Published online Mar 7, 2012. doi: 10.3748/wjg.v18.i9.986
Revised: November 10, 2011
Accepted: December 16, 2011
Published online: March 7, 2012
AIM: To investigate the seroprevalence and molecular characteristics of hepatitis E virus (HEV) in the illegal blood donors (IBDs) of central China in the early 1990s.
METHODS: A total of 546 blood samples were collected from the IBDs in Maanshan city, a questionnaire was completed by each subject, detailing the age, sex, and periods of blood or plasma donation. Anhui Province and tested for the anti-HEV antibodies. The seropositive samples were subjected to nested reverse transcription-polymerase chain reaction and sequencing to analyze HEV partial genome.
RESULTS: The prevalence of IgG and IgM HEV antibody in IBDs was 22.7% and 1.8%, and genotype 4 was the dominant circulating HEV type in IBDs. The prevalence of anti-HEV IgG was significantly related to sex (OR = 4.905, P = 0.004) and increased with age (OR = 2.78, P = 0.022), which ranged from 13.0% in those < 40 years old to 30.6% among older persons aged > 60 years. Moreover, frequency of blood donation was significantly associated with HEV seropositivity (OR = 2.06, P = 0.006). HEV partial sequences of ORF2 and obtained 3 sequences in serum samples of 10 IBDs which developed HEV specific IgM.
CONCLUSION: This study helps define one of the possible routes of transmission of sporadic HEV infection and provides guidance to screen HEV in the blood donors so as to guarantee safe blood banks in China.
- Citation: Cheng XF, Wen YF, Zhu M, Zhan SW, Zheng JX, Dong C, Xiang KX, Xia XB, Wang G, Han LF. Serological and molecular study of hepatitis E virus among illegal blood donors. World J Gastroenterol 2012; 18(9): 986-990
- URL: https://www.wjgnet.com/1007-9327/full/v18/i9/986.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i9.986
Hepatitis E virus (HEV) infection is an important public-health concern as a major cause of enterically transmitted hepatitis worldwide. Epidemiologic studies have shown that HEV is prevalent in most developing countries, such as southeast Asia, northern and central Africa, India and central America. In addition, a high incidence of sporadic HEV infection has been observed in several industrialized countries, including the United States, European countries and Japan[1-5]. Although the ingestion of contaminated drinking water contributes mainly to the spread of HEV, other routes of transmission should be considered, because some studies implicated that blood transfusion was the possible route of sporadic HEV infection in non-endemic developed countries[6-8].
Between 1992 and 1995, illegal blood donation (IBDs) occurred frequently in several provinces in central China, including Henan, Anhui and Shanxi provinces. Although commercial blood donation was eradicated by the Chinese government by the end of 1995, the practice of using contaminated blood collection equipment caused the spread of some viruses, such as hepatitis C virus (HCV) and human immunodeficiency virus (HIV)[9,10].
While many studies have reported the prevalence of HEV infection and the HEV genome characteristics in different groups in China, to date, there has been no report on the prevalence of HEV infection among the IBDs. The aim of this study was to investigate whether HEV can be transmitted by the blood transfusion route and analyze the partially conserved nucleotide sequences of HEV strains among the IBDs in Maanshan city in Anhui Province, one of the provinces with the illegal blood collection in the early 1990s.
This work has been carried out in accordance with the Declaration of Helsinki (2000) of the World Medical Association. The study was initiated after the study protocol was approved by the Institutional Review Board (IRB) of China Center for AIDS/STD Control and Prevention and the IRB of Maanshan Center of Disease Control and Prevention.
A total of 546 samples were collected between January and August in 2005 from those who donated their blood or plasma frequently from 1992 to 1995. All participants were from Dangtu District in Maanshan city. A questionnaire was completed by each subject, detailing the age, sex, and periods of blood or plasma donation.
Serum samples were tested by enzyme-liked immunosorbent assay (ELISA) for IgG and IgM with anti-HEV activity described previously [11,12]. All serum samples were assayed at a 1:20 dilution. The absorbance of each sample was read at 450 nm. The cutoff value used for the anti-HEV IgG and IgM assay was 0.152. Then the serum positive for IgM antibody against HEV were tested for HEV RNA. All patients were previously tested for HBsAg (Diasorin, United States) and anti-HCV (third generation assay, Diasorin, United States) by ELISA.
Viral RNA was extracted from serum samples using Qiagen viral RNA kit (Qiagen) according to the manufacturer’s instructions. The viral RNA was finally dissolved in 20 μL RNase-free water. Reverse transcription-polymerase chain reaction (RT-PCR) was performed using TaKaRa RNA PCR kit (TaKaRa, Japan). The primers and the PCR protocol used were adapted from a previous study[13]. The external primers were P1 (5’-CCGACAGAATTGATTTC GTCGGC-3’) and P4 (5’-CCGTAAGTCGACTGGTCGTACTC-3’). The internal primers were P2 (5’-GTTGTCTCGGCCAATGGCGAGCC-3’) and P3 (5’-TCGGCGGCGGTGAGAGAGCCA-3’). The first-round and the second-round amplifications were carried out according to the following cycling program denaturation at 9 °C for 45 s, annealing at 52 °C for 60 s, extension at 72 °C for 60 s, for 35 cycles. The size of the first-round PCR product was 307 bp, and that of the second-round PCR was 236 bp.
The PCR products were visualized on a 10 mL/L agarose gel, excised and purified using a gel extraction kit (Qiagen) and directly sequenced on an ABI model 3730 DNA Auto Sequencer (Shanghai, China). The 236 bp fragment amplified from ORF2 of HEV genome was sequenced for comparison with the corresponding regions of other known human, porcine and avian HEV strains available in the GenBank database (DNAstar). Sequences were aligned using ClustalX v1.8 (http://www-igbmc.ustrasbg.fr/BioInfo/Clustal X /Top.html), and the phylogenetic analysis was performed using the MEGA program, version 3.1 (Pennsylvania State University).
The Pearson χ2 test was used to evaluate the difference in the prevalence between groups in the univariate analyses. Odds ratios (OR) with 95% confidence intervals were used to determine whether a variable was associated with HEV infection.
A total of 546 IBDs in the 1990s were enrolled to this study. The IBDs were aged from 29 to 75 years with a mean of 51 ± 9 years. Among them, 156 (28.6%) were males and 390 (71.4%) were females. While 124 IBDs developed HEV IgG antibody, only 10 IBDs developed HEV IgM antibody, therefore, the prevalence of IgG and IgM HEV antibody in this group was 22.7% and 1.8%, respectively.
The prevalence of HEV IgG seropositivity in the 546 IBDs is showed in Table 1. In the male group, 30.8% (48/156) were positive as against 19.5% (76/390) in the female group. The prevalence of HEV IgG seropositivity was significantly higher in men than in women (OR = 4.905, P = 0.004). In addition, subjects over 60 years of age had a higher prevalence of HEV IgG seropositivity than those aged < 40 years (OR = 2.780, P = 0.022). The frequency of plasma donation was also associated with HEV infection. The odds ratio was 2.06 among those who donated more than 20 times compared with those who donated 10 or fewer times.
| HEV positive IgG n (%) | OR (95% CI) | P value | |
| Sex | |||
| Female | 76/390 (19.5) | - | 0.004 |
| Male | 48 /156 (30.8) | 1.84 (1.20, 2.80) | |
| Donation frequency | |||
| < 10 | 47/252 (18.7) | - | |
| 10-20 | 44/191 (23.0) | 1.31 (0.82, 2.07) | 0.258 |
| ≥ 20 | 33/103 (32.0) | 2.06 (1.22, 3.46) | 0.006 |
| Age (yr) | |||
| < 40 | 7/54 (13.0) | - | |
| 40-50 | 37/192 (19.3) | 1.50 (0.63, 3.60) | 0.361 |
| 50-60 | 46/189 (24.3) | 2.02 (0.85, 4.80) | 0.105 |
| ≥ 60 | 34/111 (30.6) | 2.78 (1.14, 6.79) | 0.022 |
The sex specific prevalence of HEV IgG in IBDs with different frequency of donation is showed in Table 2. HEV infection prevalence was significantly correlated with the increasing age in total participants (z = 2.91, P = 0.004) and female participants (z = 1.97, P = 0.048).
| Frequency | Hepatitis E virus positive IgG | P value | |
| Male | Female | ||
| < 10 | 15/54 (27.8) | 32/198 (16.2) | 0.052 |
| 10-20 | 18/57 (31.5) | 28/137 (20.4) | 0.097 |
| ≥ 20 | 15/45 (33.3) | 16/55 (29.1) | 0.648 |
We also examined the relationship between HEV infection and HBV or HCV coinfection in these IBDs. The results showed no significant difference in HBsAg positive status (3.2% vs 3.1%) and HCV positive status (11.3% vs 11.1%) between HEV IgG positive and negative IBDs (Table 3).
| Characteristics | HEV positive IgG (n = 124) | HEV negative IgG (n = 422) | P value |
| HBsAg positive | 4 (3.2) | 13 (3.1) | 0.935 |
| HCV antibody positive | 14 (11.3) | 47 (11.1) | 0.962 |
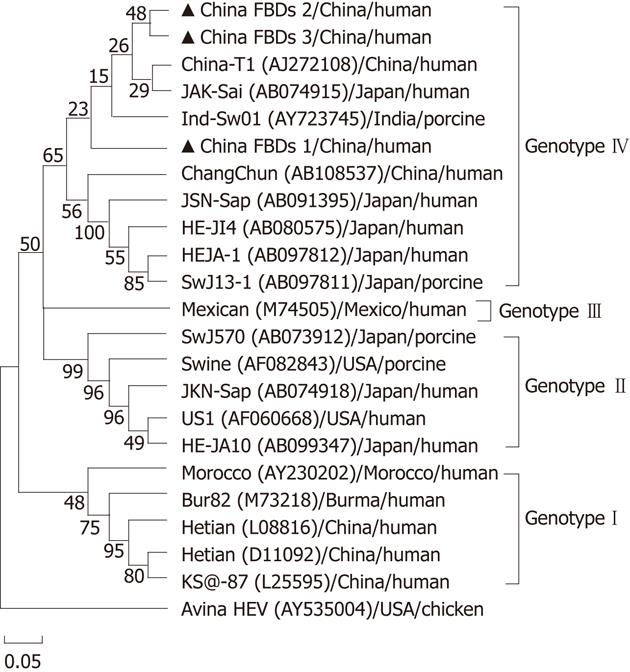
We detected HEV partial sequences of ORF2 and obtained 3 sequences in serum samples of 10 IBDs which developed HEV specific IgM. Sequence analysis demonstrated that these 3 strains were 83.3%-93.6% identical to each other. When compared with the HEV reference isolates, the strains were closely related to Chinese strain T1 with an 82.6%-89.4% nucleotide homology, and demonstrated a 91.1% sequence homology to a Japanese strain JAK-Sai. The nucleotide homology of other Japanese HEV strains with the strains from IBDs ranged from 78.6% to 85.4%. The homology of strains from Burma, Mexico and the United States was 79.2%-80.2%, 80.5%-81.4% and 77.1%-78.0%, respectively.
In the phylogenetic tree generated, the Bur82 strain (genotype 1), the Mexican strain (genotype 2), the US1/swine strain (genotype 3), and the Chinese strain T1 (genotype 4) represent major branches. Phylogenetic analyses clearly illustrate that all HEV sequences except avian strain can be divided into these four distinct genotypes and the three HEV sequences isolated in our study were genotype 4. The IBDs 1 sequence we analyzed formed an exclusive cluster, and was bound to a new subgenotype within genotype 4, which was supported by the bootstrap values obtained from 1000 replicates resampling analysis (Figure 1).
HCV and HIV infections were prevalent among the IBDs who donated blood in the early 1990s in China. However, data on HEV infection in this population have been unavailable so far. To our knowledge, the present study is the first seroepidemiological and molecular study on HEV infection in this unique population. Our results demonstrated that HEV infection had been introduced into this population in this area and that the prevalence was much higher (22.5%) than that in the normal population (4.76%) in this area[14].
Our data indicated that 30.7% of males were HEV positive compared to 19.5% of females and the difference was statistically significant (OR = 1.84, P = 0.004). In addition, we found that males with a history of blood transfusion had a high HEV seropositivity than females, suggesting that male IBDs are more likely to get infected by HEV than female donors. Our study also showed that HEV seropositivity increased with age of the first donating blood, consistent with previous studies demonstrating that age may be an important risk associated with HEV in other populations[15,16].
Although no statistically significant association was observed between HEV seropositivity and blood-borne hepatitis viruses such as HBV and HCV, our findings show that more frequent blood/plasma donation increased the risk of HEV infection, providing evidence that HEV can be transmitted by viremic blood units and have similar or overlapping routes of transmission with HCV[17].
HEV IgM antibody is known as a marker of the early seroconversion period. In this study, HEV IgM antibody was detected in 10 samples in the population. We also tested the presence of serum HEV RNA in the IBDs. HEV has been classified into four genotypes based on the full sequence heterogeneity. These include genotypes 1 (mainly prevalent in Asia and Africa), 2 (mainly prevalent in Mexico, Nigeria), 3 (mainly prevalent in the US, Japan, Argentina, and Europe), and 4 (mainly prevalent in Taiwan, Japan, and mainland China)[18]. More recently, HEV genotype 4 has been isolated from various regions of China, ranging from the south (Guangzhou and Shanghai), the centre (Henan province) to the north (Liaoning Province and Beijing), and has been found to be responsible for a significant proportion of cases of sporadic acute hepatitis in China[19-21]. Schlauder et al[22] reported that the analysis of small regions of HEV genome yields evolutionary distances similar to those produced from the full-length HEV genome. Therefore, we amplified and sequenced three HEV partial sequences in the serum samples of 10 IBDs positive for HEV IgM antibody. The three sequences share an 81.4%-88.1% identity at the nucleotide level with each other, and 79.2%-80.2%, 80.5%-81.4%, 77.1%-78.0% and 83.3%-93.6% identity with HEV genotypes 1-4, respectively. Clearly, they belong to genotype 4 and resemble HEV genotype 4 sequences but form some new subgenotypes. These results indicate that there is great genetic variability in HEV genome of genotype 4, even within a certain region or population investigated in China. Previous data showed that a substantial proportion of voluntary blood donors (3/200 or 1.5%) was positive for HEV RNA and the isolates all had a high nucleotide sequence identity (> 90%) with swine HEV, which means that HEV may be zoonotically transmitted from viremic animals to humans[23], however, in the present study, three isolates only shared a 77.5%-86.0% identity with swine HEV.
In conclusion, we report the first molecular and seroepidemiological study on HEV infection in the IBDs in China in the 1990s. The results demonstrate that HEV is widely spread in this population and confined to genotype 4. Males, individuals aged beyond 60 years, and people who donated blood more than 20 times showed a higher rate of previous HEV infection. Our study may help define one of the possible routes of transmission of sporadic HEV infection in this population and provide guidance to screen HEV in the donors to guarantee safe blood banks in China.
Hepatitis E virus (HEV) infection is an important public-health concern as a major cause of enterically transmitted hepatitis worldwide. Epidemiologic studies have shown that HEV is prevalent in most developing countries and some industrialized countries. Although commercial blood donation was eradicated by Chinese government by the end of 1995, the practice of using contaminated blood collection equipment caused the spread of some viruses such as hepatitis C virus and hepatitis I virus, but there has been no report on the prevalence of HEV infection in the illegal blood donors (IBDs).
This is a first serological and molecular study on HEV infection in IBDs. The results showed that the prevalence of HEV IgG antibody was higher (22.5%) in the IBDs than in general population, and the risks of HEV infection in IBDs were age, gender and times of donation. Additionally, phylogenetic analysis showed that 3 HEV strains isolated from IBDs belong to genotype 4. Therefore, the present study indicated that HEV is widely spread in the IBDs and the new possible modes of transmission of sporadic HEV infection in IBDs should be defined.
As it was indicated in this study that HEV is wildly spread among the IBDs, and age, gender and times of blood donation are the risk factors of HEV infection. Therefore, HEV should be detected regularly among the IBDs for the safety of transfusion.
Hepatitis E virus (HEV): Hepatitis E Virus 1 has a particle diameter of 32-34 nm, a buoyant density of 1.29 g/mL in KTar/Gly gradient, and is very labile. Serologically related smaller (27-30 nm) particles are often found in feces of patients with hepatitis E and are presumed to represent degraded viral particles. HEV has a single-stranded polyadenylated RNA genome of approximately 8 kb. Based on its physicochemical properties, it is presumed to be a calici2-like virus.
The molecular and sero-epidemiological study on HEV infection is presented in a proper way and this study could help in defining one of the possible modes of transmission of sporadic HEV infection in Chinese population and guide the screening of HEV in the donors to guarantee safe blood banks. The manuscript is well presented and of interest and the design of this study is appropriate. The study was done well and their results can contribute to knowledge of this topic.
Peer reviewers: Vladimir C Serafimoski, Profesor, Clinic of Gastroenterohepatology, Medical Faculty, Skopje, Fyrom, Vodnjanska 17, 1000 Skopje, Macedonia; Masahiro Arai, MD, PhD, Department of Gastroenterology, Toshiba General Hospital, 6-3-22 Higashi-ooi, Shinagawa-ku 140-8522 Tokyo, Japan
S- Editor Gou SX L- Editor Ma JY E- Editor Xiong L
| 1. | Emerson SU, Purcell RH. Running like water--the omnipresence of hepatitis E. N Engl J Med. 2004;351:2367-2368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 48] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 2. | Ijaz S, Arnold E, Banks M, Bendall RP, Cramp ME, Cunningham R, Dalton HR, Harrison TJ, Hill SF, Macfarlane L. Non-travel-associated hepatitis E in England and Wales: demographic, clinical, and molecular epidemiological characteristics. J Infect Dis. 2005;192:1166-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 178] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 3. | Mansuy JM, Peron JM, Abravanel F, Poirson H, Dubois M, Miedouge M, Vischi F, Alric L, Vinel JP, Izopet J. Hepatitis E in the south west of France in individuals who have never visited an endemic area. J Med Virol. 2004;74:419-424. [PubMed] |
| 4. | Sainokami S, Abe K, Kumagai I, Miyasaka A, Endo R, Takikawa Y, Suzuki K, Mizuo H, Sugai Y, Akahane Y. Epidemiological and clinical study of sporadic acute hepatitis E caused by indigenous strains of hepatitis E virus in Japan comparedwith acute hepatitis A. J Gastroenterol. 2004;39:640-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 36] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 5. | Widdowson MA, Jaspers WJ, van der Poel WH, Verschoor F, de Roda Husman AM, Winter HL, Zaaijer HL, Koopmans M. Cluster of cases of acute hepatitis associated with hepatitis E virus infection acquired in the Netherlands. Clin Infect Dis. 2003;36:29-33. [PubMed] |
| 6. | Boxall E, Herborn A, Kochethu G, Pratt G, Adams D, Ijaz S, Teo CG. Transfusion-transmitted hepatitis E in a 'nonhyperendemic' country. Transfus Med. 2006;16:79-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 244] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 7. | Tamura A, Shimizu YK, Tanaka T, Kuroda K, Arakawa Y, Takahashi K, Mishiro S, Shimizu K, Moriyama M. Persistent infection of hepatitis E virus transmitted by blood transfusion in a patient with T-cell lymphoma. Hepatol Res. 2007;37:113-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 157] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 8. | Matsubayashi K, Kang JH, Sakata H, Takahashi K, Shindo M, Kato M, Sato S, Kato T, Nishimori H, Tsuji K. A case of transfusion-transmitted hepatitis E caused by blood from a donor infected with hepatitis E virus zoonotic food-borne route. Transfusion. 2008;48:1368-1375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 227] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 9. | Wu Z, Rou K, Detels R. Prevalence of HIV infection among former commercial plasma donors in rural eastern China. Health Policy Plan. 2001;16:41-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 108] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 10. | Xu JQ, Wang JJ, Han LF, Xu C, Ruan YH, Xu ZH, Chen X, Liu ZD, Wang J, Su B. Epidemiology, clinical and laboratory characteristics of currently alive HIV-1 infected former blood donors naive to antiretroviral therapy in Anhui Province, China. Chin Med J (Engl). 2006;119:1941-1948. [PubMed] |
| 11. | Lu J, Dai X, Meng JH. Application of p166 recombinant proteins derived from different genotypes of hepatitis E virus (HEV) in anti-HEV antibody detection. Chin J Microbiol Immunol. 2006;26:369-374. [DOI] [Full Text] |
| 12. | Dong C, Dai X, Shao JS, Hu K, Meng JH. Identification of genetic diversity of hepatitis E virus (HEV) and determination of the seroprevalence of HEV in eastern China. Arch Virol. 2007;152:739-746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 13. | Zhai L, Dai X, Meng J. Hepatitis E virus genotyping based on full-length genome and partial genomic regions. Virus Res. 2006;120:57-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 94] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 14. | Chen XF, Chen J, Zhan SW, Chen DL, Xiang KX, Zhu M, Fang DC, Wen YF. pidemiological characteristics of hepatitis E and genotypes of hepatitis E virus in Maanshan area of Anhui province. Chin J Public Health Jun. 2010;26:710-712. |
| 15. | Taremi M, Khoshbaten M, Gachkar L, EhsaniArdakani M, Zali M. Hepatitis E virus infection in hemodialysis patients: a seroepidemiological survey in Iran. BMC Infect Dis. 2005;5:36. [PubMed] |
| 16. | Li RC, Ge SX, Li YP, Zheng YJ, Nong Y, Guo QS, Zhang J, Ng MH, Xia NS. Seroprevalence of hepatitis E virus infection, rural southern People's Republic of China. Emerg Infect Dis. 2006;12:1682-1688. [PubMed] |
| 17. | Mitsui T, Tsukamoto Y, Yamazaki C, Masuko K, Tsuda F, Takahashi M, Nishizawa T, Okamoto H. Prevalence of hepatitis E virus infection among hemodialysis patients in Japan: evidence for infection with a genotype 3 HEV by blood transfusion. J Med Virol. 2004;74:563-572. [PubMed] |
| 18. | Dalton HR, Bendall R, Ijaz S, Banks M. Hepatitis E: an emerging infection in developed countries. Lancet Infect Dis. 2008;8:698-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 498] [Cited by in RCA: 483] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 19. | Li K, Zhuang H, Zhu W. Partial nucleotide sequencing of hepatitis E viruses detected in sera of patients with hepatitis E from 14 cities in China. Chin Med J (Engl). 2002;115:1058-1063. [PubMed] |
| 20. | Wei S, Xu Y, Wang M, To SS. Phylogenetic analysis of hepatitis E virus isolates in southern China (1994-1998). J Clin Virol. 2006;36:103-110. [PubMed] |
| 21. | Chen Y, Tian DY, Xia NS. Epidemiology and genotypes of HEV in Wuhan. Chin J Dig Dis. 2005;6:182-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 22. | Schlauder GG, Dawson GJ, Erker JC, Kwo PY, Knigge MF, Smalley DL, Rosenblatt JE, Desai SM, Mushahwar IK. The sequence and phylogenetic analysis of a novel hepatitis E virus isolated from a patient with acute hepatitis reported in the United States. J Gen Virol. 1998;79:447-456. [PubMed] |
| 23. | Chobe LP, Lole KS, Arankalle VA. Full genome sequence and analysis of Indian swine hepatitis E virus isolate of genotype 4. Vet Microbiol. 2006;114:240-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |