Published online Nov 28, 2012. doi: 10.3748/wjg.v18.i44.6481
Revised: August 10, 2012
Accepted: August 16, 2012
Published online: November 28, 2012
AIM: To evaluate a novel biosensor-based microarray (BBM) assay for detecting rs12979860 and rs8099917 genotypes.
METHODS: Four probes specific for rs8099917C/T or rs12979860G/T detection and three sets of quality control probes were designed, constructed and arrayed on an optical biosensor to develop a microarray assay. Two sets of primers were used in a one tube polymerase chain reaction (PCR) system to amplify two target fragments simultaneously. The biosensor microarray contained probes that had been sequenced to confirm that they included the rs8099917C/T or rs12979860G/T alleles of interest and could serve as the specific assay standards. In addition to rehybridization of four probes of known sequence, a total of 40 clinical samples collected from hepatitis C seropositive patients were also tested. The target fragments of all 40 samples were amplified in a 50 μL PCR system. Ten μL of each amplicon was tested by BBM assay, and another 40 μL was used for sequencing. The agreement of the results obtained by the two methods was tested statistically using the kappa coefficient. The sensitivity of the BBM assay was evaluated using serial dilutions of ten clinical blood samples containing 103-104 white cells/μL.
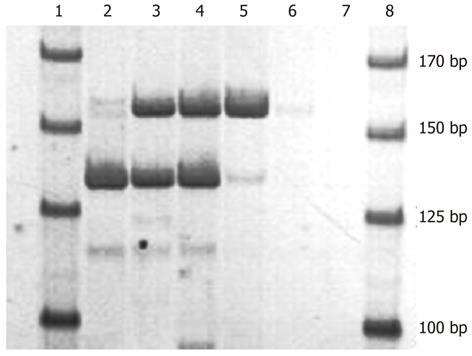
RESULTS: As shown by polyacrylamide gel electrophoresis, two target segments of the interleukin 28B-associated polymorphisms (SNPs) were successfully amplified in the one-tube PCR system. The lengths of the two amplified fragments were consistent with the known length of the target sequences, 137 and 159 bps. After hybridization of the PCR amplicons with the probes located on the BBM array, the signals of each allele of both the rs8099917 SNPs and rs12979860 SNPs were observed simultaneously and were clearly visible by the unaided eye. The signals were distinct from each other, could be interpreted visually, and accurately recorded using an ordinary digital camera. To evaluate the specificity of the assay, both the plasmids and clinical samples were applied to the microarray. First, 30 PCR amplicons of the various SNP alleles were hybridized on the BBM microarray. Full agreement between plasmids and the BBM assay was observed, with 30/30 correct matches (100%). The kappa value for the BBM assay with plasmids was 1.00 (P < 0.05). For the 40 clinical blood samples, the BBM assay hybridization and direct sequencing results were compared for each amplicon. For patient blood samples, agreement was 28/28 for rs8099917T/T, 9/11 for rs8099917T/G, 1/1 for rs8099917G/G, 24/24 for rs12979860C/C, 11/14 for rs12979860C/T, and 2/2 for rs12979860T/T. Only five clinical samples of amplicon assay and direct sequencing results were discordant and heterozygotes: 2/11 rs8099917T/G and 3/14 rs12979860C/T. The agreement of outcomes between BBM assay and direct sequencing for the detection of rs8099917 and rs12979860 was 95% and 92.5%, respectively; and the corresponding kappa values were 0.88 and 0.85 (A kappa value > 0.75 was defined as substantial agreement). The BBM assay and sequencing had similar specificities for detection and identification of the two SNPs and their alleles. The sensitivity evaluation showed that the BBM assay could detect and identify SNP sequences present in blood samples containing as few as 102 white blood cells/μL.
CONCLUSION: This biosensor microarray assay was highly specific, sensitive, rapid and easy to perform. It is compatible with clinical practice for detection of rs8099917 and rs12979860.
- Citation: Li PY, Zhou XJ, Yao L, Fang XH, Ren JN, Song JW. Novel biosensor-based microarray assay for detecting rs8099917 and rs12979860 genotypes. World J Gastroenterol 2012; 18(44): 6481-6488
- URL: https://www.wjgnet.com/1007-9327/full/v18/i44/6481.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i44.6481
Hepatitis C virus (HCV) infection is a worldwide health problem. It is estimated that about 3% of the world population are infected with HCV, including more than 170 million with chronic infection, who are at risk of developing liver cirrhosis and/or liver cancer. Approximately 3 to 4 million become infected with HCV, and more than 350 000 people die from HCV-related liver diseases each year[1]. Only about 30% patients with acute hepatitis C experience spontaneous clearance[2,3], while the remaining 70% develop persistent chronic infection and gradually progress to liver cirrhosis and/or hepatocellular carcinoma[4,5]. Chronic HCV infection that progresses to end-stage liver disease often requires a liver transplant, with high cost and extensive use of medical resources. Hepatocellular carcinoma is the seventh most common cancer worldwide and the third leading cause of cancer-related deaths. HCV thus has a very high morbidity and mortality, which result in a substantial burden on society[5].
Since the discovery of HCV in 1989, some improvement in treatment have been gained with the application of routine weekly injections of pegylated interferon (IFN) plus daily administration of oral ribavirin (Rb)[6]. Although triple therapy with IFN + Rb and boceprevir or telaprevir has produced a sustained virologic response (SVR) of approximately 70%, this regimen still requires co-administration of IFN + Rb and has a high cost in most countries[7]. At present, only 50% of patients infected with HCV genotype 1 achieved an SVR, defined as absence of HCV RNA six months after the cessation of therapy[8,9]. Even in non-genotype 1 patients, who are reported to have better responses to the combination treatment, the SVR rate was about 75%[10,11]. A number of viral and host factors may reduce adherence to the treatment. In addition to a limited treatment response to current combined therapy by some patients, treatment is often poorly tolerated due to various adverse reactions, including flu-like symptoms, severe fatigue, and neutropenia[12]. Moreover, the inconvenience of a prolonged course of weekly injections of pegylated interferon and the high costs of the antiviral drugs make some patients give up the combined therapy. Additional research to advance HCV treatment regimens is needed to increase the patient adherence and response to therapy, thereby increasing SVR rates.
It is also puzzling why approximately 30% of patients with acute HCV infection experience spontaneous viral clearance, whereas the rest become chronically infected[2,3]. Meanwhile, the global epidemiology of HCV infection shows that the different HCV genotypes clearly show geographic variation in their relative frequencies. Genotype 1, consisting of subtypes 1a and 1b, is the most prevalent genotype worldwide, with a higher prevalence of 1b in Europe and 1a in the United States[11-13]. The estimated prevalence of people with detectable anti-HCV antibodies is highest in Africa, with an overall seroprevalence of 5.3%, and most common in genotype 4. In contrast, the infection rate of HCV is lower, and genotype 2 or 3 are more frequent in Asia[11-13]. Furthermore, previous studies have revealed that the effectiveness of the combination treatment with peg-interferon and ribavirin differs among various ethnic populations[14]. In the US, chronic HCV patients of European ancestry have a higher probability of being cured than patients of African ancestry[15]. It is well documented that patients originating from East Asia are more likely to achieve SVR than patients from Europe[16]. These observations suggest that host genetic variations play a critical role in patients infected with HCV.
Four recent genome-wide association studies (GWAS) independently identified several single nucleotide polymorphisms (SNPs) in the interleukin 28B (IL28B) locus. Variability at that locus is known to be associated with the successful treatment of HCV infection[15,17-19]. Ge et al[15] identified an rs12979860 SNP residing approximately 3 kilo-bases upstream of the IL28B gene as the variant most strikingly associated with SVR, and demonstrated that patients with the CC genotype had a higher SVR rate than those with the TT genotype. Suppiah et al[17], Tanaka et al[18], and Rauch et al[19] found a strong association between another SNP, rs8099917 located 8 kilo-bases upstream of the IL28B gene, and SVR, which is in linkage disequilibrium with rs12979860. Tanaka et al[18] also found that the rs8099917 GG allele was the most significant host factor for predicting nonvirological response after adjusting for confounding factors. Their results suggest that Japanese patients with a minor GG allele may require new antiviral therapies to achieve SVRs.
These exciting discoveries show promise that detecting the IL28B genotype will help predict treatment response, thus guiding clinical decisions in the choice of treatment regimens for chronic HCV infection[7]. Such discoveries may ultimately usher in the era of personalized treatment for HCV infection[18,20].
An easily performed and specific method for the detection of IL28B genotypes is urgently needed to improve the currently available methods. Currently there are several assays for the detection of IL28B genotypes. However, due to technological limitations, these assays focus on either rs8099917 or rs12979860. Until now, there has been no assay for the simultaneous detection of rs8099917 or rs12979860. Furthermore, none of the available assays can be routinely applied in clinical practice. For example, direct sequencing, widely used in current research, is not suitable for clinical practice because of its complex technology, high cost, and time consumption.
The primary aim of this study was to develop a novel biosensor-based clinical microarray (BBM) assay stemming from of our previous research to detect the IL28B genotypes containing rs12979860 and rs8099917 SNPs, and to evaluate the specificity and sensitivity of the test[21,22].
Plasmids (Invitrogen, Shanghai, China) confirmed by direct sequencing to contain both rs8099917 and rs12979860 SNPs, were used to construct the probes that were arrayed on the BBM; and plasmids diluted to 104 copies/mL were used to determine the assay specificity. The two alleles of each SNP were tested separately. Blood samples obtained from HCV seropositive patients at the 5th Affiliated Hospital of Sun Yat-Sen University (Zhuhai, China) were used to evaluate the clinical sensitivity and specificity of the BBM assay. The study protocol was approved by the Ethics Committee of our hospital and all patients enrolled in this study provided written informed consent.
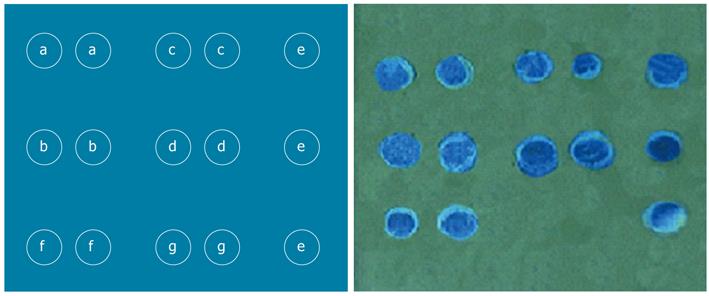
Several probes (Invitrogen, California, United States), highly specific for rs8099917 or rs12979860 polymorphisms, plus positive and negative quality control probes, were designed and synthesized. All the probe sequences included in this study were from a public database (http://www.ncbi.nlm.nih.gov/) and were based on the known human genome in the region of the IL28B gene. After optimization, the probes specific for the SNPs as well as positive and negative control probes were arrayed on the BBM as shown in Figure 1. The positive control probes were used to monitor the assay process. The negative probes served to monitor contamination. The probe sequences are summarized in Table 1.
| Names | Sequences |
| IL28B primers forward (9860) | GTGCCTGTCGTGTACTGAAC |
| IL28B primers reverse (9860) | CGCTGAGCACTGCCTGGGC |
| IL28B primers forward (9917) | ATTTGTCACTGTTCCTCCT |
| IL28B primers reverse (9917) | GCCCTAACTGATACGCTATAA |
| rs12979860 probe (C allele) | AAAAAAAAAAAGCTCCCCGAAGGCGCGAACCAGGGTTGAAT |
| rs12979860 probe (T allele) | AAAAAAAAAAAGCTCCCCGAAGGCGTGAACCAGGGTTGAAT |
| rs8099917 probe (T allele) | AAAAAAAAAACTTTCTGTGAGCAATTTCACCCAAATTGGAA |
| rs8099917 probe (G allele) | AAAAAAAAAACTTTCTGTGAGCAATGTCACCCAAATTGGAA |
Human genomic DNA was extracted from 200 μL of patient blood samples using 200 μL commercially available DNA extraction buffer (Qiagen, Dusseldorf, Germany) according to the manufacturer’s instructions. To determine the SNP genotypes, two sets of special primers were designed to amplify the rs8099917 and rs12979860 SNP fragments within the IL28B gene region (Table 1). The extracted DNA was amplified by polymerase chain reaction (PCR). The reaction mixture (25 μL) contained 5 μL DNA, 2.5 μL of 10 μmol/L buffer (Juntan, Shanghai, China), 0.5 μL deoxynucleoside triphosphates (Roche, Basel, Switzerland), 1.5 μL primers (Invitrogen, California, United States), 1.25 U hot Taq polymerase (Juntan, Shanghai, China), and 14.25 μL high-performance liquid chromatography-grade water. Simultaneous amplification of two SNPs was carried out under the following conditions: an initial denaturation at 95 °C for 10 min, 40 cycles at a denaturation temperature of 94 °C for 30 s, annealing at 56 °C for 30 s, and extension at 72 °C for 30 s, with a final prolonged extension at 72 °C for 5 min. All PCR products were visualized on a 2% agarose gel or by polyacrylamide gel electrophoresis (PAGE) electrophoresis.
All SNP amplicons were subjected to reverse hybridization and direct sequencing. A 10 μL volume of PCR amplified product was heated at 99 °C for 5 min and denatured, with subsequent cooling for 5 min on ice. Then the amplified product was placed on the surface of the BBM and incubated at 50 °C for 60 min with a prepared hybridization reaction mixture. The BBM was eluted three times at 45 °C, incubated with anti-biotin horseradish peroxidase reagent at room temperature (10-35 °C) for 10 min, rinsed three times with a buffer wash, and incubated with tetramethylbenzidine for 2 min in the dark. Finally, the residues on the BBM were washed in 0.1 × standard saline citrate and distilled water at room temperature so as to obtain a clear signal. The remaining amplified PCR product was sent for direct sequencing.
Four synthetic plasmids, each including an SNP allele (rs12979860CC, rs12979860TT, rs8099917GG, and rs8099917TT) and 40 HCV-seropositive blood samples were used to validate the specificity of the assay to detect the two SNPs. Each successfully amplified product obtained from these plasmids was evaluated by the BBM assay, and the results thus obtained were compared with the their known genotypes. For the clinical evaluation, amplicons obtained from patient blood samples were tested under the same conditions as the amplified plasmid products. The percent agreement of the BBM assay results with the results of direct sequencing was determined for both the synthetic probes and the clinical samples.
To estimate the sensitivity and detection limit of the SNP genotype assay in clinical practice, ten blood samples were used. The sensitivity calculation was based the white blood cell (WBC) count in the original sample and the amount of DNA extracted from the WBC remaining in serial dilutions of each sample. The WBC counts were determined in each sample and then diluted to 101-103 cells/μL. The DNA was extracted from 1 μL of each sample and was amplified. All samples with successful PCR amplification were evaluated for SNP genotype by the BBM assay.
The kappa coefficient was used to measure the agreement between direct sequencing and the BBM assay for the detection of SNP genotype. A kappa value > 0.75 was defined as substantial agreement, and a kappa > 0.95 as perfect agreement. A P < 0.05 was considered statistically significant.
Both fragments were amplified simultaneously in the one-tube system, and the amplicons were analyzed on PAGE electrophoresis (Figure 2). The length of the two amplified fragments was about 137 and 159 bp, being consistent with the length of the target fragments. The results indicated that these two target fragments could be amplified in a single tube PCR system.
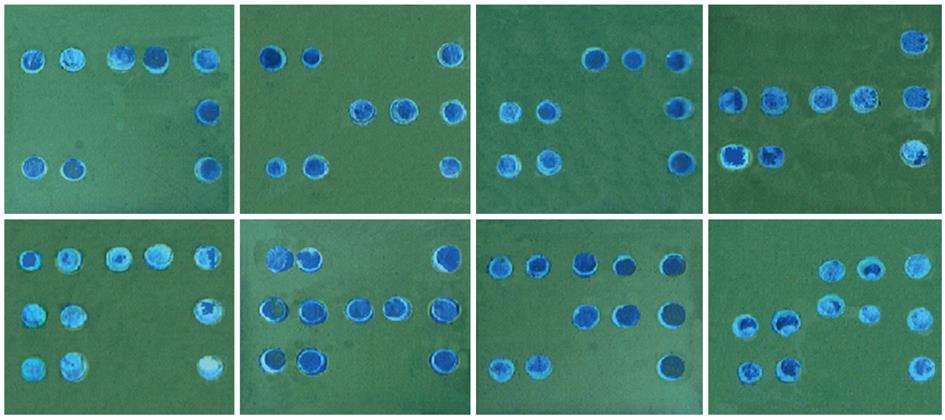
As shown in Figures 1 and 3, the signals on the chip appeared sufficiently distinct seen with the unaided eye and photographed with a normal digital camera. It was thus confirmed that no special equipment was needed to read the BBM assay results. Moreover, each allele of both rs8099917 rs12979860 was successfully detected at the same time. The BBM assay was able to simultaneously genotype the two SNPs.
As shown in Figure 3, the positive signals were displayed clearly, and there were no signals from the sites containing the negative probes. There were no ambiguous signals on the chip background, which could have interfered with precise interpretation of the results. This suggested that the assay was specific for the detection of rs8099917 and rs12979860 polymorphisms.
To confirm the BBM assay specificity for detection of each allele, we tested all the possible genotypes using the available synthetic plasmids. As shown in Figure 3, the BBM assay clearly differentiated all the genotypes and haplotypes of the two SNPs. The signals of the alleles of each SNP were clearly differentiated from each other, with no visible evidence of cross-reaction at any SNP binding site. Data obtained using 30 plasmid amplicons showed the same results for all the known genotypes of rs12979860 or rs8099917 (Tables 2 and 3), and its agreement rate with the plasmids was up to 100% in our tests. The results showed that the BBM was an accurate assay for genotype determination of these two SNPs.
| BBM assay | Plasmids with given genotype | Total | ||
| CC | CT | TT | ||
| rs12979860 | ||||
| CC | 10 | 0 | 0 | 10 |
| CT | 0 | 10 | 0 | 10 |
| TT | 0 | 10 | 10 | 10 |
| Total | 10 | 10 | 10 | 30 |
| rs8099917 | ||||
| TT | 10 | 0 | 0 | 10 |
| GT | 0 | 10 | 0 | 10 |
| GG | 0 | 0 | 10 | 10 |
| Total | 10 | 10 | 10 | 30 |
| BBM assay | Direct sequencing | Total | ||
| CC | CT | TT | ||
| rs129798601 | ||||
| CC | 24 | 0 | 0 | 24 |
| CT | 3 | 11 | 0 | 14 |
| TT | 0 | 0 | 2 | 2 |
| Total | 27 | 11 | 2 | 40 |
| rs80999172 | ||||
| TT | 28 | 0 | 0 | 28 |
| GT | 2 | 9 | 0 | 11 |
| GG | 0 | 0 | 1 | 2 |
| Total | 30 | 9 | 1 | 40 |
To validate the BBM assay for clinical application, the amplicons derived from the 40 blood samples taken from HCV-infected patients were examined by both direct sequencing and BBM. As shown in Tables 2 and 3, the percent of agreement of the BBM assay results and direct sequencing of rs8099917 and rs12979860 were 95% and 92.5%, respectively. The corresponding kappa values were 0.88 and 0.85, respectively, with a P value < 0.05. The BBM assay accurately detected the two SNPs in clinical samples. Discordance of the BBM assay and direct sequencing of PCR amplicons most often occurred with heterozygous clinical samples.
Ten clinical blood samples and a series of sequential dilutions were used to validate the sensitivity and detection limit of BBM. Blood samples with WBC counts ranging from 101 to 103 cells/μL were amplified with BBM primers, and visualized by PAGE electrophoresis. Amplicons with detectable human genomic DNA were assayed by reverse hybridization with the BBM. The hybridization signal was visualized clearly in the BBM assay in a total of 20 dilutions with WBC counts in the patient blood samples ranging from 102 to 103 cells/μL.
The combined therapy of pegylated interferon plus ribavirin is currently the standard of care for chronic HCV infection around the world[6]. Its administration in clinical practice has resulted in substantial progress in the treatment of chronic hepatitis C over the past decades. However, several persistent shortcomings of the combined therapy prevent some patients from completing the entire 48-wk course of treatment. Patient adherence is frequently compromised by an inability to tolerate adverse reactions or the many weeks of routine subcutaneous injections, and the high cost of the drugs[8,9,12]. The limited efficacy of pegylated interferon/ribavirin therapy to produce SVR, which varies unpredictably from patient to patient, makes it difficult for health care providers to make an informed choice of treatment regimens[18-20]. There is an urgent need of personalized therapy for chronic hepatitis C.
The fact that about 30% of patients achieve natural clearance following acute hepatitis C virus infection and ethnic differences in response to treatment suggests that host genetic variation plays a critical role in the drug response[2,11,17,19,20]. Four recent studies have found that two SNPs, rs8099917 and rs12979860, located near the IL28B gene were highly associated with treatment response and spontaneous clearance following acute hepatitis C infection[15,17-19]. Subsequent studies found that the two SNPs had an impact on the occurrence of various side effects of the combined therapy, treatment response and the HCV RNA genotype distribution of the infecting virus[23,24]. Other studies have shown that IL28B polymorphisms were a significant, independent predictive factor regardless of HCV genotype or HCV RNA load. The determination of IL28B polymorphisms may assist in evaluating the likelihood of response to treatment with peg-interferon and ribavirin therapy in patients chronically infected with HCV, especially for genotype 1 patients[25-28]. The recent significant findings regarding IL28b SNPs and the genotyping of HCV showed how personalizing treatment for HCV infection may be possible[24-27]. The development of an accurate assay for the detection of rs8099917 and rs12979860 polymorphisms, and progress in HCV genotyping will help both clinicians and patients choose the treatment regimen and its anticipated duration[22] and this may be an early step in the era of personalized therapy for chronic hepatitis C[20,29,30].
Host genetic diversity is of great significance in making an informed decision regarding the risk-benefit treatment and the likelihood of success for any individual treatment, so developing a simple, rapid and clinically available assay is an urgent demand. An assay that could accurately and rapidly detect the IL28b SNPs is a priority for clinical practitioners. Developing a novel and efficient assay for the detection of IL28B polymorphisms has been the focus of numerous researches.
Direct sequencing of PCR amplicons, restriction fragment length polymorphism (RFLP) and traditional microarrays are valuable research techniques. However, the technical demands of these assays make them unsuitable for routine diagnostic use in clinical practice. For example, direct sequencing is technologically complicated, time-consuming and needs special expertise[28,29]. The RFLP is almost impossible to be used for the detection of all significant SNPs since it requires a specific cutting site and restriction enzyme for the particular site[31]. Microarray assays widely used in the previous research, apart from being provided by specialized and high-cost facilities, can only detect either of the two SNPs separately[19,21-23]. No microarray assay was available currently that can detect the two SNPs simultaneously. There is a real need for a highly sensitive and specific rapid assay for the detection of IL28B polymorphisms that is easy to perform and compatible with clinical practice. The BBM described in our report may be an ideal clinical assay.
As depicted in Figure 3, the positive signals on the chip were sufficiently distinct to be completely discriminated from negative results. The results with plasmid amplicons showed that all the SNP alleles could be detected successfully and that heterozygous and homozygous alleles could be distinguished. Of 40 plasmid amplicons employed to validate the specificity of the BBM assay, a complete agreement between the results of the BBM assay and known plasmid sequences was observed, confirming the high specificity of the BBM assay in determining the rs8099917 and rs12979860 genotypes.
The clinical blood samples successfully amplified by the BBM assay primers were sent for both direct sequencing and reverse hybridization with BBM. The BBM assay showed a high agreement with direct sequencing in detecting the rs8099917 and rs12979860 polymorphisms. The agreement rates were 95% and 92.5%, and the kappa values were 0.88 and 0.85 for rs8099917 polymorphisms and rs12979860, respectively. These data indicated that the BBM assay showed a good agreement with direct sequencing and that it would be a satisfactory novel assay when used for determination of the two SNPs. The main difference between the BBM results and direct sequencing was the detection of allele heterozygotes. The BBM assay was more sensitive in detecting heterozygous alleles, which was consistent with other reverse hybridization procedures such as line probe assay[32,33]. In brief, we demonstrated that the novel BBM assay could successfully detect all genotypes of the two SNPs simultaneously, not only in the synthetic plasmids but also in the clinical samples used in this study.
To evaluate the sensitivity and detection limit of the BBM assay, serial dilutions of 10 clinical blood samples were tested. The amount of human DNA from white blood cells in 1 μL peripheral blood containing 102 WBC/μL was sufficient for SNP detection by the BBM assay. The DNA extracted from all samples was successfully amplified, and the blood samples with a starting white blood cell count > 102 WBC/μL hybridized with the probes arrayed on the BBM. As the white blood cell count of human peripheral blood is generally greater than 103 WBC/μL, the detection limit of the BBM assay ensures that clinical blood samples would be tested accurately. This novel method thus met the sensitivity criteria for clinical diagnosis, i.e., determination of IL28B polymorphisms of HCV-infected patients.
The results indicated that the BBM assay has the advantages of simple and convenient operation as well as rapid detection. After a successful PCR amplification, it took only one hour to detect the IL28B polymorphisms and less than four hours before getting the final result. More important, the signal on the chip was clear enough to be interpreted even by the naked eye, which meant that only an ordinary digital camera was needed to record the results. The results also suggest that the BBM assay might also be used in developing countries having limited access to the latest laboratory facilities. The assay is also suitable for clinical diagnostic and research laboratories where large numbers of samples are tested on a daily basis. In short, the BBM assay is more likely to be accepted for the clinical detection of IL28B polymorphisms than the other existing methods[21,22].
The BBM assay has some disadvantages to overcome in its future development. The patient population benefiting from the BBM assay is small, being limited to patients with chronic HCV infection, meanwhile, the assay is only capable of detecting the presence of known SNPs. With the identification of more SNPs associated with drug response, novel probes have to be designed and the BBM assay has to be constantly upgraded.
In conclusion, we have demonstrated that the BBM assay is simple, rapid, accurate, and suitable for clinical application. The assay is highly sensitive and specific, and can simultaneously perform the detection of rs8099917 and rs12979860 genotypes, thereby enabling further progress in the diagnosis and treatment of chronic HCV infection.
Antivirals are essential to the therapeutic management of hepatitis C virus (HCV)-infected patients. However, sustained virological response is not achieved in all patients who receive the standard combination therapy of once-weekly injections of pegylated interferon plus daily oral ribavirin or even in those treated with the triple therapy regimen. Recent research has characterized single nucleotide polymorphisms (SNPs) in the interleukin 28B (IL28B) gene as the most important host factor influencing the efficacy of HCV therapy. Many scientists believe that identifying each of the IL28B SNPs related to HCV treatment response will usher in a new era of HCV personalized therapy.
Four recent genome-wide association studies (GWAS) independently identified several SNPs in the IL28B gene locus that are associated with an individual’s ability to respond to therapy for HCV infection. Ge et al[15] characterized rs12979860 as the variant most strongly associated with SVR, and demonstrated that patients with the CC genotype have a higher SVR rate than those with the TT genotype. In contrast, studies demonstrated that rs8099917 has the strongest association with SVR. However, GWAS are capable of only testing SNPs in isolation and could not determine if interaction between these two SNPs, or more, can affect a patient’s response to HCV therapy. Thus, it is urgent to develop an accurate and easy clinical assay to test the panel of IL28B SNPs in a patient and assess the effect of multiple SNPs on SVR.
This is the first report of an assay that is capable of defining the rs8099917 and rs12979860 SNPs using clinical samples. This innovative biosensor-based microarray (BBM) assay can identify the two SNPs in less than four hours. In addition, evidence is provided to show that the new assay can accurately identify all of the SNPs and gene alleles with plasmids. There was a good concordance between BBM-detected SNPs in clinical samples and direct sequencing results of PCR amplicons and plasmids. Finally, the excellent sensitivity and reproducibility of the BBM assay supports the clinical applicability of this new detection approach.
This study aimed to develop a clinically applicable assay to accurately and rapidly detect IL28B SNPs in patient samples. The results indicate that the BBM assay is simple, rapid, accurate, and highly sensitive and specific in detecting the rs8099917 and rs12979860 genotypes. Thus, this novel assay has promise for clinical application and may facilitate accurate and timely prognosis of HCV-infected patients so that the appropriate therapies may be initiated earlier.
BBM is a novel SNP detection assay based on biosensor technology, its most obvious advantage is in situ amplification that facilitates easy interpretation of results; rs8099917 and rs12979860 are SNPs in the IL28B gene locus; SNP is a single nucleotide polymorphism.
In this work, the authors describe a new BBM assay to rapidly detect two SNPs in the IL28B gene. The interest of the paper is its description of this rapid and inexpensive technique to detect IL28B SNPs in clinical samples. The results suggest that the BBM assay is a simple, rapid, sensitive, and highly specific method that may be applied clinically to detect rs8099917 and rs12979860 without any large-scale instrumentation.
Peer reviewers: Dr. Assy Nimer, MD, Assistant Professor, Liver Unit, Ziv Medical Centre, BOX 1008, Safed 13100, Israel; Juan-Ramón Larrubia, PhD, Gastroenterology Unit and Liver Research Unit, Guadalajara University Hospital, Donante de Sangre s/n, 19002 Guadalajara, Spain
S- Editor Gou SX L- Editor Ma JY E- Editor Zhang DN
| 1. | Hepatitis C. Wkly Epidemiol Rec. 2011;86:445-447. [PubMed] |
| 2. | Thomas DL, Seeff LB. Natural history of hepatitis C. Clin Liver Dis. 2005;9:383-98, vi. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 267] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 3. | Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798-801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1706] [Cited by in RCA: 1687] [Article Influence: 105.4] [Reference Citation Analysis (0)] |
| 4. | Poynard T, Bedossa P, Opolon P. Natural history of liver fibrosis progression in patients with chronic hepatitis C. The OBSVIRC, METAVIR, CLINIVIR, and DOSVIRC groups. Lancet. 1997;349:825-832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2199] [Cited by in RCA: 2160] [Article Influence: 77.1] [Reference Citation Analysis (0)] |
| 5. | Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 2010;7:448-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 979] [Cited by in RCA: 1060] [Article Influence: 70.7] [Reference Citation Analysis (0)] |
| 6. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: management of hepatitis C virus infection. J Hepatol. 2011;55:245-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 889] [Cited by in RCA: 919] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 7. | Jesudian AB, Gambarin-Gelwan M, Jacobson IM. Advances in the treatment of hepatitis C virus infection. Gastroenterol Hepatol (. N Y). 2012;8:91-101. [PubMed] |
| 8. | Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL, Häussinger D, Diago M, Carosi G, Dhumeaux D. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975-982. [PubMed] |
| 9. | Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958-965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4736] [Cited by in RCA: 4558] [Article Influence: 189.9] [Reference Citation Analysis (0)] |
| 10. | von Wagner M, Huber M, Berg T, Hinrichsen H, Rasenack J, Heintges T, Bergk A, Bernsmeier C, Häussinger D, Herrmann E. Peginterferon-alpha-2a (40KD) and ribavirin for 16 or 24 weeks in patients with genotype 2 or 3 chronic hepatitis C. Gastroenterology. 2005;129:522-527. [PubMed] [DOI] [Full Text] |
| 11. | Antaki N, Craxi A, Kamal S, Moucari R, Van der Merwe S, Haffar S, Gadano A, Zein N, Lai CL, Pawlotsky JM. The neglected hepatitis C virus genotypes 4, 5 and 6: an international consensus report. Liver Int. 2010;30:342-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 120] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 12. | Fried MW. Side effects of therapy of hepatitis C and their management. Hepatology. 2002;36:S237-S244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 320] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 13. | Esteban JI, Sauleda S, Quer J. The changing epidemiology of hepatitis C virus infection in Europe. J Hepatol. 2008;48:148-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 309] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 14. | Wapner J. Pharmacogenomics. Gene variants affect hepatitis C treatment, but link is elusive. Science. 2010;330:579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 15. | Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2776] [Cited by in RCA: 2723] [Article Influence: 170.2] [Reference Citation Analysis (0)] |
| 16. | Yan KK, Guirgis M, Dinh T, George J, Dev A, Lee A, Zekry A. Treatment responses in Asians and Caucasians with chronic hepatitis C infection. World J Gastroenterol. 2008;14:3416-3420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 35] [Cited by in RCA: 38] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 17. | Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100-1104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1505] [Cited by in RCA: 1504] [Article Influence: 94.0] [Reference Citation Analysis (0)] |
| 18. | Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105-1109. [PubMed] |
| 19. | Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338-145, 1338-145. [PubMed] [DOI] [Full Text] |
| 20. | Iadonato SP, Katze MG. Genomics: Hepatitis C virus gets personal. Nature. 2009;461:357-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 21. | Song J, Lin J, Ni X, Kong X, Wu H, Xie Y, Mao Y. [Development of lamivudine-resistance microarray and its reliability in clinical examination]. Zhonghua Gan Zang Bing Zazhi. 2002;10:468. [PubMed] |
| 22. | Tang J, Wu B, Song J. Development and primary clinical evaluation of a biosensor based microarray for hepatitis b virus drug resistance mutation assay. Hepatol Int. 2009;3:223 [DOI 10.1007/S12072-008-9167-5]. |
| 23. | Lotrich FE, Loftis JM, Ferrell RE, Rabinovitz M, Hauser P. IL28B Polymorphism Is Associated with Both Side Effects and Clearance of Hepatitis C During Interferon-Alpha Therapy. J Interferon Cytokine Res. 2010;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Montes-Cano MA, García-Lozano JR, Abad-Molina C, Romero-Gómez M, Barroso N, Aguilar-Reina J, Núñez-Roldán A, González-Escribano MF. Interleukin-28B genetic variants and hepatitis virus infection by different viral genotypes. Hepatology. 2010;52:33-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 166] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 25. | McCarthy JJ, Li JH, Thompson A, Suchindran S, Lao XQ, Patel K, Tillmann HL, Muir AJ, McHutchison JG. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology. 2010;138:2307-2314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 261] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 26. | Sarrazin C, Susser S, Doehring A, Lange CM, Müller T, Schlecker C, Herrmann E, Lötsch J, Berg T. Importance of IL28B gene polymorphisms in hepatitis C virus genotype 2 and 3 infected patients. J Hepatol. 2011;54:415-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 180] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 27. | Kurosaki M, Tanaka Y, Nishida N, Sakamoto N, Enomoto N, Honda M, Sugiyama M, Matsuura K, Sugauchi F, Asahina Y. Pre-treatment prediction of response to pegylated-interferon plus ribavirin for chronic hepatitis C using genetic polymorphism in IL28B and viral factors. J Hepatol. 2011;54:439-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 97] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 28. | Kawaoka T, Hayes CN, Ohishi W, Ochi H, Maekawa T, Abe H, Tsuge M, Mitsui F, Hiraga N, Imamura M. Predictive value of the IL28B polymorphism on the effect of interferon therapy in chronic hepatitis C patients with genotypes 2a and 2b. J Hepatol. 2011;54:408-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 29. | Imazeki F, Yokosuka O, Omata M. Impact of IL-28B SNPs on control of hepatitis C virus infection: a genome-wide association study. Expert Rev Anti Infect Ther. 2010;8:497-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 30. | Clark PJ, Thompson AJ, McHutchison JG. IL28B genomic-based treatment paradigms for patients with chronic hepatitis C infection: the future of personalized HCV therapies. Am J Gastroenterol. 2011;106:38-45. [PubMed] |
| 31. | Nakamoto S, Kanda T, Imazeki F, Wu S, Arai M, Fujiwara K, Yokosuka O. Simple assay based on restriction fragment length polymorphism associated with IL28B in chronic hepatitis C patients. Scand J Gastroenterol. 2011;46:955-961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 32. | Hussain M, Fung S, Libbrecht E, Sablon E, Cursaro C, Andreone P, Lok AS. Sensitive line probe assay that simultaneously detects mutations conveying resistance to lamivudine and adefovir. J Clin Microbiol. 2006;44:1094-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 55] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 33. | Degertekin B, Hussain M, Tan J, Oberhelman K, Lok AS. Sensitivity and accuracy of an updated line probe assay (HBV DR v.3) in detecting mutations associated with hepatitis B antiviral resistance. J Hepatol. 2009;50:42-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |