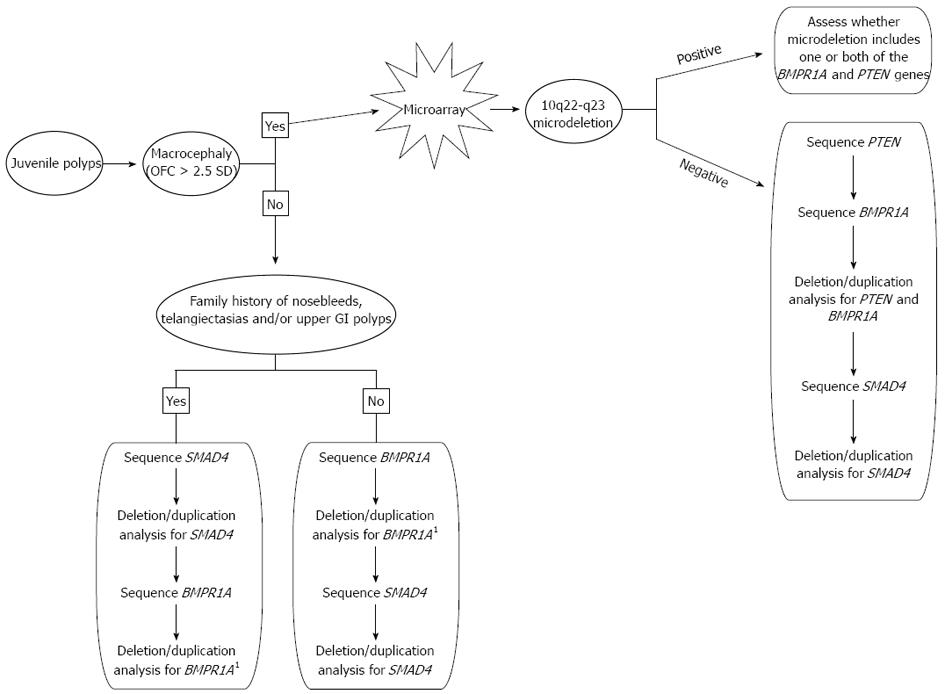
Genetic testing plays a critical role in establishing the correct diagnosis for patients who have features of JPS, PHTS, or both since this will have an impact on the surveillance and management of the patient. We propose the following algorithm to achieve a genetic diagnosis in the most timely and cost-effective manner (Figure 2). Macrocephaly of greater than 2.5 SD is a common feature seen in PHTS and is not commonly associated with JPS so it is a reasonable starting point in making a diagnosis. Additionally, in patients with JPS, a mutation in SMAD4 is more likely when there is family history of polyps when compared to BMPR1A. A SMAD4 mutation is also more likely when there is a positive family history of nosebleeds and/or telangiectasias, as SMAD4 mutations are also associated with hereditary hemorrhagic telangiectasia syndrome, along with features of JPS. Immunohistochemistry for SMAD4 may also be done in some centers, and if positive, guide genetic testing[36]. BMPR1A is located more proximal to the centromere on chromosome 10 than PTEN and there are several genes located between these two. Therefore, if a deletion is detected in either BMPR1A or PTEN, it is important to assess the precise location of this deletion in the event it could represent a larger 10q23 microdeletion. For this reason, we recommend a microarray analysis (if one has not already been completed) if a deletion is detected in either BMPR1A or PTEN. Both PHTS and JPS are inherited in an autosomal dominant pattern and both can either be inherited from a parent or occur as a de novo event. Once a genetic diagnosis is established in a presenting patient, parental studies may be critical to assess if either parent is at risk for medical complications that are associated with these conditions.
Figure 2 Algorithm for genetic testing and diagnosis of individuals with juvenile polyps.
1If a deletion in BMPR1A is found, ask the testing laboratory if a microarray is indicated based on the location of the deletion. OFC: Occipital-frontal circumference. GI: Gastrointestinal.
Our report highlights the phenotypic diversity of deletions including chromosome 10q23 and involving PTEN and BMPR1A. These patients are at risk for cardiac, endocrine, gastrointestinal and neurodevelopmental abnormalities. They have a heightened risk of accelerated polyposis, in some cases conforming to the traditional definition of JPI and, in addition harboring an increased risk of gastrointestinal malignancy that appears greater than if there is a mutation or deletion in either gene alone. Multidisciplinary assessment of these patients is an early prerequisite for care.