Published online Oct 21, 2012. doi: 10.3748/wjg.v18.i39.5635
Revised: June 14, 2012
Accepted: July 18, 2012
Published online: October 21, 2012
We describe a patient with a Homo sapiens mutL homolog 1 (MLH1)-associated Lynch syndrome with previous diagnoses of two distinct primary cancers: a sigmoid colon cancer at the age of 39 years, and a right colon cancer at the age of 50 years. The mutation identified in his blood and buccal cells, c.1771delG, p.Asp591Ilefs*25, appears to be a de novo event, as it was not transmitted by either of his parents. This type of de novo event is rare in MLH1 as only three cases have been reported in the literature so far. Furthermore, the discordant results observed between replication error phenotyping and immunohistochemistry highlight the importance of the systematic use of both pre-screening tests in the molecular diagnosis of Lynch syndrome.
-
Citation: Airaud F, Küry S, Valo I, Maury I, Bonneau D, Ingster O, Bezieau S. A
de novo germlineMLH1 mutation in a Lynch syndrome patient with discordant immunohistochemical and molecular biology test results. World J Gastroenterol 2012; 18(39): 5635-5639 - URL: https://www.wjgnet.com/1007-9327/full/v18/i39/5635.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i39.5635
Lynch syndrome or hereditary non-polyposis colorectal cancer syndrome (MIM#120435)[1] is a familial form of cancer mainly involving the colon, rectum or endometrium and, more rarely, the small bowel, urinary tract, ovaries, stomach, brain or skin. The pathology is transmitted in an autosomal dominant mode of inheritance with an incomplete penetrance. It is linked to mutations in the genes of the DNA mismatch repair system (MMR), including Homo sapiens mutL homolog 1 (MLH1), Homo sapiens mutS homolog 2 (MSH2), and to a lesser extent, Homo sapiens mutS homolog 6 (MSH6) and PostMeiotic Segregation increased 2 (PMS2). International scientific community defined the clinical and familial criteria which had to be fulfilled to consider a likely diagnosis of Lynch syndrome, and thereby the value of a mutation screening of MMR genes in a patient. The initial recommendations following the Amsterdam (1991)[2] and Amsterdam II (1999) criteria[3] were too restrictive. New criteria were therefore proposed at the Bethesda conference in 2004[4], in order to increase the sensitivity of mutation carrier detection. These recommendations include an age at diagnosis younger than 50 years, or younger than 60 years when the tumor has a microsatellite instability-high phenotype (MSI-high), which is the hallmark of tumors linked to a defective MMR gene system[5,6].
Here we report on the unique case of a de novo mutation encountered by our Oncogenetic Laboratory through its diagnostic activity in Lynch syndrome. In our molecular diagnostic strategy, replication error (RER) phenotyping is performed systematically before sequencing of the MMR genes whenever tumor tissue is available, and this is done concomitantly with immunohistochemistry in order to direct the sequencing as far as possible. Sequencing of either MLH1, MLH2 or both genes is initiated each time a tumor is found to be MSI-high or -low (low microsatellite instability: one system with replication error), even when the immunohistochemistry results are ambiguous or show no extinction of a MMR protein. The sequencing step is also initiated under three specific circumstances: (1) the finding of an microsatellite stable (MSS) phenotype with validated Amsterdam criteria; (2) when no tumor material is available; and (3) in the case of a discrepancy between the immunohistochemistry results and the RER phenotype.
Whenever necessary, DNA samples are sent to another laboratory within the French MMR network (Groupe Génétique et Cancer MMR) for further testing of MSH6.
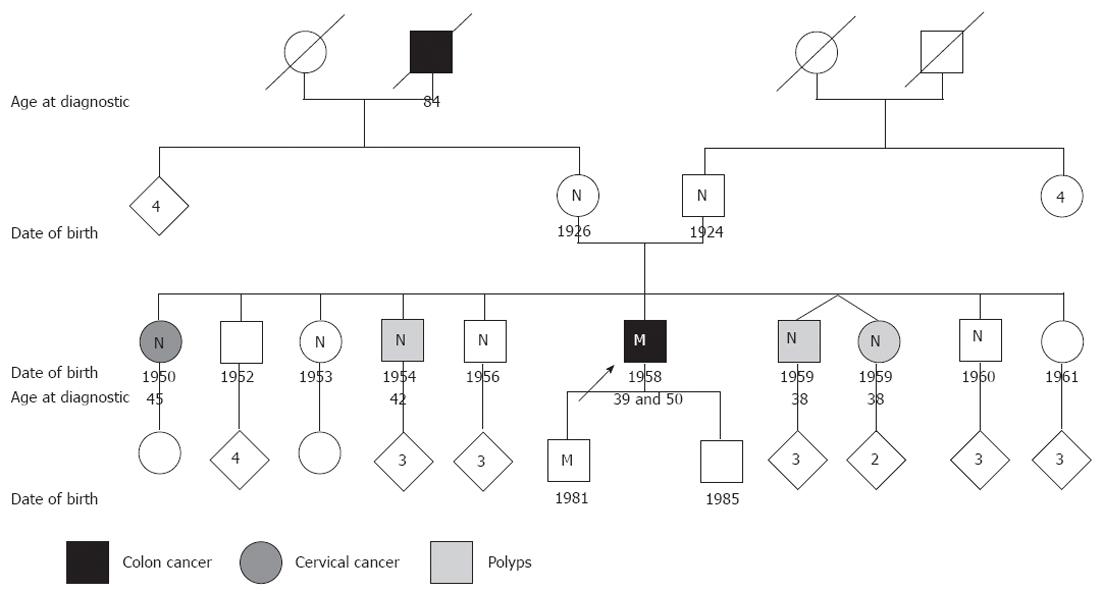
The patient is a 54 year-old man who had previously presented two distinct primary cancers, a sigmoid colon cancer at the age of 39 years, and a right colon cancer at the age of 50 years. He is the sixth of a family of 10 siblings, three of whom have had polyps removed: two brothers and one sister, at the ages of 38, 42 and 38 years respectively. A fourth sibling, another of his sisters, was diagnosed with cervical cancer at 45 years of age, but it is not in the tumor spectrum for Lynch syndrome. Among the older generations of the patient’s family, the maternal grandfather died of colon cancer at the age of 84 years and the mother’s medical records report an ovarian cyst and ovariectomy (Figure 1).
The right colon tumor had been removed by a colectomy covering 30 cm of the right colon and 5 cm of the ileum. The tumor measured 7 cm × 5 cm × 1.5 cm and was located in the caecum, invading the terminal ileum. The pathology examination revealed a grade 3 stage for the poorly differentiated, burgeoning and stenosing adenocarcinoma, and cancer staging was classified as pT3N0Mx. Immunochemistry and molecular biology analyses were performed on formalin-fixed paraffin-embedded material from this tumor.
The immunohistochemistry analysis for the MMR proteins MLH1, MSH2 and MSH6[7] was performed but did not demonstrate a loss of expression, albeit a very weak expression was detected for MLH1 and MSH2.
RER phenotyping was then performed and showed an instable phenotype for four of the five markers routinely tested in our laboratory (BAT25, BAT26, NR21, NR22 and NR24)[8,9], with stability noted only for the NR24 system. The tumor was thus classified MSI-high, and the patient was considered for mutational analysis of MMR genes, according to the Bethesda criteria.
In keeping with routine practice at our laboratory, we started with the search for large rearrangements using a commercial Multiplex Ligation-dependent Probe Amplification (MLPA®) kit for MLH1 and MSH2 (P003-B1 kit, MRC-HOLLAND, Amsterdam, The Netherlands)[10]. This revealed the isolated loss of MLH1 exon 16. In such cases of single exon deletion or duplication, we systematically verify the hybridizing sequence of the MLPA probes to exclude a false result caused by a single nucleotide polymorphism (SNP). However, in the present case, the sequence analysis showed a single nucleotide deletion of a G in position 1771 (NM_000249.3: c.1771delG, p.Asp591Ilefs*25). This mutation had previously been described in a Taiwanese Lynch family[11]. It had been classified as deleterious because of the occurrence of a premature stop codon at position 615, which disrupts the interacting protein domains to Pms2 and exonuclease 1[12].
From this point on, we set out to investigate the status of the mutation within the tumor. Finding the mutation in a homozygous or hemizygous state would have meant that the second hit in the carcinogenetic process was a loss of heterozygosity, and that the mutation was the first hit. Unfortunately, the poor quality of the DNA extracted from the Formaldehyde-fixed Paraffin-embedded material did not enable us to get readable sequences.
We confirmed the presence of mutation c.1771delG (p.Asp591Ilefs*25) in a second independent sample (DNA extracted from a buccal swab spotted onto a FTA paper). Once the mutation had been confirmed, a presymptomatic test was offered to the patient’s family. In this context, we tested both parents but did not identify any parental origin for the mutation as both parents tested negative for the mutation, in both blood and buccal cell samples. We then performed the complete sequencing of MLH1 to exclude any other mutational event that could have been the true cause of the patient’s personal and familial history of cancer, but we did not identify anything else.
These results led us to question the patient’s paternity, given that a meta-analysis performed in 2005 reported false paternity in a median of 3.7% of births[13]. Nevertheless, a genetic fingerprinting kit (AmpFℓSTR® SGM Plus®; Applied) confirmed the paternal and maternal status of the supposed parents without ambiguity.
In order to further investigate the possibility of a mosaicism, we performed a haplotyping study using two frequent SNP within the MLH1 gene (rs1800734 and rs2241031) chosen for their high heterozygosity frequencies and their belonging to different ancestral haplotypes. However, the lack of proband informativity meant that this analysis did not enable us to identify the maternal or paternal origin of the chromosome harboring the mutation. However, because we had tested two different types of tissues of different origins, i.e., mesodermic for blood cells and ectodermic for buccal cells, we were able to exclude mosaicism with quite a high level of confidence. We were finally able to conclude that the Lynch syndrome-causing mutation observed in our patient was a de novo event.
Up to now, we have been able to test seven of the patient’s nine siblings and have not identified any of them as a carrier of the mutation, even those who have had polyps or cervical cancer. This could be considered as a further argument for a de novo mutation, since it does not follow the Mendelian transmission ratio of 1:2 and it is not concordant with polyp antecedents.
Finally, a presymptomatic test in our patient’s 30 year-old son revealed the presence of mutation c.1771delG (p.Asp591Ilefs*25) in a heterozygous state, thereby highlighting the transmission (and conservation) of this de novo mutation by the proband.
Here we report on a French Lynch family in whom we have identified a frame-shift mutation that induces a premature stop codon in a crucial part of the MLH1 gene. This mutation has never been reported in the open access MMR mutation database (INSIGHT, Newfoundland etc.), or in the French MMR network database (unpublished data). Moreover, we have never found this mutation in 592 chromosomes of patients of similar geographic origin who have been tested in our laboratory as part of routine Lynch syndrome screening.
In contrast with certain other genes, such as NF1, which exhibit a de novo mutation rate of about 50%, this event in MLH1 is relatively rare (1% to 5%) according to the study recently published by Win et al[14]. To our knowledge, a de novo point mutation in MLH1 has only been described three times until now. The first occurrence was a c.2101C>T (p.Gln701X) mutation in exon 18, which was detected in a 35 year-old man[15]. The second was a c.666dupA (p.Asn222Lysfs*4) mutation in exon 8 found in a 31 year-old man[16]. The third mutation was a nonsense mutation in exon 13, c.1459C>T (p.Arg487X) identified in a 36 year-old patient[14]. In all three cases the patients had no family history of colorectal cancer and seemed to develop cancer younger than inherited mutation carriers. In addition to these three single nucleotide mutations, two large deletions have already been published, one of the entire MLH1 gene and one of exon 15, once again in a young man without any family history of cancer[14,17]. De novo mutations seem to be more frequent in MSH2, for which four different mutations have already been described[14,18,19], including the recurrent mutation c.942+3A>T. The latter can even be considered as a kind of mutation hotspot, as it has been proved to occur de novo with a relatively high frequency[20]. The nucleotide implicated in this mutation is part of the BAT26 homopolymer containing 26 adenines. This particular context is hypothesized to be responsible for misalignment during replication or recombination. Even though the de novo MLH1 mutation described here arose in two families of different ethnic origin, a similar explanation cannot be considered.
This case report confirms the relevance of preceding MMR gene sequencing by the combination of the two prescreening tests (RER phenotyping and immunohistochmistry) in the molecular diagnostic strategy in Lynch syndrome, especially for young patients without familial antecedents. Indeed, it is worth noting that most of these patients would not have been considered as candidates for mutation analysis according to the Amsterdam I and II criteria; this might be the reason why de novo events in MLH1 were not described prior to the implementation of the Bethesda criteria. It is also interesting to point out the discordance between immunohistochemistry and RER phenotyping results for our patient’s tumor, which confirms the benefit of a dual approach for the screening of Lynch syndrome patients. Indeed, MSI-high phenotypes with conservation of protein expression have already been described and can easily be explained when they concern a missense mutation that does not occur in the epitope of the antibody used. Inversely, extinction of a protein associated with an MSS or MSI-low phenotype can also be encountered, especially when the MSH6 gene is affected[21]. In glioblastoma, in the context of Turcot syndrome, changes in microsatellite profiles have also been described as more subtle than those in colorectal tumors[22] and thus have to be considered very carefully.
In conclusion, the frequency of de novo mutations in MMR genes may be higher than actually observed in diagnostic laboratories because once a mutation is identified, the parents of the proband are not systematically analyzed in routine practice. This may be because they are deceased, as the average age at molecular diagnosis of our index cases is 53 years, or because they do not wish to be tested
Moreover, we show here that the combined use of molecular biology and immunohistochemistry should be recommended when screening patients with suspected Lynch syndrome. This combined strategy should help to avoid missing a tumor linked to a deficiency in a MMR gene, and also to orientate the subsequent sequencing to one of these genes in a more precise and therefore cost-effective manner.
Peer reviewers: Dr. Francesco Franceschi, Department of Internal Medicine, Catholic University of Rome, Policlinico A. Gemelli, Largo A. Gemelli, 8, 00168 Rome, Italy; Joerg Trojan, Professor, Department of Medizinische Klinik 1, Klinikum der Johann Wolfgang Goethe-Universität, Theodor-Stern-Kai 7, 60590 Frankfurt, Germany; Peter Laszlo Lakatos, MD, PhD, 1st Department of Medicine, Semmelweis University, Koranyi S 2A, H1083 Budapest, Hungary; Jae J Kim, MD, PhD, Professor, Department of Medicine, Samsung Medical Center, Irwon-dong, Gangnam-gu, Seoul 135-710, South Korea
S- Editor Wu X L- Editor A E- Editor Li JY
| 1. | Lynch HT, de la Chapelle A. Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet. 1999;36:801-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 2. | Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC). Dis Colon Rectum. 1991;34:424-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1357] [Cited by in RCA: 1277] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 3. | Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453-1456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1765] [Cited by in RCA: 1691] [Article Influence: 65.0] [Reference Citation Analysis (0)] |
| 4. | Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2154] [Cited by in RCA: 2223] [Article Influence: 105.9] [Reference Citation Analysis (1)] |
| 5. | Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248-5257. [PubMed] |
| 6. | Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044-2058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 945] [Cited by in RCA: 857] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 7. | Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, Walsh-Vockley C, Petersen GM, Walsh MD, Leggett BA. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol. 2002;20:1043-1048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 207] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 8. | Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073-2087.e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1677] [Cited by in RCA: 1545] [Article Influence: 103.0] [Reference Citation Analysis (0)] |
| 9. | Suraweera N, Duval A, Reperant M, Vaury C, Furlan D, Leroy K, Seruca R, Iacopetta B, Hamelin R. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123:1804-1811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 483] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 10. | Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1811] [Cited by in RCA: 1853] [Article Influence: 80.6] [Reference Citation Analysis (0)] |
| 11. | Tang R, Hsiung C, Wang JY, Lai CH, Chien HT, Chiu LL, Liu CT, Chen HH, Wang HM, Chen SX. Germ line MLH1 and MSH2 mutations in Taiwanese Lynch syndrome families: characterization of a founder genomic mutation in the MLH1 gene. Clin Genet. 2009;75:334-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 12. | Boland CR, Fishel R. Lynch syndrome: form, function, proteins, and basketball. Gastroenterology. 2005;129:751-755. [PubMed] [DOI] [Full Text] |
| 13. | Bellis MA, Hughes K, Hughes S, Ashton JR. Measuring paternal discrepancy and its public health consequences. J Epidemiol Community Health. 2005;59:749-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 147] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 14. | Win AK, Jenkins MA, Buchanan DD, Clendenning M, Young JP, Giles GG, Goldblatt J, Leggett BA, Hopper JL, Thibodeau SN. Determining the frequency of de novo germline mutations in DNA mismatch repair genes. J Med Genet. 2011;48:530-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 15. | Stulp RP, Vos YJ, Mol B, Karrenbeld A, de Raad M, van der Mijle HJ, Sijmons RH. First report of a de novo germline mutation in the MLH1 gene. World J Gastroenterol. 2006;12:809-811. [PubMed] |
| 16. | Plasilova M, Zhang J, Okhowat R, Marra G, Mettler M, Mueller H, Heinimann K. A de novo MLH1 germ line mutation in a 31-year-old colorectal cancer patient. Genes Chromosomes Cancer. 2006;45:1106-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 17. | Smith L, Tesoriero A, Mead L, Royce S, Grubb G, Young J, Giles G, Jenkins M, Macrae F, Hopper JL. Large genomic alterations in hMSH2 and hMLH1 in early-onset colorectal cancer: identification of a large complex de novo hMLH1 alteration. Clin Genet. 2006;70:250-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 18. | Kraus C, Kastl S, Günther K, Klessinger S, Hohenberger W, Ballhausen WG. A proven de novo germline mutation in HNPCC. J Med Genet. 1999;36:919-921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 19. | Morak M, Laner A, Scholz M, Madorf T, Holinski-Feder E. Report on de-novo mutation in the MSH2 gene as a rare event in hereditary nonpolyposis colorectal cancer. Eur J Gastroenterol Hepatol. 2008;20:1101-1105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 20. | Desai DC, Lockman JC, Chadwick RB, Gao X, Percesepe A, Evans DG, Miyaki M, Yuen ST, Radice P, Maher ER. Recurrent germline mutation in MSH2 arises frequently de novo. J Med Genet. 2000;37:646-652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 62] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 21. | Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, van Lier B, Arts P, Wieskamp N, del Rosario M. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109-1112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 599] [Cited by in RCA: 616] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 22. | Zhao YS, Hu FL, Wang F, Han B, Li DD, Li XW, Zhu S. Meta-analysis of MSH6 gene mutation frequency in colorectal and endometrial cancers. J Toxicol Environ Health A. 2009;72:690-697. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |