Published online Nov 28, 2012. doi: 10.3748/wjg.v18.i44.6437
Revised: August 23, 2012
Accepted: August 26, 2012
Published online: November 28, 2012
AIM: To identify hepatitis B virus polymerase gene mutations during antiviral therapy using lamivudine-adefovir sequential monotherapy followed by lamivudine-adefovir combination therapy.
METHODS: The patient cohort included four adult chronic hepatitis B patients who had undergone sequential monotherapy, first with lamivudine (LMV) and then, after developing viral breakthrough, with adefovir (ADV) therapy. All of the patients had non-response or viral breakthrough after LMV-ADV sequential monotherapy, which resulted in the switching of their antiviral regimen to LMV-ADV combination therapy. Eleven serum samples from the four patients who showed non-response to rescue LMV-ADV combination therapy were collected sequentially at a time before the antiviral treatment and then during the LMV monotherapy, ADV monotherapy, and LMV-ADV combination therapy. For the genotypic analysis, the whole 1310-bp polymerase gene region was amplified, cloned and sequenced.
RESULTS: All patients had been previously treated with 100 mg of LMV once daily for a 15- to 26-mo period. The emergence of resistance mutations to LMV, such as rtM204V/I and/or rtL180M, were found in all patients. Their antiviral regimens were switched to ADV monotherapy as the second line treatment. All patients had viral breakthrough or non-response after the LMV-ADV sequential monotherapy. ADV-resistant mutations were detected after 13 to 19 mo of LMV-ADV sequential monotherapy. The rtA181V/T mutations were predominantly identified during the ADV treatment in the LMV-resistant patients. Twenty-seven of 38 clones were combined with an amino acid change at rt181; three clones had mutations in rt236 and one clone had a combined mutation. The rtA181V/T mutations were not suppressed by the LMV-ADV combination therapy. Thirty-nine of 64 clones showed an rtA181V/T mutation and six clones showed combined mutations in rt181 and rt236. Mutations in rt204 re-emerged during the combination treatment. The rt181 and rt204 mutations did not co-exist in one clone.
CONCLUSION: Add-on lamivudine therapy with adefovir for adefovir resistance may not suppress the pre-existing adefovir-resistant mutation that develops during lamivudine-adefovir sequential monotherapy.
- Citation: Ko SY, Kim BK, Kwon SY, Kim KH, Kim JH, Choe WH, Lee CH. Clonal evolution of hepatitis B virus polymerase gene mutations during lamivudine-adefovir combination treatment. World J Gastroenterol 2012; 18(44): 6437-6446
- URL: https://www.wjgnet.com/1007-9327/full/v18/i44/6437.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i44.6437
Chronic infection with hepatitis B virus (HBV) can result in cirrhosis and hepatocellular carcinoma. The ultimate goal of antiviral therapy for chronic hepatitis is to prevent its devastating complications of decompensated liver cirrhosis and hepatocellular carcinoma. In recent years, the treatment of chronic hepatitis B (CHB) has been improved by introducing nucleos(t)ide analogs (NAs) such as lamivudine (LMV), adefovir (ADV), entecavir (ETV), telbivudine (LDT), and tenofovir (TDF)[1-5]. However, drug-resistant HBV mutants frequently arise during treatment with NAs, leading to treatment failure and progression to liver disease[6]. ADV, a phosphate acyclic nucleotide analog of adenosine monophosphate, is a potent inhibitor of HBV reverse transcriptase in the wild-type HBV, and in lamivudine-, telbivudine-, and entecavir-resistant mutants[7,8]. Switching to ADV has become the most widely-used option since the emergence of LMV-resistant mutations, especially in Asian countries[8]. The 5-year cumulative probability of genotypic resistance and viral breakthrough was 65.6% and 61.8%, respectively, in LMV-resistant CHB patients[3]. Unfortunately, ADV resistance occurs more frequently in the second-line treatment of lamivudine-resistant patients than in treatment-naive patients[4]. According to large-scale clinical studies of ADV, the rtA181V and rtN236T mutations associated with resistance to ADV were identified in the HBV DNA polymerase gene[5]. In vitro studies demonstrated that those two mutations decreased susceptibility to ADV by 4.3- to 23-fold[8,9]. Other minor mutations resistant to ADV, such as rtS85A, rtT184S, rtQ215S, rtP237H, rtN238D, and rtM250L, have been reported, but the significance of these mutations is unclear[4,10,11]. In 2006, it was reported that a novel ADV-resistant HBV variant rtI233V (substitution of isoleucine by valine) was reported to be a cause of primary resistance to ADV[12,13]. Studies also showed that rtI233V was sensitive to TDF in vitro. However, this result was not confirmed in recent studies. To our knowledge, little is known about clonal analysis in patients who successively received LMV monotherapy, ADV monotherapy, and LMV-ADV combination therapy. We investigated the clonal evolution of genetic mutations in the HBV polymerase gene in non-responders to LMV-ADV sequential monotherapy followed by LMV-ADV combination therapy.
From January 2003 to July 2007, four adult chronic hepatitis B patients who showed non-response during LMV-ADV combination therapy were identified at Konkuk University Medical Center in Seoul, South Korea. All patients had undergone sequential monotherapy, first with LMV and then, after developing viral breakthrough, with ADV therapy. All of the patients had non-response or viral breakthrough after the LMV-ADV sequential monotherapy which resulted in the switching of their antiviral regimen to LMV-ADV combination therapy. The patients were then followed from when they were switched to an antiviral regimen. These patients gave written informed consent for the treatment of CHB and testing for antiviral resistance. Approval for this study was obtained from the Institutional Review Board at the Konkuk University Medical Center.
Before ADV plus LMV, the clinical and laboratory data of each patient were analyzed, including the quantification of serum HBV DNA levels. After the initiation of LMV-ADV combination therapy, clinical, biochemical, and virological parameters, such as aspartate aminotransferase, alanine aminotransferase (ALT), and serum HBV DNA levels, were monitored every month up to three months. A clinical diagnosis of liver cirrhosis was based on imaging findings, using methods such as abdominal computed tomography or magnetic resonance imaging, together with compatible clinical features such as esophageal varices or thrombocytopenia. The severity of liver cirrhosis was classified according to the Child-Pugh score.
Viral breakthrough was defined as a ≥ 1 log10 IU/mL increase in HBV DNA from the nadir on two consecutive occasions after an initial virologic response or an initial decline in HBV DNA by ≥ 1 log10 IU/mL. Non-response was defined as a decrease in serum HBV DNA ≤ 2 log10 IU/mL after at least 24 wk of therapy.
Serial samples were collected from each patient at the time of initiation of each antiviral agent, every three months during treatment and at the time of viral breakthrough and stored frozen at -80 °C. The serum HBV DNA levels were quantified by real-time polymerase chain reaction, with a lower limit of detection of 2.6 log10copies/mL (approximately 400 copies/mL). The HBV DNA quantification was also performed before cloning and sequencing using the COBAS TaqMan HBV test (Roche Molecular Systems Inc, Pleasanton, CA, United States).
Using the stored serum samples, HBV DNA polymerase mutations were analyzed using a matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS)-based genotyping assay, termed a restriction fragment mass polymorphism (RFMP; GeneMatrix, Yongin, South Korea)[14]. LMV-resistant mutations at rt180 or rt204, and ADV-resistant mutations at rt181 or rt236, were detected using this method.
For the genotypic analysis, the entire 1310-bp polymerase gene region was amplified, cloned, and sequenced. To analyze the complete sequence of HBV reverse transcriptase (RT) from four patients with resistant mutations during LMV-ADV combination therapy, we isolated HBV DNA from the patients’ sera at the time of viral breakthrough during the antiviral therapy. DNA was extracted using a QIAamp MinElute virus spin kit (Valencia, CA, United States) according to the manufacturer’s protocol. To generate HBV1.2mers harboring patient-derived RT mutations, we amplified the RT region of the HBV genome using the following primers: forward, 5’-AAT CTT CTC GAG GAC TGG GGA CCC TGC ACC-3’ (the Xho I site is underlined) and reverse, 5’-GAG CAG CCA TGG GAA GGA GGT GTA TTT CCG-3’ (the Nco Isite is underlined)[15]. The purified products were digested with Xho Iand Nco I, after which the wild-type RT sequence of the HBV1.2mer was swapped for the patient-derived sequence. All clones were confirmed by sequencing.
Four patients were included in this study. The age of the patients was between 39 and 58 years; all were men who had cirrhosis. All patients had been previously treated with 100 mg of LMV once daily for a 15- to 26-mo period. The emergence of resistant mutations to LMV, such as rtM204V/I and/or rtL180M, was found in all patients. Their antiviral regimens were switched to ADV monotherapy (10 mg/d) as the second line treatment and no patient required a dose reduction. All patients had viral breakthrough or non-response after the LMV-ADV sequential monotherapy (Table 1). ADV-resistant mutations, including rtA181V/T or rtN236T, were detected after 13 to 19 mo of LMV-ADV sequential monotherapy. LMV was added to the ongoing ADV treatment as a salvage therapy.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | |
| Baseline characteristics | ||||
| Gender | Male | Male | Male | Male |
| Age (yr) | 39 | 58 | 51 | 42 |
| Liver disease | Cirrhosis | Cirrhosis | Cirrhosis | Cirrhosis |
| Initial viral load (copies/mL) | 3.0 × 107 | 3.3 × 107 | 7.0 × 108 | 5.3 × 108 |
| HBeAg | + | + | + | + |
| During LMV mono-therapy | ||||
| Duration of LMV (mo) | 26 | 22 | 15 | 25 |
| LMV-resistant mutants1 | M204I + L180M | M204V | M204I + L180M | M204I + L180M |
| Response to LMV | VT | VT | VT | VT |
| During ADV mono-therapy | ||||
| Duration of ADV (mo) | 18 | 16 | 19 | 13 |
| ADV-resistant mutants1 | A181V | A181V/T | A181V | A181V |
| Response to ADV | NR | NR | NR | NR |
| During LMV-ADV combination therapy | ||||
| Viral load at the start of | ||||
| LMV-ADV (copies/mL) | 8.0 × 107 | 8.0 × 107 | 1.2 × 105 | 3.1 × 106 |
| Duration of LMV-ADV (mo) | 9 | 13 | 11 | 15 |
| Resistant mutants1 | A181V | A181T + M204I | A181V | A181V |
| Response to LMV-ADV | NR | NR | NR | NR |
| Current treatment | LMV + ADV | LMV + TDF | LMV + TDF | LMV + TDF |
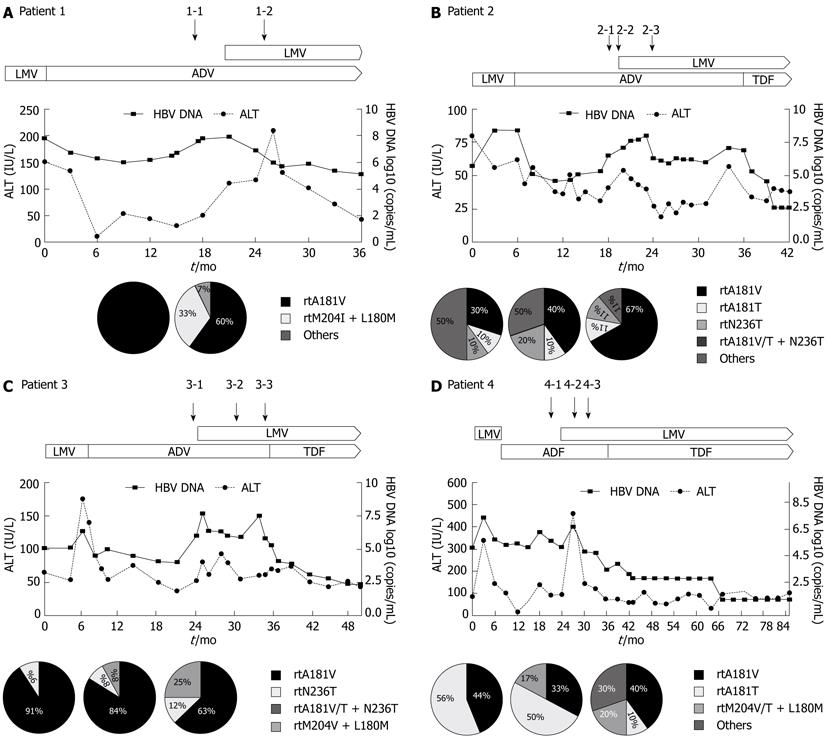
The outcome of the LMV-ADV combination therapy in the patients with ADV resistance is demonstrated in Table 1. The duration of the combination therapy was 9 to 15 mo. Patient 1 had viral breakthrough after 26 mo of LMV monotherapy. LMV was stopped and the patient was switched to ADV. Non-response occurred after 18 mo of ADV monotherapy. Patient 2 experienced non-response after 16 mo of second-line ADV monotherapy. Patient 3 had viral breakthrough after 15 mo of LMV monotherapy. Non-response occurred after 19 mo of second-line ADV monotherapy. Patient 4 experienced non-response after 13 mo of second-line ADV monotherapy. One patient continued to receive LMV-ADV combination therapy and three patients were switched to TDF-LMV combination therapy as a salvage therapy.
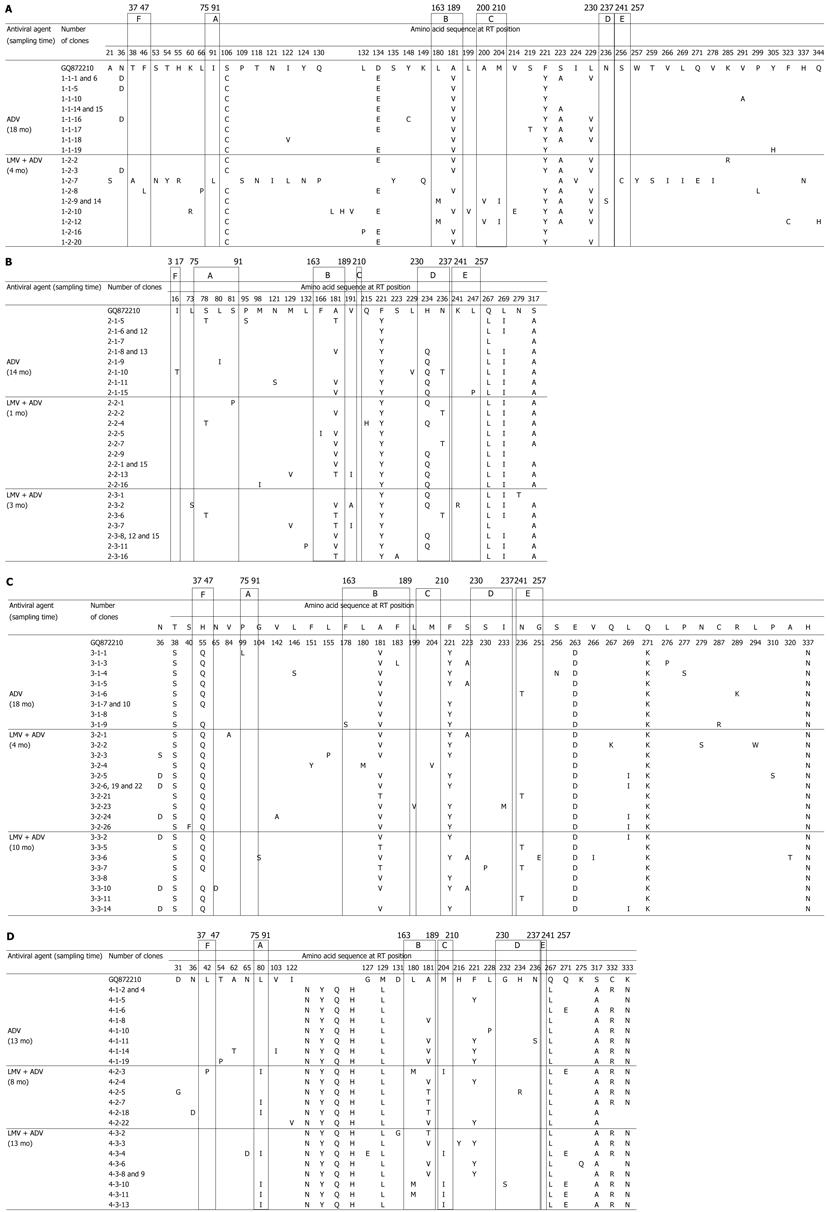
Clonal analyses were performed in four patients harboring antiviral resistant mutants at the time of viral breakthrough during the antiviral therapy. From eleven serum samples, we obtained and analyzed a total of 38 clones of the HBV polymerase gene during the ADV monotherapy and a total 64 of clones during the LMV-ADV combination therapy. The sequences were compared against the sequence from genotype C HBV (NCBI GenBank accession no. GQ872210), a wild type HBV genome isolated from the serum of a 25-year-old HBeAg-positive asymptomatic HBV carrier in our hospital.
During the ADV monotherapy, 27 of 38 clones were combined with an amino acid change at rt181. Four clones had a single mutation at rt236, one of which showed an rtA181T + N236T double mutation. During the treatment period with LMV-ADV combination therapy, 39 of 64 clones showed an rtA181V/T mutation. The viral quasispecies evolved with the extension of the combination therapy, followed by LMV-ADV sequential monotherapy (Figure 1). In all patients, clonal analyses during the LMV-ADV combination therapy revealed the maintenance of the rtA181V/T mutant that emerged during the ADV monotherapy (Figure 2). The combined mutation of rtA181V/T + N236T was detected in seven clones. The LMV-resistant mutation rtM204I reemerged in two clones. The combined LMV-resistant mutation rtM204I/V + L1801M was also detected in seven clones. However, the rt181 and rt204 mutations did not co-exist in the same clone.
The clinical course of four patients in whom the clonal analyses were performed during the antiviral treatment is illustrated in Figure 2. In patient 1, the previously noted resistant mutations to LMV, rtM204I, and rtL180M were suppressed, but a resistant mutation to ADV, rtA181V, was detected after the second-line ADV monotherapy. That ADV-resistant mutation remained detectable after 4 mo of combination therapy and a resistant mutation to LMV, rtM204I, was also detected. In patient 2, a resistant mutation to LMV, rtM204V, was suppressed, but a resistant mutation to ADV, rtA181V/T, was detected after the LMV-ADV sequential monotherapy. After 3 mo of LMV-ADV combination therapy, a resistant mutation to ADV remained detectable. In patient 3, the previously noted resistant mutations to LMV, rtM204I and rtL180M were suppressed, but a resistant mutation to ADV, rtA181V/T, was detected after 18 mo of second-line ADV monotherapy. After 10 mo of LMV-ADV combination therapy, a resistant mutation to ADV, rtA181V, remained detectable. In patient 4, resistant mutations to LMV, rtM204V and rtL180M, were suppressed, but a resistant mutation to ADV, rtA181V, was detected after the LMV-ADV sequential monotherapy. After 13 mo of LMV-ADV combination therapy, a resistant mutation to ADV remained detectable, and a resistant mutation to LMV, rtM204I, was also detected.
Sequential NA monotherapy can promote the selection for multidrug-resistant HBV mutants, but little is known about the multidrug-resistant HBV based on clonal analysis. We analyzed the full-length genomic sequence of HBV polymerase in patients who successively received LMV monotherapy, ADV monotherapy, and LMV-ADV combination therapy. This study demonstrated that an ADV mutant, rtA181V/T, was not suppressed by LMV-ADV combination therapy.
Seven drugs are approved by the Food and Drug Administration in the United States for the treatment of HBV infection: interferon alpha, pegylated interferon α-2a, LMV, ADV, ETV, LDT, and TDF[1]. The treatment of chronic hepatitis B has been improved by the introduction of these NAs. However, drug-resistant HBV mutants frequently arise after the extended use of NAs, leading to treatment failure and progression to liver disease[16]. The drug resistant mutations selected with one agent may affect the efficacy of the other NAs[17]. Several major HBV-evolutionary NA-resistance pathways have been characterized[17]. The rtM204V/I pathway is responsible for the resistance to the L-nucleosides, such as LMV, LDT, and ETV[17]. The rtN236T pathway is responsible for ADV and TDF resistance[17]. The rtA181T/V pathway is associated with resistance to LMV and ADV, and is a potential multidrug resistance pathway[17].
LMV resistance increases progressively over the course of treatment; 14%-32% of patients become resistant to the drug each year after the treatment is initiated, and more than 80% are resistant after 48 mo of treatment[6,18]. The resistance to LMV was mediated primarily by mutations rtM204I/V ± rtL180M and rtA181T/V[6,19]. Viral replication levels may be increased by compensatory mutations, such as rtL80V/I, rtI169T, rtV173L, rtT184S/G, rtS202I, and rtQ215S[6,20-23].
In treatment-naive patients, the cumulative percentage of patients who showed ADV resistance has been reported to be 0% in year 1, 3% in year 2, 11% in year 3, 18% in year 4, and 28% in year 5[4]. The cumulative percentage of patients who showed ADV resistance after ADV monotherapy in the LMV-resistant patients has been reported to be 18% in year 1 and 25% in year 2[4,14]. The resistance to ADV was initially associated with the rtA181T/V and rtN236T substitutions[6,24-26]. The rtN236T mutation does not significantly affect the sensitivity to LMV, LDT, or ETV, but does decrease the efficacy of TDF in vitro[6,27]. The rtA181T mutation has been observed in patients who developed viral resistance to LMV or to ADV following LMV breakthrough[19,28]. The other mutation, rtA181V, has been observed in patients who developed ADV resistance[4,14,29-31]. Villet et al[24] suggested that a single amino acid change at position rt181 induced cross-resistance to LMV and ADV. They also reported that the rtA181V/T substitution induced a decreased susceptibility not only to the L-nucleoside LMV, but also to the alkyl phosphonates ADV and TDF[6,24].
According to a recently reported meta-analysis, the therapeutic potential of the combination of ADV with LMV is beneficial for treating LMV-resistant CHB patients with ADV-associated mutations[32]. Switching to ADV monotherapy has been widely used in CHB patients with an LMV-resistant mutation in South Korea and has been reported to be cost-effective so far[3,33]. However, LMV-ADV sequential monotherapy has led to the frequent development of ADV resistance[4,5,14]. Combination therapy has been a practical treatment modality for patients who had an ADV-resistant mutation after LMV-ADV sequential monotherapy because TDF was not available. However, to date, the best treatment option for those with ADV resistance has not been identified.
In the present study, we observed the emergence of mutants harboring the rtA181V/T mutation in patients who were treated with LMV during ADV monotherapy[4]. The emergence of the rtA181V/T mutant was associated with non-response or viral breakthrough during LMV-ADV sequential monotherapy and LMV-ADV combination therapy. We performed the RFMP assay to detect mutations resistant to antiviral agents. This assay enables better quantitative detection of mixed populations, but the result revealed the possibility of the presence of substitutions at other sites in the HBV reverse transcriptase. To analyze the complete sequence, we sequenced all of the clones.
In the present study, clonal analyses during LMV-ADV combination therapy revealed the maintenance of an rtA181V/T mutant that emerged during ADV monotherapy. The LMV-resistant mutations rtM204I/V and/or rtL180M reemerged during the LMV-ADV combination therapy. These results showed that LMV-ADV sequential monotherapy may induce the risk of the emergence of ADV-resistant strains in patients who were treated with LMV. Our observation also showed that add-on LMV therapy for ADV resistance might not suppress the pre-existing ADV-resistant mutation that emerged during the sequential treatment with ADV monotherapy for LMV resistance. This result was consistent with the study reported by Villet et al[24], who showed that a single amino acid change at position rt181 might induce cross-resistance to LMV and ADV.
This study demonstrated that a poor response to LMV-ADV combination therapy did not always occur due to the co-localization of each drug resistance mutation in the same genome. Antiviral agents with a low or modest antiviral potency and low genetic barrier of resistance, such as LMV or ADV, may strengthen viral fitness while not being strong enough to suppress the drug mutation, and the combination of stronger drugs, such as ETV and TDF, with a high antiviral potency and high genetic barrier of resistance, would be more effective in the treatment of non-responders to LMV-ADV sequential monotherapy. Viral fitness, defined as the ability to produce infectious progeny in a defined environment, was proposed to be one of the most important factors in the process of selecting resistant mutants[34-36]. We recently reported that TDF plus LMV is a useful therapeutic option for patients with resistance or non-response to ADV, particularly in patients with cirrhosis or who had previously received sequential LMV-ADV monotherapy. TDF-containing therapy was shown to result in a rapid viral load reduction, with HBV becoming undetectable in ADV-resistant patients. At three months, a viral load reduced to a level of 4 log10 copies/mL was achieved in all six patients (100%), and fell to below the limit of detection in three patients (50%)[37].
Our study has some limitations: the small number of patients and the variations in the initial HBV DNA levels. These limitations may make establishing this combination as an optimal rescue therapy for patients with ADV-resistant HBV difficult. To verify the efficacy and safety of this regimen, further large cohort studies are warranted.
In conclusion, sequential LMV and ADV monotherapy may favor the emergence of HBV variants with cross-resistance to analogs of different chemical classes. The emergence of rtA181V/T with or without rtM204V/I resulted in viral breakthrough during ADV monotherapy and LMV-ADV combination treatment, as well as a poor response to ADV. Therefore, LMV-ADV combination therapy is not effective in patients with ADV-resistant mutations after LMV-ADV sequential monotherapy. This result may be attributed to an ADV mutation, rtA181V/T, which is responsible for cross-resistance to LMV and ADV. Therefore, careful monitoring and more effective antiviral agents should be considered in chronic hepatitis B patients with multiple resistance mutations.
The treatment of chronic hepatitis B has been improved by introducing nucleos(t)ide analogs such as lamivudine, adefovir, entecavir, telbivudine, and tenofovir. However, drug-resistant hepatitis B virus (HBV) mutants frequently arise during treatment with nucleos(t)ide analogs. Adefovir resistance occurs more frequently in the second-line treatment of lamivudine-resistant patients than in treatment-naïve patients.
Little is known about clonal analysis in patients who successively received lamivudine monotherapy, adefovir monotherapy, and lamivudine-adefovir combination therapy. This research hotspot is to demonstrate HBV polymerase gene mutations during antiviral therapy using lamivudine-adefovir sequential monotherapy followed by lamivudine- adefovir combination therapy.
According to a recently reported meta-analysis, the therapeutic potential of the combination of adefovir with lamivudine is beneficial for treating lamivudine-resistant chronic hepatitis B patients with adefovir-associated mutations. However, lamivudine-adefovir sequential monotherapy has led to the frequent development of adefovir resistance. In the present study, clonal analyses during lamivudine-adefovir combination therapy revealed the maintenance of an rtA181V/T mutant that emerged during adefovir monotherapy. The lamivudine-resistant mutations rtM204I/V and/or rtL180M reemerged during lamivudine-adefovir combination therapy.
The study results suggest that add-on lamivudine therapy with adefovir for adefovir resistance may not suppress the pre-existing adefovir-resistant mutation that develops during the lamivudine-adefovir sequential monotherapy.
Combination therapy of lamivudine and adefovir has been widely used in lamivudine-resistant chronic hepatitis B. However, a molecular mechanism is not clearly known by which non-response to combination therapy occurs. This is an interesting study to demonstrate the clonal evolution of HBV drug-resistant mutations during sequential and combination therapy in lamivudine-resistant chronic hepatitis B.
Peer reviewers: Sang Hoon Ahn, MD, PhD, Associate Professor, Department of Internal Medicine, Institute of Gastroenterology and Hepatology, Yonsei University College of Medicine, Severance Hospital, Seoul 120-752, South Korea; Evelyne Schvoerer, MD, PhD, HDR, Virology, University of Strasbourg, France, 3 Rue de Koeberlé, F- 67000 Strasbourg, France
S- Editor Lv S L- Editor Rutherford A E- Editor Lu YJ
| 1. | Dienstag JL. Hepatitis B virus infection. N Engl J Med. 2008;359:1486-1500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 689] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 2. | Liaw YF, Sung JJ, Chow WC, Farrell G, Lee CZ, Yuen H, Tanwandee T, Tao QM, Shue K, Keene ON, Dixon JS, Gray DF, Sabbat J. Lamivudine for patients with chronic hepatitis B and advanced liver disease. N Engl J Med. 2004;351:1521-1531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1739] [Cited by in RCA: 1740] [Article Influence: 82.9] [Reference Citation Analysis (0)] |
| 3. | Lee JM, Park JY, Kim do Y, Nguyen T, Hong SP, Kim SO, Chon CY, Han KH, Ahn SH. Long-term adefovir dipivoxil monotherapy for up to 5 years in lamivudine-resistant chronic hepatitis B. Antivir Ther. 2010;15:235-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 4. | Lee YS, Suh DJ, Lim YS, Jung SW, Kim KM, Lee HC, Chung YH, Lee YS, Yoo W, Kim SO. Increased risk of adefovir resistance in patients with lamivudine-resistant chronic hepatitis B after 48 weeks of adefovir dipivoxil monotherapy. Hepatology. 2006;43:1385-1391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 254] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 5. | Delaney WE. Progress in the treatment of chronic hepatitis B: long-term experience with adefovir dipivoxil. J Antimicrob Chemother. 2007;59:827-832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 6. | Zoulim F, Locarnini S. Hepatitis B virus resistance to nucleos(t)ide analogues. Gastroenterology. 2009;137:1593-1608.e1-e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 517] [Cited by in RCA: 539] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 7. | Danta M, Dusheiko G. Adefovir dipivoxil: review of a novel acyclic nucleoside analogue. Int J Clin Pract. 2004;58:877-886. [PubMed] |
| 8. | Liaw YF, Leung N, Kao JH, Piratvisuth T, Gane E, Han KH, Guan R, Lau GK, Locarnini S. Asian-Pacific consensus statement on the management of chronic hepatitis B: a 2008 update. Hepatol Int. 2008;2:263-283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 666] [Cited by in RCA: 743] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 9. | Yoo BC, Kim JH, Kim TH, Koh KC, Um SH, Kim YS, Lee KS, Han BH, Chon CY, Han JY. Clevudine is highly efficacious in hepatitis B e antigen-negative chronic hepatitis B with durable off-therapy viral suppression. Hepatology. 2007;46:1041-1048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 96] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 10. | Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. 2009;50:661-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2125] [Cited by in RCA: 2171] [Article Influence: 135.7] [Reference Citation Analysis (0)] |
| 11. | Locarnini S. Molecular virology of hepatitis B virus. Semin Liver Dis. 2004;24 Suppl 1:3-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 136] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 12. | Schildgen O, Sirma H, Funk A, Olotu C, Wend UC, Hartmann H, Helm M, Rockstroh JK, Willems WR, Will H. Variant of hepatitis B virus with primary resistance to adefovir. N Engl J Med. 2006;354:1807-1812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 166] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 13. | Chang TT, Lai CL. Hepatitis B virus with primary resistance to adefovir. N Engl J Med. 2006;355:322-323; author reply 323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 14. | Yeon JE, Yoo W, Hong SP, Chang YJ, Yu SK, Kim JH, Seo YS, Chung HJ, Moon MS, Kim SO. Resistance to adefovir dipivoxil in lamivudine resistant chronic hepatitis B patients treated with adefovir dipivoxil. Gut. 2006;55:1488-1495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 167] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 15. | Kwon SY, Park YK, Ahn SH, Cho ES, Choe WH, Lee CH, Kim BK, Ko SY, Choi HS, Park ES. Identification and characterization of clevudine-resistant mutants of hepatitis B virus isolated from chronic hepatitis B patients. J Virol. 2010;84:4494-4503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 16. | Lok AS, Lai CL, Leung N, Yao GB, Cui ZY, Schiff ER, Dienstag JL, Heathcote EJ, Little NR, Griffiths DA. Long-term safety of lamivudine treatment in patients with chronic hepatitis B. Gastroenterology. 2003;125:1714-1722. [PubMed] [DOI] [Full Text] |
| 17. | Locarnini S. Primary resistance, multidrug resistance, and cross-resistance pathways in HBV as a consequence of treatment failure. Hepatol Int. 2008;2:147-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 115] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 18. | Lai CL, Dienstag J, Schiff E, Leung NW, Atkins M, Hunt C, Brown N, Woessner M, Boehme R, Condreay L. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for patients with chronic hepatitis B. Clin Infect Dis. 2003;36:687-696. [PubMed] [DOI] [Full Text] |
| 19. | Yeh CT, Chien RN, Chu CM, Liaw YF. Clearance of the original hepatitis B virus YMDD-motif mutants with emergence of distinct lamivudine-resistant mutants during prolonged lamivudine therapy. Hepatology. 2000;31:1318-1326. [PubMed] [DOI] [Full Text] |
| 20. | Ogata N, Fujii K, Takigawa S, Nomoto M, Ichida T, Asakura H. Novel patterns of amino acid mutations in the hepatitis B virus polymerase in association with resistance to lamivudine therapy in japanese patients with chronic hepatitis B. J Med Virol. 1999;59:270-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 21. | Tenney DJ, Levine SM, Rose RE, Walsh AW, Weinheimer SP, Discotto L, Plym M, Pokornowski K, Yu CF, Angus P. Clinical emergence of entecavir-resistant hepatitis B virus requires additional substitutions in virus already resistant to Lamivudine. Antimicrob Agents Chemother. 2004;48:3498-3507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 427] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 22. | Delaney WE, Yang H, Westland CE, Das K, Arnold E, Gibbs CS, Miller MD, Xiong S. The hepatitis B virus polymerase mutation rtV173L is selected during lamivudine therapy and enhances viral replication in vitro. J Virol. 2003;77:11833-11841. [PubMed] |
| 23. | Shaw T, Bartholomeusz A, Locarnini S. HBV drug resistance: mechanisms, detection and interpretation. J Hepatol. 2006;44:593-606. [PubMed] [DOI] [Full Text] |
| 24. | Villet S, Pichoud C, Billioud G, Barraud L, Durantel S, Trépo C, Zoulim F. Impact of hepatitis B virus rtA181V/T mutants on hepatitis B treatment failure. J Hepatol. 2008;48:747-755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 160] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 25. | Angus P, Vaughan R, Xiong S, Yang H, Delaney W, Gibbs C, Brosgart C, Colledge D, Edwards R, Ayres A. Resistance to adefovir dipivoxil therapy associated with the selection of a novel mutation in the HBV polymerase. Gastroenterology. 2003;125:292-297. [PubMed] [DOI] [Full Text] |
| 26. | Villeneuve JP, Durantel D, Durantel S, Westland C, Xiong S, Brosgart CL, Gibbs CS, Parvaz P, Werle B, Trépo C. Selection of a hepatitis B virus strain resistant to adefovir in a liver transplantation patient. J Hepatol. 2003;39:1085-1089. [PubMed] [DOI] [Full Text] |
| 27. | Brunelle MN, Jacquard AC, Pichoud C, Durantel D, Carrouée-Durantel S, Villeneuve JP, Trépo C, Zoulim F. Susceptibility to antivirals of a human HBV strain with mutations conferring resistance to both lamivudine and adefovir. Hepatology. 2005;41:1391-1398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 211] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 28. | Yatsuji H, Noguchi C, Hiraga N, Mori N, Tsuge M, Imamura M, Takahashi S, Iwao E, Fujimoto Y, Ochi H. Emergence of a novel lamivudine-resistant hepatitis B virus variant with a substitution outside the YMDD motif. Antimicrob Agents Chemother. 2006;50:3867-3874. [PubMed] [DOI] [Full Text] |
| 29. | Qi X, Xiong S, Yang H, Miller M, Delaney WE. In vitro susceptibility of adefovir-associated hepatitis B virus polymerase mutations to other antiviral agents. Antivir Ther. 2007;12:355-362. [PubMed] |
| 30. | Fung SK, Chae HB, Fontana RJ, Conjeevaram H, Marrero J, Oberhelman K, Hussain M, Lok AS. Virologic response and resistance to adefovir in patients with chronic hepatitis B. J Hepatol. 2006;44:283-290. [PubMed] [DOI] [Full Text] |
| 31. | Yim HJ, Hussain M, Liu Y, Wong SN, Fung SK, Lok AS. Evolution of multi-drug resistant hepatitis B virus during sequential therapy. Hepatology. 2006;44:703-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 209] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 32. | Chen EQ, Wang LC, Lei J, Xu L, Tang H. Meta-analysis: adefovir dipivoxil in combination with lamivudine in patients with lamivudine-resistant hepatitis B virus. Virol J. 2009;6:163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 33. | Moon JH, Cho M, Yoon KT, Bae JH, Heo J, Kim GH, Kang DH, Song GA. [The efficacy of adefovir dipivoxil monotherapy and the incidence of genotypic resistance to adefovir dipivoxil in patients with lamivudine-resistant chronic hepatitis B infection]. Korean J Hepatol. 2008;14:503-512. [PubMed] [DOI] [Full Text] |
| 34. | Litwin S, Toll E, Jilbert AR, Mason WS. The competing roles of virus replication and hepatocyte death rates in the emergence of drug-resistant mutants: theoretical considerations. J Clin Virol. 2005;34 Suppl 1:S96-S107. [PubMed] [DOI] [Full Text] |
| 35. | Villet S, Billioud G, Pichoud C, Lucifora J, Hantz O, Sureau C, Dény P, Zoulim F. In vitro characterization of viral fitness of therapy-resistant hepatitis B variants. Gastroenterology. 2009;136:168-176.e2. [PubMed] [DOI] [Full Text] |
| 36. | Durantel D. Fitness and infectivity of drug-resistant and cross-resistant hepatitis B virus mutants: why and how is it studied? Antivir Ther. 2010;15:521-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |