Published online Oct 21, 2012. doi: 10.3748/wjg.v18.i39.5581
Revised: June 29, 2012
Accepted: July 9, 2012
Published online: October 21, 2012
AIM: To evaluate the potential of thioredoxin (TXN) and thioredoxin-interacting protein (TXNIP) expression as biomarkers for predicting gastric cancer recurrence.
METHODS: TXN and TXNIP expression levels were acquired from gene expression microarray data for 65 human gastric cancer tissues. We determined whether each gene expression level was associated with cancer recurrence and investigated the relationship between the two genes. For validation, the expression levels of TXN and TXNIP were measured by quantitative real-time reverse transcription polymerase chain reaction in 68 independent stage III gastric cancer patients. The correlation between gene expression and cancer prognosis was evaluated. Immunohistochemical staining was performed to investigate the protein expression levels of TXN and TXNIP and to characterize the expression patterns of each protein.
RESULTS: TXN was a prognosis-related gene (P = 0.009), whereas TXNIP, a TXN inhibitor, demonstrated a negative correlation with TXN in the gene expression microarray data. In the 68 stage III patients, the expression levels of both TXN and TXNIP had a statistically significant effect on recurrence-free survival (RFS, P = 0.008 and P = 0.036, respectively). The low TXN and high TXNIP expression group exhibited a better prognosis than the other groups, and the high TXN and low TXNIP expression group exhibited a poorer prognosis (P < 0.001 for RFS and P = 0.001 for overall survival). More than half of the patients in the simultaneously high TXN and low TXNIP expression group experienced a recurrence within 1 year after curative surgery, and the 5-year survival rate of the patients in this group was 29%, compared with 89% in the low TXN and high TXNIP expression group. The TXN protein was overexpressed in 65% of the gastric cancer tissues, whereas the TXNIP protein was underexpressed in 85% of the cancer cells. In a correlation analysis, TXN and TXNIP were highly correlated with many oncogenes and tumor suppressors as well as with genes related to energy, protein synthesis and autophagy.
CONCLUSION: TXN and TXNIP are promising prognostic markers for gastric cancer, and performing personalized adjuvant treatment based on TXN and TXNIP expression levels would be an effective practice in the treatment of gastric cancer.
- Citation: Lim JY, Yoon SO, Hong SW, Kim JW, Choi SH, Cho JY. Thioredoxin and thioredoxin-interacting protein as prognostic markers for gastric cancer recurrence. World J Gastroenterol 2012; 18(39): 5581-5588
- URL: https://www.wjgnet.com/1007-9327/full/v18/i39/5581.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i39.5581
Gastric cancer is the second leading cause of global cancer mortality, and it has the highest mortality rates in East Asia, including South Korea, Japan and China[1]. The standard treatment for gastric cancer is surgery. However, relapses occur in many patients who undergo curative resection, even after adjuvant therapy[2,3]. Even among individuals with the same stage of cancer, gastric cancer patients present with diverse clinical manifestations and prognoses. The clinical diversity of gastric cancer arises from its molecular biological diversity, which is caused by changes in different genes. Molecular markers are important in predicting patient outcome and in personalizing treatments according to the individual biology of the patient. Importantly, in the adjuvant treatment setting, uniform adjuvant treatment after curative resection of gastric cancer has been performed regardless of the individual molecular prognostic markers of the cancer. However, cancer researchers have identified biomarkers that can predict recurrence and survival[4-8], and doctors should consider using biomarker-driven personalized adjuvant treatment.
Thioredoxin (TXN) is a low-molecular-weight redox protein and a putative oncoprotein that provides growth and survival advantages to tumor cells through the activation of redox-sensitive transcription factors, such as nuclear factor kappa B (NF-κB), p53, and activator protein-1 (AP-1)[9,10]. TXN inhibits apoptosis via apoptosis signaling kinase-1 (ASK-1) and phosphatase and tensin homolog (PTEN)[11]. TXN also induces hypoxia-inducible factor-1α (HIF-1α), which increases the production of vascular endothelial growth factor and leads to tumor angiogenesis and drug resistance[12]. The overexpression of TXN has been found in several cancers, including lung[13], pancreatic[14], cervical[15], and colorectal cancers[16] as well as hepatomas[17]. The increased expression of TXN in tumors has been associated with decreased patient survival in several cancers and with resistance to anticancer drugs[18,19]. The TXN-interacting protein (TXNIP), also known as vitamin D3 up-regulated protein-1 (VDUP1) and TXN-binding protein-2 (TBP-2), inhibits the interaction between TXN and other factors. TXNIP overexpression inhibits TXN activity, which in turn inhibits tumor cell proliferation and cell cycle progression[20,21]. Recently, it was reported that knockout of the TXNIP gene in a mouse model induced Helicobacter pylori (H. pylori)-related gastric cancer[22]. However, there is little information regarding the prognostic value of the expression level of TXN and TXNIP in gastric cancer.
In this study, we evaluated the use of TXN expression combined with TXNIP expression as prognostic markers to individualize the postoperative treatment strategy following gastric cancer removal.
The previously generated gene expression data from gastric cancer patients are available in the NCBI’s GEO public database (microarray data accession number, GSE13861)[23]. Sixty-five gastric cancer patients underwent curative surgery as a primary treatment, with clinical data obtained from the Yonsei University Severance Hospital (Table 1). Sixty-five surgically removed frozen gastric adenocarcinoma tissues and 19 normal surrounding tissue samples were used for the microarray experiments. The total RNA was extracted from the fresh-frozen tissues using a mirVana RNA Isolation Labeling Kit (Ambion, Austin, TX, United States). For the labeling and hybridization, 500 ng of total RNA was used, according to the manufacturer’s protocols (Human-HT12 v.3 Expression BeadChip, Illumina, San Diego, CA, United States). The microarray data were normalized using the quantile normalization method in the Linear Models for Microarray Data package in the R language environment. The expression level of each gene was transformed into a log2 base prior to further analysis. The random variance t test was applied to identify the differentially expressed genes between the two tissue types. The gene expression differences were considered significant if the P value was less than 0.001. Cluster analysis was performed with Cluster 3.0 and TreeView[24]. Univariate analysis was performed by dividing the patients into two groups based on the median value of each gene expression level to search for prognostic genes.
| Characteristics | Microarray(n = 65) | qRT-PCR(n = 68) | TMA(n = 328) |
| Age | |||
| mean (range), yr | 63 (32-83) | 56 (26-82) | 57 (25-82) |
| Sex, n (%) | |||
| Male/female | 46 (71)/19 (29) | 36 (53)/32 (47) | 204 (62)/124 (38) |
| Follow up duration | |||
| Mean (95% CI), mo | 41.7 (41-42) | 89.5 (79-100) | 99.8 (97.5-102) |
| Histological type, n (%) | |||
| Intestinal | 23 (35) | 14 (21) | 100 (30) |
| Diffuse | 42 (65) | 54 (79) | 228 (70) |
| TNM stage, n (%) | |||
| I | 12 (18) | 0 | 101 (31) |
| II | 11 (17) | 0 | 79 (24) |
| III | 26 (40) | 68 (100) | 110 (33) |
| IV | 16 (25) | 0 | 38 (12) |
| Location, n (%) | |||
| Cardia | 5 (8) | 8 (12) | 25 (8) |
| Non cardia | 60 (92) | 60 (88) | 303 (92) |
| Adjuvant chemotherapy, n (%) | |||
| Yes | 49 (75) | 59 (87) | 230 (70) |
| No | 16 (25) | 9 (13) | 98 (30) |
Paraffin-embedded cancer tissues were collected from gastric adenocarcinoma patients who underwent curative surgery between 1999 and 2007 as a primary treatment at Gangnam Severance Hospital. The clinical data of the patients were reviewed to obtain age, sex, tumor location, tumor differentiation, and stage based upon the American Joint Committee on Cancer 2002 criteria. The patients were followed up for more than 36 mo after surgery or until recurrence or death within 36 mo after surgery.
Sixty-eight stage III gastric cancer tissues were chosen to validate the microarray data (Table 1). The total RNA was extracted according to the manufacturer’s instructions (RecoverAll™ Total Nucleic Acid Isolation; Applied Biosystems, Foster City, CA, United States). The TXN and TXNIP genes were assayed using quantitative reverse transcription-polymerase chain reaction (qRT-PCR) with TaqMan gene-specific primers (Applied Biosystems, Foster City, CA, United States). Real-time RT-PCR amplification was performed using the 7900HT Fast Real-Time PCR System with a 384-well block module (Applied Biosystems, Foster City, CA, United States). The cycling conditions were as follows: 48 °C for 30 min and 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and at 60 °C for 60 s. The relative amounts of mRNA were calculated from the threshold cycle (CT) number using the expression of β-2 microglobulin as an endogenous control. All of the experiments were performed in triplicate, and the values were averaged.
Paraffin-embedded tissue microarray blocks of gastric cancer tissue specimens were created from tissues from 328 patients. Each block had 3-mm cores of gastric cancer tissue. The 4-μm thick sections were deparaffinized and processed to block endogenous peroxidase activity. Next, an antigen retrieval step was performed. Subsequently, primary anti-TXN (Polyclonal, 1:500, Abcam, Cambridge, MA, United States) and anti-TXNIP antibodies (Polyclonal, 1:100 Sigma, St. Louis, MO, United States) were applied to the sections. The sections were then incubated with a secondary antibody (HRP-rabbit/mouse), and the stains were developed using a NovaRED substrate kit (VECTOR Laboratory, Burlingame, CA, United States). The samples were then counterstained with Harris hematoxylin.
The TXN and TXNIP protein expression levels were evaluated by two pathologists. Over-expression was defined as staining higher in more than 50% of cancer cells compared to the matching normal cells, regardless of cytoplasmic or nuclear location. Underexpression was defined as no staining or staining positivity lower than that of the matching normal tissue, and normal expression was defined as a level of staining positivity similar to that of the matching normal tissue. For slides that were heterogeneously stained within a tumor, we graded the highest intensity within the tumor.
The BRB-Array Tools system was used for the analysis of the microarray data. Statistical analysis were primarily performed with PASW Statistics 17.0 (SPSS Inc., Chicago, IL, United States). A χ2 test was used to compare the difference between the groups. Kaplan-Meier plots and a log-rank test were used to estimate patient survival. Associations between the expression levels of the two targets and in-trans correlation were analyzed using the Pearson correlation coefficient. A P value of less than 0.05 was considered statistically significant, and all tests were two tailed.
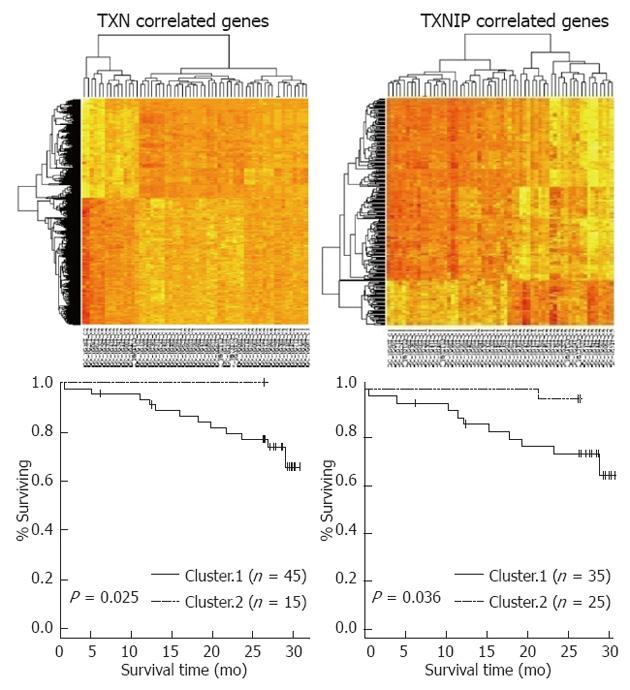
First, a hierarchical clustering analysis was applied to the gene expression data from the 65 human primary tumor tissue samples. Unsupervised clustering revealed 2 distinctive subtypes with clear differences in overall gene expression patterns[23]. Recurrence-free survival (RFS) was found to differ significantly between the 2 clusters (P = 0.001 by the log rank test), indicating that the molecular features of these tumors reflected in gene expression patterns might be strong independent predictors of clinical outcomes.
We next sought to identify genes whose expression was unique to the poor prognostic subgroup by cross-comparing gene lists. Between the two groups, 1258 genes were differentially expressed (P < 0.001) and presented more than a 1.5-fold difference between the two groups. Next, a univariate analysis identified 84 prognostic genes (P < 0.01). Of the 84 genes, TXN was a prognosis-related gene (P = 0.009 by univariate analysis) and was up-regulated in the poor prognostic group. From the correlation analysis between the expression levels of TXN and associated genes, we determined the relative correlations. TXNIP, a TXN inhibitor, demonstrated a negative correlation with TXN (r = -0.295, P = 0.024). The survival analysis based on the hierarchical clustering of TXN- or TXNIP-related genes demonstrated that each cluster influenced patient survival (P = 0.025 and P = 0.036, respectively) (Figure 1).
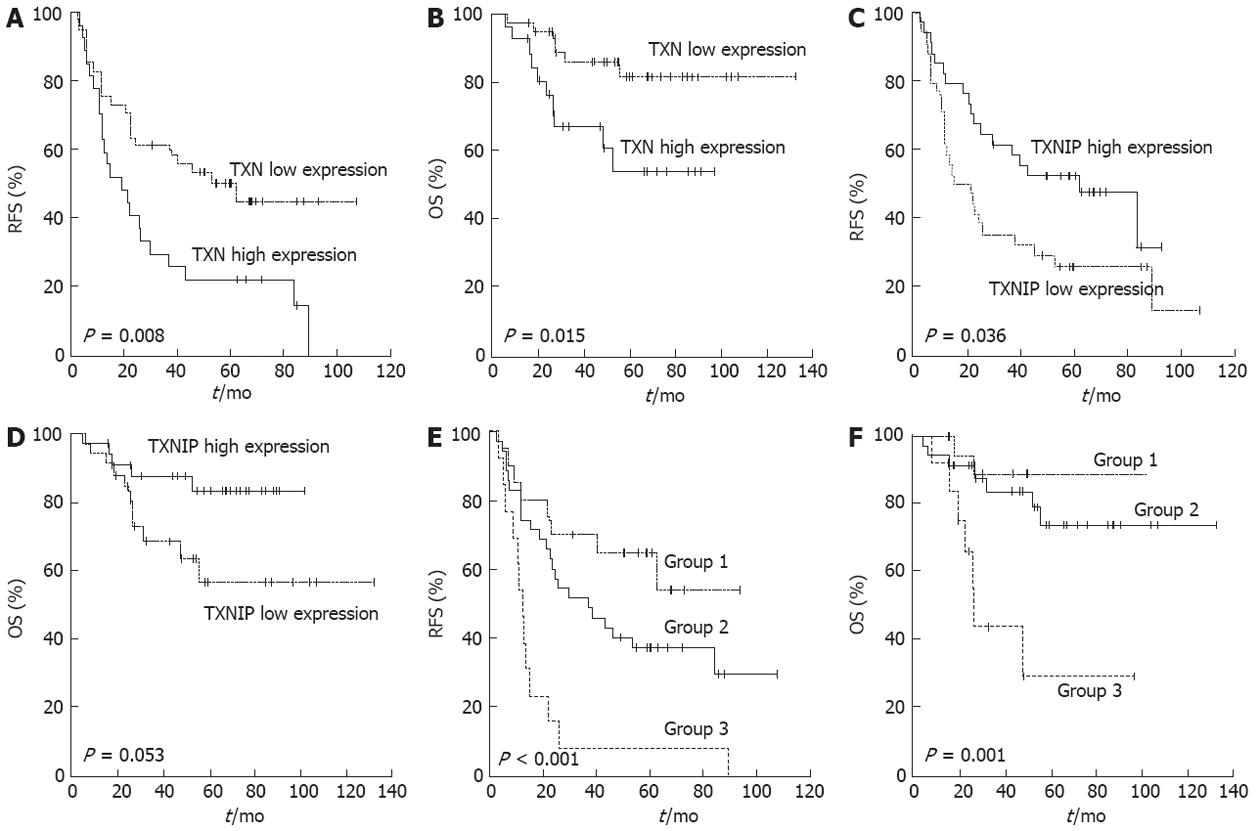
To determine if TXN and TXNIP gene expression was associated with prognosis among patients with the same stage of cancer, 68 stage III patients were randomly selected for qRT-PCR analysis (Table 1). The median follow-up duration after curative resection was 89.5 mo. By the last follow-up visit, 44 patients had experienced a recurrence, and 16 patients had died of gastric cancer. The patients were divided into high or low expression groups according to the TXN and TXNIP expression levels. RFS and overall survival (OS) were analyzed between the two groups (Figure 2A-D). Both TXN and TXNIP expression influenced RFS, and these associations were significant (P = 0.008 and P = 0.036, respectively). The patients in the high TXN expression or low TXNIP expression groups presented a poor prognosis. TXN expression also influenced OS (P = 0.015), but TXNIP expression did not have a significant relationship with OS (P = 0.053).
Stage III patients were classified into the following three combination groups based on the TXN and TXNIP expression levels: Group 1, simultaneously low TXN and high TXNIP levels (n = 20); Group 2, neither Group 1 nor Group 3 (n = 35); and Group 3, simultaneously high TXN and low TXNIP levels (n = 13). As expected, Group 1 exhibited a better prognosis than the other groups, and Group 3 presented with a poor prognosis (Figure 2E and F; P < 0.001 for RFS and P = 0.001 for OS). More than half of the Group 3 patients experienced a recurrence within 1 year after curative surgery, and their 5-year survival rate was one-third (29%) that of Group 1 (89%) (Table 2). There was no relationship between the TXN/TXNIP expression levels and either tumor stage or histological cell type.
| Group | Mean RFS,(95% CI), mo | 3-yr recurrenc-e-free rate (%) | 5-yr surviv-al rate (%) |
| 1: Low TXN and high TXNIP | Non applicable | 70 | 89 |
| 2: Neither group 1 nor group 3 | 36.7 (15.5-57.9) | 51 | 73 |
| 3: High TXN and low TXNIP | 11.9 (9.5-14.2) | 7 | 29 |
| P value by log rank test | P < 0.001 | P = 0.001 |
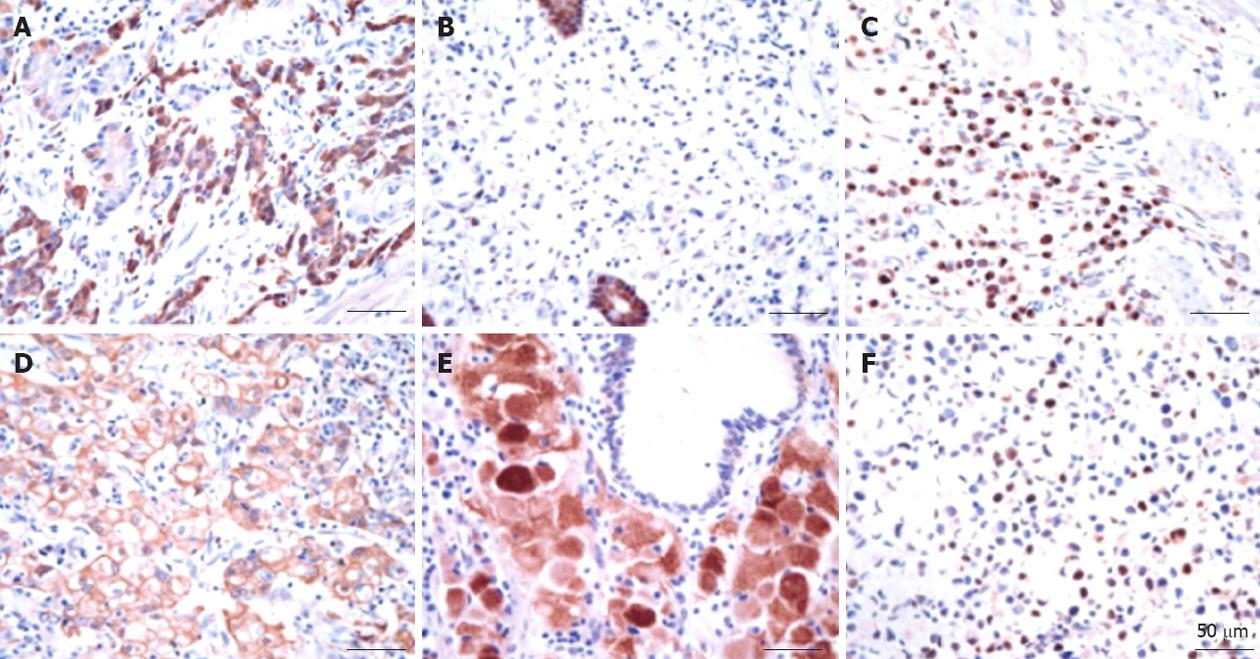
The gastric glands were relatively well stained with the anti-TXN antibody regardless of the presence of cancerous and non-cancerous lesions based on the immunohistochemical assay results. Thioredoxin was overexpressed in approximately 65% of cancer tissues regardless of the histological type and TNM stage (Table 3). Thioredoxin staining was observed in the cytoplasm or nucleus or in both areas of the cancer cells. The histological location of thioredoxin in the cytoplasm or the nucleus in cancer tissues varied among patients, and there was heterogeneity in the staining intensity within the individual samples. In contrast to the staining with the anti-thioredoxin antibody, the staining of the gastric cancer tissue with the anti-TXNIP antibody was weak. TXNIP was underexpressed in almost 85% of the cancer cells and was found in the cytoplasm or nucleus of the cancer cells. There was a tendency for TXNIP to be underexpressed in two poor prognosis groups: diffuse histology type (P = 0.009) and high-stage gastric cancer (P = 0.011) (Table 3). Representative immunohistochemical staining results are shown in Figure 3.
| Characteristic (n) | TXN overexpressionn (%) | TXNIP underexpressionn (%) |
| Total (328) | 213 (65) | 278 (85) |
| Histologic type | ||
| Diffuse type (228) | 149 (65) | 201 (88) |
| Intestinal type (100) | 64 (64) | 77 (77) |
| P = 0.9 | P = 0.012 | |
| Stage | ||
| Stage I/II (180) | 116 (64) | 142 (79) |
| Stage III/IV (148) | 97 (66) | 136 (92) |
| P = 0.767 | P = 0.009 |
Approximately 7000 genes were significantly correlated with TXN according to the bioinformatic in-trans correlation analysis of TXN expression with whole mRNA-expressing genes (P < 0.001). These findings suggest that TXN plays an important role in gastric cancer. Glutaredoxin 2 (GLRX2) and peroxiredoxin 4 (PRDX4) were highly correlated with TXN (r = 0.76, P < 0.001 and r = 0.72, P < 0.001, respectively; Table 4), and this correlation explains the function of TXN as a reactive oxygen species (ROS) scavenger with these known redox molecules. Ribosomal proteins and mitochondrial ribosomal proteins (i.e., RPL6, RPL29, MRPL22, MRPL42, MRPS17 and ATP5S) were highly correlated with TXN (P < 0.001; Table 4), which suggests the active involvement of TXN in protein synthesis and mitochondrial ATP synthesis for energy production. TXNIP was negatively correlated with TXN and known poor prognostic markers, such as AURKA, ERBB3, CCNB1, and many genes that are significantly correlated with TXN (Table 4). These results indirectly support the functional role of the TXNIP in the inhibition of TXN.
| Symbol | Gene name | Correlation with TXN | Correlation with TXNIP | ||
| r | P value | r | P value | ||
| TXN | Thioredoxin | 1.000 | 0.000 | -0.295 | 0.022 |
| RPL6 | Ribosomal protein L6 | 0.779 | < 0.001 | -0.335 | 0.009 |
| GLRX2 | Glutaredoxin 2 | 0.764 | < 0.001 | -0.314 | 0.015 |
| RPL29 | Ribosomal protein L29 | 0.757 | < 0.001 | -0.360 | 0.005 |
| MRPL22 | Mitochondrial ribosomal protein L22 | 0.751 | < 0.001 | -0.225 | 0.085 |
| MRPL42 | Mitochondrial ribosomal protein L42 | 0.734 | < 0.001 | -0.181 | 0.166 |
| MRPS17 | Mitochondrial ribosomal protein S17 | 0.732 | < 0.001 | -0.387 | 0.002 |
| ATP5S | ATP synthase-coupling factor B | 0.728 | < 0.001 | -0.182 | 0.165 |
| PRDX4 | Peroxiredoxin 4 | 0.722 | < 0.001 | -0.321 | 0.012 |
| AURKA | Aurora kinase A | 0.611 | < 0.001 | -0.486 | < 0.001 |
| ERO1L | ERO1-like | 0.586 | < 0.001 | -0.200 | 0.125 |
| HIG2 | Chromosome 7 open reading frame 68 | 0.574 | < 0.001 | -0.089 | 0.498 |
| CCNB1 | Cyclin B1 | 0.563 | < 0.001 | -0.503 | < 0.001 |
| ERBB3 | HER3 | 0.004 | 0.975 | -0.536 | < 0.001 |
| TXNIP | Thioredoxin interacting protein | -0.295 | 0.022 | 1.000 | 0.000 |
| ATG12 | ATG12 autophagy related 12 homolog | -0.482 | < 0.001 | 0.072 | 0.582 |
| ATG16L2 | ATG16 autophagy related 16-like 2 | -0.722 | < 0.001 | 0.216 | 0.098 |
| ATG10 | Autophagy related 10 homolog | -0.756 | < 0.001 | 0.266 | 0.040 |
We demonstrated that TXN and TXNIP are poor and good prognostic gastric cancer markers, respectively. In stage III patients who underwent curative gastrectomy, the 3-year recurrence-free rate (P < 0.001) and the 5-year survival rate (P = 0.001) were significantly different between patients with different TXN and TXNIP expression levels (Table 2). High TXN and low TXNIP patients had a definitively poor prognosis. Most of these patients experienced a recurrence within 2 years and died within 4 years. Thus, it is necessary to perform intensive treatment and plan post-adjuvant treatment for these patients.
The gene expression microarray data imply that TXN plays an important role in gastric cancer. TXN functions as a ROS scavenger with known redox molecules, such as GLRX2 and PRDX4, and TXN is actively involved in protein synthesis and mitochondrial ATP synthesis for energy production. Hypoxia-induced genes [i.e., ERO1L and hypoxia inducible gene 2 (HIG2)] were significantly correlated with TXN. Tumors with high TXN expression also exhibited elevated ERO1L and HIG2 levels; therefore, these tumors are in a relatively high hypoxic state compared with tumors with low TXN expression. Additionally, autophagy-related genes (i.e., ATG10, ATG16L2, and ATG12) were negatively correlated with TXN expression, which indirectly suggests that TXN is involved in autophagic inhibition in a hypoxic cancer state. Autophagy is associated with cancer pathogenesis and chemotherapy resistance; therefore, the function of TXN in autophagy should be elucidated through additional studies.
Grogan et al[25] demonstrated that TXN was localized to tumor cells and was overexpressed in gastric cancer tissues compared with the levels in normal gastric mucosa. TXN overexpression was typically found in both the nucleus and the cytoplasm of neoplastic cells. These findings are consistent with our immunohistochemical staining results. Furthermore, high TXN expression was observed regardless of the gastric cancer stage or cell type.
Our study confirmed that TXNIP was significantly underexpressed in gastric cancer tissues compared with normal tissues; furthermore, it was expressed at the lowest levels in cancer patients with poor prognoses. TXNIP is a known potent tumor suppressor whose expression is markedly decreased in various human cancers, including gastric cancer. Knockout of the TXNIP gene in a mouse model was associated with H. pylori-related gastric cancer[22]. In an in vitro experiment, TXNIP overexpression in pancreatic cells resulted in a higher basal level of apoptosis and an increased sensitivity to cisplatin and oxaliplatin[26]. In microarray data analysis, TXNIP expression was negatively correlated with the expression of TXN expression, known as poor prognostic cancer biomarkers[27-29], and many genes that are significantly correlated with TXN. These results indirectly confirm the functional role of the TXNIP in TXN inhibition. Additionally, autophagy-related genes were correlated with TXNIP, and these data support that TXNIP induces cancer cell autophagy.
In this study, we analyzed the gene expression profile of human gastric cancer to identify potential biomarkers that could be used to classify patients according to prognosis after curative resection. We found that TXN and TXNIP were significantly associated with prognosis. TXN up-regulation and the simultaneous down-regulation of TXNIP were associated with a poor prognosis in gastric cancer patients. Bioinformatic analysis revealed that TXN and TXNIP were highly correlated with many oncogenes and tumor suppressor genes and demonstrated that TXN and TXNIP were associated with genes related to energy, protein synthesis and the modulation of autophagy under hypoxic or other stressful conditions. One of the limitations of this study is that we did not elucidate how TXN and TXNIP affect the recurrence of gastric cancer after curative resection. Further investigation of TXN and TXNIP in gastric cancer would likely identify unknown pathogenic mechanisms. In addition, because our results were derived from retrospective assessment, it is necessary to utilize TXN and TXNIP gene expression-based prediction in a prospective randomized trial(s) to validate the true clinical relevance of TXN and TXNIP. It will be beneficial to identify effective anti-tumor treatments other than the current standard adjuvant chemotherapy for gastric cancer patients with potent recurrence factor, high TXN and low TXNIP expression. Furthermore, TXN-targeted agents or those that up-modulate TXNIP could be used in targeted therapy in the treatment of gastric cancer patients who are selected based on biomarker gene signatures.
In conclusion, TXN and TXNIP are promising prognostic markers for gastric cancer, and performing personalized adjuvant treatment based on TXN and TXNIP expression levels would be an effective practice in the treatment of gastric cancer.
The standard of treatment for gastric cancer is surgery. After curative resection, even among individuals with the same stage of cancer, diverse recurrence patterns are present. However, uniform adjuvant treatment after curative resection has been performed. Authors evaluated the use of thioredoxin (TXN) expression combined with thioredoxin-interacting protein (TXNIP) expression as prognostic markers to individualize the postoperative treatment strategy following gastric cancer removal.
TXN is putative oncoprotein that provides growth and survival advantages to tumor cells through the activation of redox-sensitive transcription factors. TXNIP is a potent tumor suppressor and inhibits the interaction between TXN and other factors. They have been recognized as cancer-related markers in diverse malignancies.
High TXN and low TXNIP expression in human gastric cancer tissue definitively related with poor prognosis of gastric cancer patients who had undergone curative resection.
The establishment of adjuvant treatment strategy based on our discovery would be useful to accomplish better outcomes after surgical treatment. The patients with high TXN and low TXNIP expressions in cancer tissue require novel treatment and should be more carefully monitored after surgery.
Redox (reduction-oxidation) reactions: Many important biological processes involve redox reactions. Free radicals are a part of redox molecules and can become harmful to the human body if they do not reattach to the redox molecule or an antioxidant. Unsatisfied free radicals can trigger the mutation of cells they encounter and are thus cancer causing.
This study investigated the potential of TXN and TXNIP genes as a prognostic marker after curative resection of gastric cancer. Patients with high TXN and low TXNIP expression in cancer tissue manifested significantly more recurrence and shorter survival. However, it is necessary to validate through the prospective trial and to elucidate the functional mechanism of TXN and TXNIP.
Peer reviewers: Dr. Ki Baik Hahm, Professor, Department of Gastroenterology, Gachon Graduate School of Medicine, 7-45 Songdo-dong, Yeonsu-gu, Incheon 406-840, South Korea; Hitoshi Tsuda, MD, PhD, Diagnostic Pathology Section, Clinical Laboratory Division, National Cancer Center Hospital, 5-1-1 Tsukiji, Chuo-ku, Tokyo 104-0045, Japan
S- Editor Gou SX L- Editor A E- Editor Lu YJ
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25542] [Article Influence: 1824.4] [Reference Citation Analysis (7)] |
| 2. | Gallo A, Cha C. Updates on esophageal and gastric cancers. World J Gastroenterol. 2006;12:3237-3242. [PubMed] |
| 3. | Bang YJ, Kim YW, Yang HK, Chung HC, Park YK, Lee KH, Lee KW, Kim YH, Noh SI, Cho JY. Adjuvant capecitabine and oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): a phase 3 open-label, randomised controlled trial. Lancet. 2012;379:315-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1267] [Cited by in RCA: 1291] [Article Influence: 99.3] [Reference Citation Analysis (0)] |
| 4. | Bao G, Qiao Q, Zhao H, He X. Prognostic value of HMGB1 overexpression in resectable gastric adenocarcinomas. World J Surg Oncol. 2010;8:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 5. | Santini D, Vincenzi B, Fratto ME, Perrone G, Lai R, Catalano V, Cass C, Ruffini PA, Spoto C, Muretto P. Prognostic role of human equilibrative transporter 1 (hENT1) in patients with resected gastric cancer. J Cell Physiol. 2010;223:384-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 6. | Ooki A, Yamashita K, Kikuchi S, Sakuramoto S, Katada N, Watanabe M. Phosphatase of regenerating liver-3 as a prognostic biomarker in histologically node-negative gastric cancer. Oncol Rep. 2009;21:1467-1475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 7. | Kim JS, Kim MA, Kim TM, Lee SH, Kim DW, Im SA, Kim TY, Kim WH, Yang HK, Heo DS. Biomarker analysis in stage III-IV (M0) gastric cancer patients who received curative surgery followed by adjuvant 5-fluorouracil and cisplatin chemotherapy: epidermal growth factor receptor (EGFR) associated with favourable survival. Br J Cancer. 2009;100:732-738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 8. | Kim YJ, Kim MA, Im SA, Kim TM, Kim DW, Yang HK, Heo DS, Lee KU, Choe KJ, Kim NK. Metastasis-associated protein S100A4 and p53 predict relapse in curatively resected stage III and IV (M0) gastric cancer. Cancer Invest. 2008;26:152-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 9. | Ueno M, Masutani H, Arai RJ, Yamauchi A, Hirota K, Sakai T, Inamoto T, Yamaoka Y, Yodoi J, Nikaido T. Thioredoxin-dependent redox regulation of p53-mediated p21 activation. J Biol Chem. 1999;274:35809-35815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 309] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 10. | Abate C, Patel L, Rauscher FJ, Curran T. Redox regulation of fos and jun DNA-binding activity in vitro. Science. 1990;249:1157-1161. [PubMed] |
| 11. | Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, Kawabata M, Miyazono K, Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 1998;17:2596-2606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1888] [Cited by in RCA: 1909] [Article Influence: 70.7] [Reference Citation Analysis (0)] |
| 12. | Welsh SJ, Bellamy WT, Briehl MM, Powis G. The redox protein thioredoxin-1 (Trx-1) increases hypoxia-inducible factor 1alpha protein expression: Trx-1 overexpression results in increased vascular endothelial growth factor production and enhanced tumor angiogenesis. Cancer Res. 2002;62:5089-5095. [PubMed] |
| 13. | Gasdaska PY, Oblong JE, Cotgreave IA, Powis G. The predicted amino acid sequence of human thioredoxin is identical to that of the autocrine growth factor human adult T-cell derived factor (ADF): thioredoxin mRNA is elevated in some human tumors. Biochim Biophys Acta. 1994;1218:292-296. [PubMed] |
| 14. | Nakamura H, Bai J, Nishinaka Y, Ueda S, Sasada T, Ohshio G, Imamura M, Takabayashi A, Yamaoka Y, Yodoi J. Expression of thioredoxin and glutaredoxin, redox-regulating proteins, in pancreatic cancer. Cancer Detect Prev. 2000;24:53-60. [PubMed] |
| 15. | Fujii S, Nanbu Y, Nonogaki H, Konishi I, Mori T, Masutani H, Yodoi J. Coexpression of adult T-cell leukemia-derived factor, a human thioredoxin homologue, and human papillomavirus DNA in neoplastic cervical squamous epithelium. Cancer. 1991;68:1583-1591. [PubMed] |
| 16. | Berggren M, Gallegos A, Gasdaska JR, Gasdaska PY, Warneke J, Powis G. Thioredoxin and thioredoxin reductase gene expression in human tumors and cell lines, and the effects of serum stimulation and hypoxia. Anticancer Res. 1996;16:3459-3466. [PubMed] |
| 17. | Kawahara N, Tanaka T, Yokomizo A, Nanri H, Ono M, Wada M, Kohno K, Takenaka K, Sugimachi K, Kuwano M. Enhanced coexpression of thioredoxin and high mobility group protein 1 genes in human hepatocellular carcinoma and the possible association with decreased sensitivity to cisplatin. Cancer Res. 1996;56:5330-5333. [PubMed] |
| 18. | Ungerstedt JS, Sowa Y, Xu WS, Shao Y, Dokmanovic M, Perez G, Ngo L, Holmgren A, Jiang X, Marks PA. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2005;102:673-678. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 399] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 19. | Kim SJ, Miyoshi Y, Taguchi T, Tamaki Y, Nakamura H, Yodoi J, Kato K, Noguchi S. High thioredoxin expression is associated with resistance to docetaxel in primary breast cancer. Clin Cancer Res. 2005;11:8425-8430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 156] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 20. | Junn E, Han SH, Im JY, Yang Y, Cho EW, Um HD, Kim DK, Lee KW, Han PL, Rhee SG. Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J Immunol. 2000;164:6287-6295. [PubMed] |
| 21. | Nishinaka Y, Nishiyama A, Masutani H, Oka S, Ahsan KM, Nakayama Y, Ishii Y, Nakamura H, Maeda M, Yodoi J. Loss of thioredoxin-binding protein-2/vitamin D3 up-regulated protein 1 in human T-cell leukemia virus type I-dependent T-cell transformation: implications for adult T-cell leukemia leukemogenesis. Cancer Res. 2004;64:1287-1292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 106] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Kwon HJ, Won YS, Nam KT, Yoon YD, Jee H, Yoon WK, Nam KH, Kang JS, Han SU, Choi IP. Vitamin D₃ upregulated protein 1 deficiency promotes N-methyl-N-nitrosourea and Helicobacter pylori-induced gastric carcinogenesis in mice. Gut. 2012;61:53-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Cho JY, Lim JY, Cheong JH, Park YY, Yoon SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17:1850-1857. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 257] [Cited by in RCA: 281] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 24. | Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863-14868. [PubMed] |
| 25. | Grogan TM, Fenoglio-Prieser C, Zeheb R, Bellamy W, Frutiger Y, Vela E, Stemmerman G, Macdonald J, Richter L, Gallegos A. Thioredoxin, a putative oncogene product, is overexpressed in gastric carcinoma and associated with increased proliferation and increased cell survival. Hum Pathol. 2000;31:475-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 139] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 26. | Baker AF, Koh MY, Williams RR, James B, Wang H, Tate WR, Gallegos A, Von Hoff DD, Han H, Powis G. Identification of thioredoxin-interacting protein 1 as a hypoxia-inducible factor 1alpha-induced gene in pancreatic cancer. Pancreas. 2008;36:178-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 27. | Dar AA, Zaika A, Piazuelo MB, Correa P, Koyama T, Belkhiri A, Washington K, Castells A, Pera M, El-Rifai W. Frequent overexpression of Aurora Kinase A in upper gastrointestinal adenocarcinomas correlates with potent antiapoptotic functions. Cancer. 2008;112:1688-1698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 93] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 28. | Sithanandam G, Anderson LM. The ERBB3 receptor in cancer and cancer gene therapy. Cancer Gene Ther. 2008;15:413-448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 186] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 29. | Begnami MD, Fregnani JH, Nonogaki S, Soares FA. Evaluation of cell cycle protein expression in gastric cancer: cyclin B1 expression and its prognostic implication. Hum Pathol. 2010;41:1120-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |