Published online Nov 7, 2012. doi: 10.3748/wjg.v18.i41.5879
Revised: May 28, 2012
Accepted: June 28, 2012
Published online: November 7, 2012
AIM: To construct formulae for predicting the likelihood of ribavirin-induced anemia in pegylated interferon α plus ribavirin for chronic hepatitis C.
METHODS: Five hundred and sixty-one Japanese patients with hepatitis C virus genotype 1b who had received combination treatment were enrolled and assigned randomly to the derivation and confirmatory groups. Single nucleotide polymorphisms at or nearby ITPA were genotyped by real-time detection polymerase chain reaction. Factors influencing significant anemia (hemoglobin concentration < 10.0 g/dL at week 4 of treatment) and significant hemoglobin decline (declining concentrations > 3.0 g/dL at week 4) were analyzed using multiple regression analyses. Prediction formulae were constructed by significantly independent factors.
RESULTS: Multivariate analysis for the derivation group identified four independent factors associated with significant hemoglobin decline: hemoglobin decline at week 2 [P = 3.29 × 10-17, odds ratio (OR) = 7.54 (g/dL)], estimated glomerular filtration rate [P = 2.16 × 10-4, OR = 0.962 (mL/min/1.73 m2)], rs1127354 (P = 5.75 × 10-4, OR = 10.94) and baseline hemoglobin [P = 7.86 × 10-4, OR = 1.50 (g/dL)]. Using the model constructed by these factors, positive and negative predictive values and predictive accuracy were 79.8%, 88.8% and 86.2%, respectively. For the confirmatory group, they were 83.3%, 91.0% and 88.3%. These factors were closely correlated with significant anemia. However, the model could not be constructed, because no patients with rs1127354 minor genotype CA/AA had significant anemia.
CONCLUSION: Reliable formulae for predicting the likelihood of ribavirin-induced anemia were constructed. Such modeling may be useful in developing individual tailoring and optimization of ribavirin dosage.
-
Citation: Tsubota A, Shimada N, Abe H, Yoshizawa K, Agata R, Yumoto Y, Ika M, Namiki Y, Nagatsuma K, Matsudaira H, Fujise K, Tada N, Aizawa Y. Several factors including
ITPA polymorphism influence ribavirin-induced anemia in chronic hepatitis C. World J Gastroenterol 2012; 18(41): 5879-5888 - URL: https://www.wjgnet.com/1007-9327/full/v18/i41/5879.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i41.5879
Development and availability of nonstructural (NS) 3 serine protease inhibitors (PIs), such as telaprevir and boceprevir, further improve treatment outcome in combination with pegylated interferon (peg-IFN) α and ribavirin (RBV) for chronic hepatitis C virus (HCV) genotype 1 infection, while the addition of novel antiviral agents increases the frequency and severity of adverse effects (including anemia), medication costs and the complexity of treatment regimens[1-3]. Triple combination therapy with PI, RBV and peg-IFN α will be the first-line treatment for the HCV genotype 1 infection until the establishment of combination with NS3/4A PIs and NS5B polymerase or NS5A inhibitors[4]. Meanwhile, conventional peg-IFN α plus RBV combination will be in demand for easy-to-treat patients who are infected with HCV genotype 2 or 3 or low viral loads and those who contraindicate or are intolerant of triple combination therapy. Accordingly, peg-IFN α plus RBV combination will assume a crucial role in the treatment of HCV infection for the foreseeable future.
In RBV-based treatment, hemolytic anemia is common and one of the major critical adverse effects[1-3,5-7] and therefore makes it difficult for patients to tolerate treatment continuation, resulting in early dose reduction or premature withdrawal that may diminish the treatment efficacy. So far, many factors have been reported to be significantly associated with the significant anemia that could necessitate dose reduction or discontinuation[8-20]. Specifically, host genetic variants at the inosine triphosphatase (ITPA) gene located on chromosome 20 (20p13 region) that lead to ITPA deficiency or low activity have an overwhelming impact on protection against RBV-induced hemolytic anemia, and decrease the need for RBV dose reduction at week 4 of treatment and throughout the treatment course[15-18]. However, there are few reports that provide a convenient prediction model or scoring system for pretreatment screening or early identification of clinically significant anemia that has been defined previously and used generally[15].
To modify RBV dose prior to treatment or during the early treatment phase and continue treatment as long as possible, the present study focused on the construction of a convenient and useful model for predicting the likelihood of clinically significant anemia and quantitative decline in the hemoglobin (Hb) concentration from baseline at week 4 of treatment in peg-IFN α plus RBV treatment for chronic hepatitis C patients infected with HCV genotype 1b. Easy identification of candidates at a high risk for clinically significant anemia may facilitate intensive safety monitoring in combination treatment.
Between 2006 and 2010, 561 chronic hepatitis C patients infected with HCV genotype 1b were consecutively enrolled in this study at Katsushika Medical Center and Kashiwa Hospital, The Jikei University School of Medicine, and Shinmatsudo Central General Hospital. Patients received peg-IFN α-2b at a dose of 1.5 µg/kg or peg-IFN α-2a at a dose of 180 µg once weekly and RBV at a dose of 600-1000 mg twice daily for 48 wk. The dose of RBV was adjusted according to body weight (BW); 600 mg for ≤ 60 kg, 800 mg for > 60 kg to ≤ 80 kg, and 1000 mg for > 80 kg. Leading inclusion criteria were chronic hepatitis C that were diagnosed by laboratory, virology and histology; HCV genotype 1b confirmed by the conventional polymerase chain reaction (PCR)-based method; acquisition of written informed consent to the provision of genetic material; availability of genetic DNA for genotyping single nucleotide polymorphisms (SNPs); absence of liver cancer, liver failure or other forms of liver disease; and lack of concurrent treatment with any other antiviral or immunomodulatory agent. The study protocol was conducted in accordance with the provisions of the Declaration of Helsinki and Good Clinical Practice guidelines and was approved by the Institutional Review Boards of all participating sites.
Clinical and laboratory data were assessed at baseline, once weekly during the first 4 wk, and thereafter every 4 wk. As described previously[15], significant anemia was provisionally defined as Hb concentrations of < 10.0 g/dL at week 4 of treatment, and significant Hb decline was defined as a decline in Hb concentration of > 3.0 g/dL at week 4 of treatment. The reasons for choosing this time point (the end of 4 wk after treatment initiation) were as follows: (1) dose of RBV or peg-IFN was not reduced in most patients, and thus Hb dynamics would not be affected by treatment modification and could be evaluated in an unbiased manner; and (2) Hb decline within the first 4 wk is most prominent throughout the treatment period and reaches a nadir after approximately 4-6 wk[6,11].
At baseline, creatinine clearance (Ccr; mL/min) was estimated by using the Cockcroft-Gault formula[21]: Ccr (for male) = [(140 - age) × BW (kg)]/(72 × Scr) (Scr, serum creatinine; Ccr × 0.85 for female). Estimated glomerular filtration rate (GFR, mL/min/1.73 m2) was calculated according to the formula proposed by the Japanese Society of Nephrology: GFR (for male) =194 × Scr-1.094× Age-0.287 (GFR × 0.739 for female). Apparent clearance of ribavirin (CL/F, L/h) was determined as follows[9]: CL/F =32.3 × BW × (1 - 0.0094 × age) × (1 - 0.42 × gender)/Scr (gender = 0 for male and 1 for female; Scr is in µmol/L). All liver biopsy specimens were reviewed by using the established ranking system for staging of fibrosis and grading of necroinflammation activity with some modification[22].
Virological data were assessed by monitoring serum HCV RNA levels every 4 wk during and off treatment. Viral loads were measured using a quantitative PCR assay (Amplicor HCV Monitor version 2.0 or Amplicor HCV version 2.0; Roche Diagnostics, Basel, Switzerland). The presence or absence of serum HCV RNA was assessed using a qualitative PCR assay (Amplicor HCV version 2.0). Virological response (VR) was defined as undetectable HCV RNA by the end of treatment. Rapid virological response and slow virological response (SVR) were defined as undetectable HCV RNA at week 4 of treatment and 24 wk post-treatment. VR with relapse was defined as VR during treatment but reappearance of HCV RNA during the follow-up period. Nonvirological response (NVR) was defined as persistent presence of HCV RNA throughout the treatment.
Genomic DNA was extracted from whole blood using the MagNA Pure LC and the DNA Isolation Kit (Roche Diagnostics). Genetic polymorphisms, rs1127354 at the ITPA exon 2[15,17,18] and rs6051702 at the C20orf194[15,18], were genotyped by real-time detection PCR using the TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA, United States). Another functional (splicing variant-related) SNP at the ITPA intron 2, rs727010, was not examined because no polymorphisms were observed in the Asian genetic population, as registered in the HapMap Project database and reported previously[17,18,23].
Mantel-Haenszel, Pearson χ2 test or Mann-Whitney test was used to compare frequencies in categorical data or differences in continuous data between two groups, respectively. Time-course changes in Hb decline from baseline were evaluated by using repeated measures analysis of variance. Possible variables influencing significant anemia and significant Hb decline included baseline characteristics (Table 1). Variables that reached statistical significance (P < 0.05) or marginal significance (P < 0.10) in univariate comparisons were subsequently entered into multiple logistic regression analysis using forward and backward stepwise selection method to identify significantly independent factors associated with each anemic event. Based on the final-step results, score (S) was constructed by the exposure of some set of independent factors (x1, x2, ···, xp):
| Variable | Overall cohort (n = 561) | Derivation group (n = 374) | Confirmatory group (n = 187) |
| Demographic feature | |||
| Age (yr) | 59.1 ± 10.9 | 59.1 ± 10.8 | 59.5 ± 11.3 |
| Sex (female/male) | 302/259 | 201/173 | 101/86 |
| Weight (kg) | 59.8 ± 11.4 | 59.9 ± 11.5 | 58.5 ± 10.9 |
| BMI (kg/m2) | 23.2 ± 3.3 | 23.2 ± 3.4 | 23.2 ± 3.2 |
| Height (cm) | 160.3 ± 9.0 | 160.4 ± 9.1 | 158.5 ± 8.7 |
| BSA (m2) | 1.62 ± 0.18 | 1.62 ± 0.18 | 1.59 ± 0.18 |
| Laboratory data | |||
| ALT (IU/L) | 63 ± 54 | 63 ± 56 | 58 ± 47 |
| GGT (IU/L) | 58 ± 63 | 56 ± 60 | 58 ± 72 |
| Albumin (g/dL) | 4.1 ± 0.4 | 4.1 ± 0.4 | 4.1 ± 0.3 |
| Creatinine (mg/dL) | 0.70 ± 0.16 | 0.70 ± 0.17 | 0.70 ± 0.17 |
| WBC count (× 103/mL) | 5.0 ± 1.5 | 5.0 ± 1.5 | 5.1 ± 1.4 |
| Hemoglobin (g/dL) | 13.7 ± 1.5 | 13.7 ± 1.5 | 13.8 ± 1.3 |
| Platelet count (× 104/mL) | 16.7 ± 5.7 | 16.5 ± 5.7 | 17.0 ± 6.2 |
| Estimated calculation value | |||
| Ccr (mL/min) | 91.8 ± 27.9 | 91.6 ± 28.0 | 87.9 ± 25.6 |
| GFR (mL/min/1.73 m2) | 77.5 ± 16.8 | 79.6 ± 17.0 | 78.2 ± 15.9 |
| CL/F (L/h) | 11.3 ± 5.4 | 11.2 ± 5.3 | 10.9 ± 5.1 |
| Liver histopathology | |||
| Stage of fibrosis 0-1/2/3-4 | 221/131/161 | 146/89/107 | 75/42/54 |
| Grade of inflammation 1/2/3 | 267/222/19 | 170/156/14 | 97/66/5 |
| SNP genotype | |||
| rs1127354 CC/CA/AA | 431/114/16 | 289/75/10 | 142/39/6 |
| rs6051702 AA/AC/CC | 388/158/15 | 260/104/10 | 128/54/5 |
| Treatment | |||
| Ribavirin dosage (mg/kg/d) | 11.4 ± 1.5 | 11.3 ± 1.9 | 11.4 ± 1.9 |
| Peg-IFN α-2a/-2b | 82/479 | 52/322 | 30/157 |
| Virology | |||
| Viral load (log10 IU/mL) | 6.2 ± 0.8 | 6.2 ± 0.8 | 6.2 ± 0.8 |
S = β0 + β1x1 + β2x2 + ··· + βpxp (β0: Intercept, β1, β2, ···, βp: Regression coefficients).
The model could be expressed as: P = 1/[1 + exp (- S)], where P > 0.5 was development of anemic events and P < 0.5 was non-development of anemic events.
Hosmer-Lemeshow goodness of fit test and likelihood-ratio χ2 test were used and positive/negative predictive values and predictive accuracy were calculated to evaluate the fitness of the model. Split-group validation was used to develop and validate the best fitness of the model. Patients were randomly divided into two groups in the ratio of 2:1 by using a computer-generated random number list: 66.7% of the patients (374 patients) were assigned to the derivation group and 33.3% (187 patients) to the confirmatory group. The reproducibility of the resulting model based on data from the derivation group was assessed with data from the validation group. Receiver operating characteristic (ROC) curves were generated with every cut-off point of predicted probability of significant Hb decline corresponding to each Hb decline at week 2. For a balanced optimization of both sensitivity and false-positive rate [= (1 - specificity)], an optimal cut-off point value was determined by maximizing Youden’s index (= sensitivity + specificity - 1). The area under the ROC curve (AUC) was calculated to assess the degree of discrimination provided by the two parameters. To formulate a predictive value of quantitative Hb decline at weeks 2 and 4, the association between Hb decline and baseline variables was also analyzed using multiple linear regression analysis. The fitness of the model was evaluated by using values of R and R2 and Durbin-Watson test. The correlation between predictive and measured values in Hb decline was assessed by Spearman’s ρ. All P values for statistical tests were two tailed and values < 0.05 denoted the presence of a statistically significant difference. All data analyses were performed using the SPSS statistical package for Windows, version 17.0 (SPSS, Chicago, IL, United States).
Baseline characteristics of the study population are summarized in Table 1. There were no significant differences in the patient profiles between the groups. The mean (SD) and median (25th to 75th quartiles) of Hb decline from baseline at week 2 and 4 of treatment are shown in Table 2. The changes at each time point were not statistically different between the groups. Significant Hb decline was observed in 113 of 374 (30%) derivation group patients and 58 of 187 (31%) confirmatory group patients. Significant anemia was observed in 51 of 374 (14%) patients and 30 of 187 (16%) patients, respectively. Incidence of these anemic events was similar between the groups. Most of the patients complained of dyspnea on effort, easy fatigability or lightheadedness. None received erythropoiesis-stimulating agents throughout the treatment period.
| Week 2 of treatment | Week 4 of treatment | |
| Overall cohort | ||
| mean (SD), g/dL | -1.12 (1.13) | -2.31 (1.39) |
| Median (25th–75th quartile), g/dL | -1.05 (-1.8 to -0.3) | -2.3 (-3.2 to -1.3) |
| Derivation group | ||
| mean (SD), g/dL | -1.09 (1.11) | -2.27 (1.40) |
| Median (25th–75th quartile), g/dL | -1.0 (-1.8 to -0.3) | -2.3 (-3.1 to -1.3) |
| Confirmatory group | ||
| mean (SD), g/dL | -1.18 (1.17) | -2.33 (1.37) |
| Median (25th–75th quartile), g/dL | -1.1 (-1.95 to -0.4) | -2.3 (-3.3 to -1.35) |
Of the overall patients, 255 (45%) achieved SVR, 165 (29%) had VR with relapse, and 141 (25%) showed NVR. Of the 374 derivation group patients, SVR was 45% (167 patients), VR with relapse was 30% (111 patients) and NVR was 26% (96 patients). Of the 187 confirmatory group patients, they were 47% (88 patients), 29% (54 patients) and 24% (45 patients), respectively. Treatment outcome was almost equal among the overall cohort and split groups.
To construct the prediction model for significant Hb decline, baseline variables were statistically analyzed in the derivation group (Table 3). Patients who showed significant Hb decline were more likely to be male (P = 0.0163), have higher BW (P = 0.00518), higher body mass index (BMI; P = 0.00593), larger body surface area (BSA; P = 0.0139), higher albumin (P = 0.00688), higher creatinine (P = 4.71 × 10-4), higher Hb (P = 7.75 × 10-8), lower GFR (P = 5.69 × 10-4), and SNP rs1127354 major genotype CC (P = 8.04 × 10-10).
| Variable | P value | OR (95% CI) | |
| Univariate analysis | Multivariate analysis | ||
| Age (yr) | 0.110 | ||
| Sex (female vs male) | 0.0163 | ||
| Weight (kg) | 5.18 × 10-3 | ||
| BMI | 5.93 × 10-3 | ||
| Height (cm) | 0.153 | ||
| BSA (m2) | 0.0139 | ||
| ALT (IU/L) | 0.114 | ||
| GGT (IU/L) | 0.118 | ||
| Albumin (g/dL) | 6.88 × 10-3 | ||
| Creatinine (mg/dL) | 4.71 × 10-4 | ||
| WBC count (× 103/mL) | 0.147 | ||
| Hemoglobin (g/dL) | 7.75 × 10-8 | 1.29 × 10-9 | 1.89 (1.54-2.32) |
| Platelet count (× 104/mL) | 0.558 | ||
| Ccr (mL/min) | 0.140 | ||
| GFR (mL/min/1.73 m2) | 5.69 × 10-4 | 6.46 × 10-4 | 0.959 (0.942-0.977) |
| CL/F (L/h) | 0.814 | ||
| Stage of fibrosis | 0.641 | ||
| Grade of inflammation | 0.570 | ||
| rs1127354 (CC vs CA/AA) | 8.04 × 10-10 | 1.60 × 10-7 | 28.26 (8.10-98.62) |
| rs6051702 (AA vs AC/CC) | 0.372 | ||
| RBV dosage (mg/kg/d) | 0.419 | ||
| Peg-IFN α (2a vs 2b) | 0.360 | ||
| Viral load (log10 IU/mL) | 0.355 | ||
Multiple logistic regression analysis identified three independent variables that were significantly associated with significant Hb decline (Table 3): baseline Hb [P = 1.29 × 10-9, odds ratio (OR) = 1.89 (g/dL), 95% confidence interval (CI): 1.54-2.32], SNP rs1127354 (P = 1.60 × 10-7, OR = 28.26, 95%CI: 8.10-98.62), and GFR [P = 6.46 × 10-4, OR = 0.959 (mL/min/1.73 m2), 95%CI: 0.942-0.977]. The model was expressed as: S = -9.369 + 0.635 × baseline Hb + 3.342 × SNP rs1127354 (where genotype CC was 1 and CA/AA was 0) -0.041 × GFR. P values were 0.401 and 9.79 × 10-24 in the Hosmer-Lemeshow test and likelihood-ratio χ2 test, respectively. Positive and negative predictive values and predictive accuracy were 67.5%, 79.9% and 77.0%, respectively. To validate the prediction model, it was used for the confirmatory group. Positive and negative predictive values and predictive accuracy were 76.7%, 79.7% and 79.0%, respectively. For the overall cohort, these values were 70.8%, 79.9% and 77.7%, respectively. Significant Hb decline was not associated with treatment outcome in the overall cohort [SVR, 40% (69/171); VR, 32% (55/171); and NVR 27% (47/171)] or split groups.
Female (P = 0.00896) and older (P = 0.0443) patients, and those with lower albumin (P = 0.0197), lower white blood cell count (P = 0.0226), lower baseline Hb (P = 5.34 × 10-13), lower Ccr (P = 1.06 × 10-4), lower GFR (P = 2.69 × 10-4), lower CL/F (P = 6.59 × 10-5), lower BW (P = 0.00309), smaller BSA (P = 0.0254), and rs1127354 major genotype CC (P = 2.76 × 10-5) were more likely to have significant anemia than those who did not. In multiple logistic regression analysis, the model could not be constructed by these variables, because no patients with rs1127354 minor genotype CA/AA suffered from significant anemia in this study population (Figure 1). All patients with significant anemia had rs1127354 major genotype CC. When SNP rs1127354 was excluded from the multivariate analysis, baseline Hb [P = 1.67 × 10-9, OR = 0.376 (g/dL), 95%CI: 0.274-0.517] and GFR [P = 0.00233, OR = 0.962 (mL/min/1.73 m2), 95%CI: 0.938-0.986] were significantly independent variables. Significant anemia was not associated with treatment outcome in the overall cohort [SVR, 31% (25/81); VR, 36% (29/81); and NVR 33% (27/81)] or split groups.
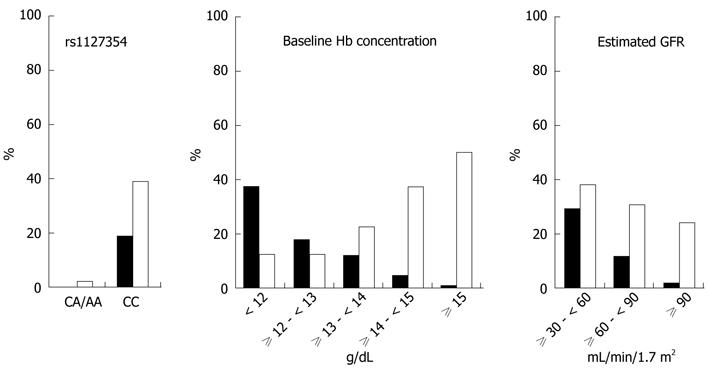
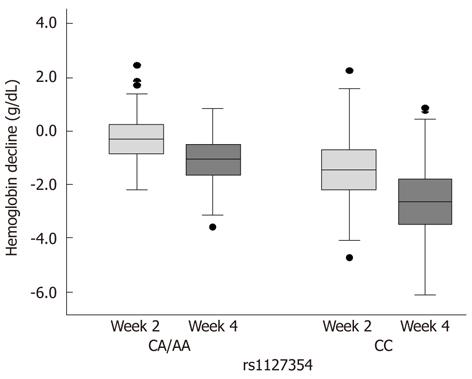
Figure 1 shows the incidence rates of significant anemia and significant Hb decline in the overall cohort according to the three significantly independent factors. Specifically, SNP rs1127354 had an overwhelming impact on the anemic events. In 431 patients with major genotype CC, significant anemia and significant Hb decline developed in 81 (19%) and 168 (39%) patients, respectively. In contrast, none (0%) and three (2%) of 130 patients with minor genotype CA/AA showed each anemic event, respectively, as described above. Positive predictive values of SNP rs1127354 alone for the likelihood of significant anemia and significant Hb decline were 14.3% and 39.1%, respectively. Negative predictive values were 100% and 97.7%, respectively. Values of predictive accuracy were 35.7% and 53.5%, respectively. Figure 2 depicts time-course changes in qualitative Hb decline from baseline according to SNP rs1127354 genotypes. The SNP genotype significantly influenced Hb decline at week 2 as well as week 4 (P = 5.437 × 10-9).
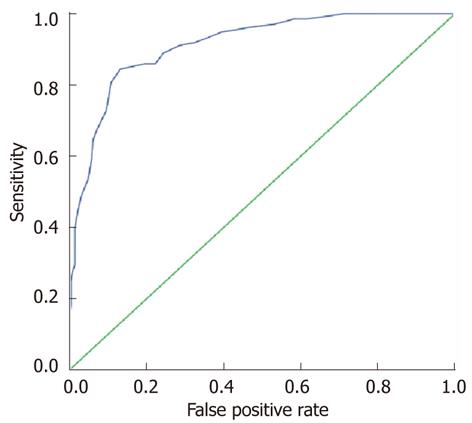
Hb decline from baseline at week 2 of treatment, an on-treatment factor, significantly influenced significant Hb decline (P = 1.96 × 10-33). An ROC curve was depicted to identify an optimal cut-off point for prediction of significant Hb decline by using Hb decline at week 2 (Figure 3). The AUC was 0.913 (95%CI: 0.885-0.941, P = 4.08 × 10-43). The maximal value of Youden’s index was 0.713. The sensitivity and false-positive rate were 0.844 and 0.131, respectively. The optimal cut-off point of Hb decline at week 2 was 1.45 g/dL.
When this variable, together with baseline variables, was incorporated into multiple logistic regression analysis to generate a statistic model for predicting significant Hb decline, the re-performed analysis using the derivation group data identified four significantly independent variables: Hb decline at week 2 [P = 3.29 × 10-17, OR = 7.54 (g/dL), 95%CI: 4.71-12.05], GFR [P = 2.16 × 10-4, OR = 0.962 (mL/min/1.73 m2), 95%CI: 0.942-0.982], rs1127354 (P = 5.75 × 10-4, OR = 10.94, 95%CI: 2.80-42.71), baseline Hb [P = 7.86 × 10-4, OR = 1.50 (g/dL), 95%CI: 1.18-1.90]. The model was expressed as: S = -8.285 - 2.020 × Hb decline at week 2 -0.039 × GFR + 2.393 × SNP rs1127354 (where genotype CC was 1 and CA/AA was 0) + 0.405 × baseline Hb. P values were 0.587 and 1.59 × 10-58 in the Hosmer-Lemeshow test and likelihood-ratio χ2 test, respectively. Positive and negative predictive values and predictive accuracy were 79.8%, 88.8% and 86.2%, respectively. These values were 83.3%, 91.0% and 88.3% in the confirmatory group, and 81.3%, 89.0% and 86.7% in the overall cohort.
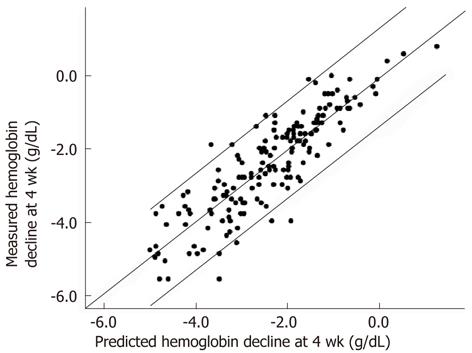
To predict qualitative Hb decline value at week 4 of treatment, the multiple linear regression model was constructed using data from the derivation group. The statistic model was expressed as: ŷ = 0.784 - 0.748 × Hb decline at week 2 - 0.878 × SNP rs1127354 (where genotype CC was 1 and CA/AA was 0) - 0.178 × baseline Hb + 0.012 × GFR (R = 0.842, R2 = 0.709, adjusted R2 = 0.706, Durbin-Watson test = 1.984, P = 2.42 × 10-7). There was a significant correlation between predicted values in the model and measured values in the confirmatory group (Spearman’s ρ = 0.880, P = 1.16 × 10-56; Figure 4).
Next, qualitative Hb decline value at week 2 was estimated by significantly independent variables in the derivation group. The model was expressed as: ŷ = 2.922 - 1.067 × SNP rs1127354 (where genotype CC was 1 and CA/AA was 0) - 0.276 × baseline Hb + 0.008 × GFR (R = 0.528, R2 = 0.279, adjusted R2 = 0.274, Durbin-Watson test = 0.537, P = 4.49 × 10-31). The correlation between predicted values in the model and measured values in the confirmatory group was statistically significant but relatively weak (Spearman’s ρ = 0.566, P = 2.41 × 10-17).
As mentioned in the introduction section, peg-IFN α plus RBV combination will be in demand for the foreseeable future. Patients at a high risk of developing RBV-induced hemolysis will expose themselves to a more increased risk for treatment-induced anemia in triple combination treatment. Identifying such high-risk patients and predicting the severity of anemia in individuals may provide an early decision to commence treatment with normal or reduced dosage and to keep the dose reduction to a minimum to lessen the disadvantages of anemia with adequate exposure to RBV continuing. To date, many studies have proposed factors that could influence the probability of clinically significant anemia in RBV-based treatment: age, sex[11,12], race, pre-existing cirrhosis[14], baseline Hb concentration[11,20], Ccr[14,20], CL/F[8,9], drug exposure[12-14], plasma RBV concentration[10], Hb decline at week 2 of treatment[12,14,20], and SNPs at the ITPA[15-18], C20orf194[15] and nucleoside transporter genes[19]. However, the definition of anemia or end point of analysis varied a little among previous studies, possibly leading to alteration of significant predictors. Despite these useful predictors, there is no convenient prediction model or formula for estimating the likelihood of clinically significant anemia that has been defined previously and used generally[15]. This study provided relevant numerical expressions constructed by independent variables for predicting the differentially defined anemia: Hb concentration < 10.0 g/dL (significant anemia) and a decline in Hb concentration > 3.0 g/dL (significant Hb decline) at week 4 of treatment and qualitative Hb decline at week 2 and 4. This is believed to be the first report to construct the prediction models by using reliable factors: the ITPA SNP rs1127354, baseline Hb concentration, estimated GFR, and quantitative Hb decline at week 2 of treatment, irrespective of the different definitions of anemia. The significant baseline factors that were shown in this study appear to influence treatment-induced anemia in triple combination treatment (under investigation, data not shown).
Two functional ITPA variants conferring ITPA deficiency or reduced activity are known to contribute most to protection against RBV-induced hemolytic anemia[15-18]. Inosine triphosphate (ITP) is hydrolyzed by ITPA to inosine monophosphate. Therefore, ITPA deficiency or low activity causes the accumulation of ITP in red blood cells (RBCs)[24-26]. The accumulated ITP may compete with the accumulated triphosphate form of RBV that could mediate oxidative damage to the RBC membrane and extravascular destruction[25-27], thereby protecting RBCs against RBV-induced hemolysis. As also shown in this study, one functional SNP rs1127354 is prominently associated with differentially defined anemia. Of note, however, the SNP was not always a factor of the top significance. The combined ITPA activity variable with another functional SNP rs7270101 is a stronger determinant of anemia than either ITPA SNP alone in European-Americans[16], whereas rs7270101 is not polymorphous in the Japanese population as registered in the HapMap database and reported by others[17,18,23]. One SNP, rs6051702 at the C20orf194 located near the ITPA, linked to the ITPA SNPs, also confers protection against anemia in European-Americans[15], while the association was statistically significant but weak in one Japanese cohort[18]. This Japanese study population showed no significant association (Table 3), supporting that rs1127354 is a single causal variant responsible for protection against anemia in the Japanese genetic cohort[17].
Certainly, the ITPA SNP rs1127354 minor variant A is a strong protective allele for anemia. In this overall cohort, none (0%) and three (3%; who had genotype CA) of patients with minor variant A had significant anemia and significant Hb decline, respectively (Figure 1). Therefore, negative predictive value of minor variant A was 100% and 97.7%, respectively. The noticeable distinction was in excellent agreement with other studies[15,18]. With respect to the likelihood of these anemic events, patients with minor variant A may be monitored less intensively and recommended to receive normal RBV doses, even in patients with relatively low baseline Hb, or more aggressive dose escalation strategies irrespective of baseline Hb. It is noteworthy that genotype AA patients with predicted ITPA deficiency, including seven patients with baseline Hb < 13.0 g/dL (range, 11.7-12.9 g/dL), showed no or little change in the Hb concentration (data not shown), although the number was small.
As shown in this study and another[18], however, only 25% of the Japanese population has minor variant A. The remaining 75% have major genotype CC. Positive predictive values of major genotype CC alone for significant anemia and significant Hb decline were low (14.3% and 39.1%, respectively), and values of predictive accuracy were low (35.7% and 53.5%, respectively). The range of Hb decline varied widely among individuals with genotype CC, indicating that some of them showed little or no change in Hb decline. Even in minor genotype CA carriers, it also varied widely and was similar to that of genotype CC patients (Figure 2). These findings strongly suggest that any factors other than the strong predictor ITPA SNP could affect hemolysis positively or negatively. Therefore, it is highly unlikely that the ITPA SNP (except genotype AA) is used alone to determine clinical decision making for treatment modification. In fact, several factors independently and strongly influenced treatment-induced anemia as well as the ITPA SNP in this study.
The clearance rate of RBV from the body is of critical importance for influencing treatment outcome and RBV-induced anemia, because the clearance parameters, such as CL/F and Ccr, reflect plasma/serum RBV concentrations at week 4 of treatment, which means the steady phase of treatment[8-10,14,20,28]. Higher or lower values of the parameters are correlated closely with lower or higher plasma/serum concentrations, respectively. Higher plasma/serum concentrations lead to an increased risk for progression of anemia as well as the higher probability of achieving SVR. Indeed, this study confirmed that the clearance rate is associated significantly and independently with RBV-induced anemia irrespective of the different definitions. This study also analyzed which of three parameters estimated by the formulae were the most stable for predicting clinically significant anemia. These formulae are composed by age, sex, BW and serum creatinine. Age and sex have been reported to affect treatment-induced anemia and dose reduction, and could reflect reactivity to treatment, tolerance and pharmacological metabolism[11,12,29]. Japan is one of the countries with the longest living people and the world’s fastest aging society, therefore, the clearance rate should especially be taken into account in RBV-based treatment of Japanese patients. The reason that estimated GFR remained an independent factor in the final model may be that the formula has been built up based on data from the Japanese population.
Higher baseline Hb concentration was significantly associated with the likelihood of significant Hb decline. Conversely, lower baseline Hb concentration was linked to significant anemia. These findings may be a matter of course. However, most of this study population received treatment without RBV dose reduction as scheduled, suggesting that kinetics of Hb decline within the first 4 wk of treatment might be delayed in patients with lower baseline Hb concentration. A certain threshold of Hb concentration might limit the progression of anemia independent of baseline Hb concentration. At least in Japanese patients, the two different definitions of anemia, significant Hb decline and significant anemia, should be separately analyzed and discussed.
In this multivariate analysis, qualitative Hb decline at week 2 of treatment was most highly predictive of significant Hb decline, compared to the strong predictor ITPA SNP rs1127354 and other baseline factors. Previous studies have shown that Hb decline of 2.0 g/dL at week 2 of treatment was predictive of Hb concentration < 10 g/dL or < 8.5 g/dL during the treatment[12,20]. In another study, Hb decline of 1.5 g/dL at week 2 was predictive of Hb decline ≥ 2.5 g/dL at week 4[14]. In this ROC analysis, the best cutoff value for Hb decline at week 2 was 1.45 g/dL. Taken together, Hb decline at week 2 is an excellent early predictor of subsequent Hb decline and could identify candidates for early intervention to maintain RBV dosing and adequate exposure. Indeed, the formula including this on-treatment variable improved positive and negative predictive values and predictive accuracy for significant anemia and significant Hb decline. When considered along with other independent baseline factors predictive of qualitative Hb decline at week 4, the final model yielded high significant values that represented goodness of fit. Using such a timely on-treatment variable and formula, more exact identification of patients prone to clinically significant anemia, early intervention with RBV dose reduction, and more careful monitoring may be indicated to reduce anemia-related adverse effects and avoid premature discontinuation of RBV.
ITPA SNP rs1127354, baseline Hb concentration and estimated GFR influenced Hb decline at week 2 significantly and independently, as well as that at week 4. However, it appears to be difficult to predict qualitative Hb decline at week 2 by using the multiple linear regression model. The point for attention is that the models and formulae did not perfectly predict the likelihood of the anemia, strongly suggesting the possibility that other unidentified factors associated with early occurring anemia might be lost, such as rare SNPs, brittleness of the RBC membrane against intracellular triphosphate form of RBV, or intracellular concentration of ITP.
In conclusion, convenient formulae for qualitatively or quantitatively predicting the likelihood of differentially defined anemia could be generated by significant independent factors in RBV-based treatment for chronic HCV infection. Such trial modeling may be useful in guiding clinical decision making on treatment modification: identifying the predisposition to develop RBV-induced anemia before treatment initiation or at the early treatment phase, and developing the individual tailoring and optimization of RBV dosage to maximize the treatment efficacy and minimize RBV-related adverse effects.
We thank physicians and staff members at Shinmatsudo Central General Hospital and Katsushika Medical Center and Kashiwa Hospital, The Jikei University School of Medicine for their help.
In ribavirin (RBV)-based treatment for chronic hepatitis C, hemolytic anemia is a major adverse effect and makes it difficult to continue treatment as scheduled. Many factors have been reported to influence clinically significant anemia that could modify or discontinue treatment. However, the definition of anemia or end point of analysis varied somewhat among studies, leading to alteration of significant predictors. Despite these useful predictors, there is no convenient prediction model for estimating the probability of clinically significant anemia.
Host genetic variation at the inosine triphosphatase (ITPA) gene that leads to ITPA deficiency or low activity are known to contribute greatly to protection against RBV-induced hemolytic anemia. However, it is highly unlikely that the ITPA single nucleotide polymorphism (SNP) alone is used to determine clinical decision making for treatment modification. Any factors other than the strong predictor ITPA SNP could affect hemolytic anemia positively or negatively.
This study provided relevant numerical expressions constructed by using significantly independent factors for predicting the differentially defined anemia. The reliable factors were the ITPA SNP rs1127354, baseline hemoglobin (Hb) concentration, estimated glomerular filtration rate and quantitative Hb decline at week 2 of treatment, irrespective of the different definitions of anemia. These factors independently and strongly influenced RBV-induced anemia, as well as the ITPA SNP. The ITPA SNP was not always a factor of major significance.
Such modeling may be useful in guiding clinical decision making on treatment modification: more exactly identifying candidates at a high risk for clinically significant anemia or predicting the severity of anemia in individuals before treatment initiation or at the early treatment phase, and developing the individual tailoring and optimization of RBV dosage to maximize the treatment efficacy and minimize RBV-related adverse effects with adequate exposure to RBV continuing.
Pegylated interferon plus RBV still assumes an important role in the treatment of chronic hepatitis C. The manuscript is well written and the study has investigated a crucial point of anti-HCV treatment. Interestingly, the authors observed patients who showed significant Hb decline and significant anemia, respectively, and showed that factors associated with anemia differed according to the definitions.
Peer reviewers: Dr. Hisato Nakajima, Department of Gastroenterology and Hepatology, The Jikei University School of Medicine, 3-25-8, Nishi-Shinbashi, Minato-ku, Tokyo 105-8461, Japan; Dr. Evelyne Schvoerer, CHRU Strasbourg, 3 rue Koeberlé, 67000 Strasbourg, France
S- Editor Wu X L- Editor Kerr C E- Editor Zhang DN
| 1. | McHutchison JG, Everson GT, Gordon SC, Jacobson IM, Sulkowski M, Kauffman R, McNair L, Alam J, Muir AJ. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360:1827-1838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 851] [Cited by in RCA: 809] [Article Influence: 50.6] [Reference Citation Analysis (0)] |
| 2. | McHutchison JG, Manns MP, Muir AJ, Terrault NA, Jacobson IM, Afdhal NH, Heathcote EJ, Zeuzem S, Reesink HW, Garg J. Telaprevir for previously treated chronic HCV infection. N Engl J Med. 2010;362:1292-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 570] [Cited by in RCA: 535] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 3. | Kwo PY, Lawitz EJ, McCone J, Schiff ER, Vierling JM, Pound D, Davis MN, Galati JS, Gordon SC, Ravendhran N. Efficacy of boceprevir, an NS3 protease inhibitor, in combination with peginterferon alfa-2b and ribavirin in treatment-naive patients with genotype 1 hepatitis C infection (SPRINT-1): an open-label, randomised, multicentre phase 2 trial. Lancet. 2010;376:705-716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 519] [Cited by in RCA: 517] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 4. | Asselah T, Marcellin P. Direct acting antivirals for the treatment of chronic hepatitis C: one pill a day for tomorrow. Liver Int. 2012;32 Suppl 1:88-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 5. | Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958-965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4736] [Cited by in RCA: 4558] [Article Influence: 189.9] [Reference Citation Analysis (0)] |
| 6. | Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL, Häussinger D, Diago M, Carosi G, Dhumeaux D. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975-982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4847] [Cited by in RCA: 4747] [Article Influence: 206.4] [Reference Citation Analysis (0)] |
| 7. | Hadziyannis SJ, Sette H, Morgan TR, Balan V, Diago M, Marcellin P, Ramadori G, Bodenheimer H, Bernstein D, Rizzetto M. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med. 2004;140:346-355. [PubMed] |
| 8. | Jen JF, Glue P, Gupta S, Zambas D, Hajian G. Population pharmacokinetic and pharmacodynamic analysis of ribavirin in patients with chronic hepatitis C. Ther Drug Monit. 2000;22:555-565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 138] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 9. | Kamar N, Chatelut E, Manolis E, Lafont T, Izopet J, Rostaing L. Ribavirin pharmacokinetics in renal and liver transplant patients: evidence that it depends on renal function. Am J Kidney Dis. 2004;43:140-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 62] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Lindahl K, Schvarcz R, Bruchfeld A, Ståhle L. Evidence that plasma concentration rather than dose per kilogram body weight predicts ribavirin-induced anaemia. J Viral Hepat. 2004;11:84-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 76] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 11. | Takaki S, Tsubota A, Hosaka T, Akuta N, Someya T, Kobayashi M, Suzuki F, Suzuki Y, Saitoh S, Arase Y. Factors contributing to ribavirin dose reduction due to anemia during interferon alfa2b and ribavirin combination therapy for chronic hepatitis C. J Gastroenterol. 2004;39:668-673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 66] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 12. | Nomura H, Tanimoto H, Kajiwara E, Shimono J, Maruyama T, Yamashita N, Nagano M, Higashi M, Mukai T, Matsui Y. Factors contributing to ribavirin-induced anemia. J Gastroenterol Hepatol. 2004;19:1312-1317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 62] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 13. | Snoeck E, Wade JR, Duff F, Lamb M, Jorga K. Predicting sustained virological response and anaemia in chronic hepatitis C patients treated with peginterferon alfa-2a (40KD) plus ribavirin. Br J Clin Pharmacol. 2006;62:699-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 66] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 14. | Reau N, Hadziyannis SJ, Messinger D, Fried MW, Jensen DM. Early predictors of anemia in patients with hepatitis C genotype 1 treated with peginterferon alfa-2a (40KD) plus ribavirin. Am J Gastroenterol. 2008;103:1981-1988. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 45] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 15. | Fellay J, Thompson AJ, Ge D, Gumbs CE, Urban TJ, Shianna KV, Little LD, Qiu P, Bertelsen AH, Watson M. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature. 2010;464:405-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 371] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 16. | Thompson AJ, Fellay J, Patel K, Tillmann HL, Naggie S, Ge D, Urban TJ, Shianna KV, Muir AJ, Fried MW. Variants in the ITPA gene protect against ribavirin-induced hemolytic anemia and decrease the need for ribavirin dose reduction. Gastroenterology. 2010;139:1181-1189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 161] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 17. | Ochi H, Maekawa T, Abe H, Hayashida Y, Nakano R, Kubo M, Tsunoda T, Hayes CN, Kumada H, Nakamura Y. ITPA polymorphism affects ribavirin-induced anemia and outcomes of therapy--a genome-wide study of Japanese HCV virus patients. Gastroenterology. 2010;139:1190-1197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 143] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 18. | Sakamoto N, Tanaka Y, Nakagawa M, Yatsuhashi H, Nishiguchi S, Enomoto N, Azuma S, Nishimura-Sakurai Y, Kakinuma S, Nishida N. ITPA gene variant protects against anemia induced by pegylated interferon-α and ribavirin therapy for Japanese patients with chronic hepatitis C. Hepatol Res. 2010;40:1063-1071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Doehring A, Hofmann WP, Schlecker C, Zeuzem S, Sarrazin C, Berg T, Müller T, Herrmann E, Geisslinger G, Lötsch J. Role of nucleoside transporters SLC28A2/3 and SLC29A1/2 genetics in ribavirin therapy: protection against anemia in patients with chronic hepatitis C. Pharmacogenet Genomics. 2011;21:289-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 20. | Hiramatsu N, Kurosaki M, Sakamoto N, Iwasaki M, Sakamoto M, Suzuki Y, Sugauchi F, Tamori A, Kakinnuma S, Matsuura K. Pretreatment prediction of anemia progression by pegylated interferon alpha-2b plus ribavirin combination therapy in chronic hepatitis C infection: decision-tree analysis. J Gastroenterol. 2011;46:1111-1119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 21. | Cockcroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron. 1976;16:31-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10672] [Cited by in RCA: 11005] [Article Influence: 224.6] [Reference Citation Analysis (1)] |
| 22. | Desmet VJ, Gerber M, Hoofnagle JH, Manns M, Scheuer PJ. Classification of chronic hepatitis: diagnosis, grading and staging. Hepatology. 1994;19:1513-1520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1582] [Cited by in RCA: 1506] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 23. | Tsubota A, Shimada N, Yoshizawa K, Furihata T, Agata R, Yumoto Y, Abe H, Ika M, Namiki Y, Chiba K. Contribution of ribavirin transporter gene polymorphism to treatment response in peginterferon plus ribavirin therapy for HCV genotype 1b patients. Liver Int. 2012;32:826-836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 24. | Fraser JH, Meyers H, Henderson JF, Brox LW, McCoy EE. Individual variation in inosine triphosphate accumulation in human erythrocytes. Clin Biochem. 1975;8:353-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 25. | Bierau J, Lindhout M, Bakker JA. Pharmacogenetic significance of inosine triphosphatase. Pharmacogenomics. 2007;8:1221-1228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 26. | Stepchenkova EI, Tarakhovskaya ER, Spitler K, Frahm C, Menezes MR, Simone PD, Kolar C, Marky LA, Borgstahl GE, Pavlov YI. Functional study of the P32T ITPA variant associated with drug sensitivity in humans. J Mol Biol. 2009;392:602-613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 50] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 27. | De Franceschi L, Fattovich G, Turrini F, Ayi K, Brugnara C, Manzato F, Noventa F, Stanzial AM, Solero P, Corrocher R. Hemolytic anemia induced by ribavirin therapy in patients with chronic hepatitis C virus infection: role of membrane oxidative damage. Hepatology. 2000;31:997-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 328] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 28. | Tsubota A, Hirose Y, Izumi N, Kumada H. Pharmacokinetics of ribavirin in combined interferon-alpha 2b and ribavirin therapy for chronic hepatitis C virus infection. Br J Clin Pharmacol. 2003;55:360-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 54] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 29. | Sezaki H, Suzuki F, Kawamura Y, Yatsuji H, Hosaka T, Akuta N, Kobayashi M, Suzuki Y, Saitoh S, Arase Y. Poor response to pegylated interferon and ribavirin in older women infected with hepatitis C virus of genotype 1b in high viral loads. Dig Dis Sci. 2009;54:1317-1324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 62] [Article Influence: 3.9] [Reference Citation Analysis (0)] |