Published online Jul 14, 2012. doi: 10.3748/wjg.v18.i26.3458
Revised: May 9, 2012
Accepted: May 13, 2012
Published online: July 14, 2012
AIM: To investigate the gene knock-down effect by the phosphoinositide-3-kinase, catalytic, alpha polypeptide (PIK3CA)-targeted double-stranded RNA (dsRNA) and its effect on cell proliferation and cycle distribution in SW948.
METHODS: Two PIK3CA-targeted dsRNAs were constructed and transfected into SW948 cells. Transfections were performed using lipofectamineTM 2000. The transfection effectiveness was calculated basing on the rate of fluorescence cell of SW948 at 6 h after transfection. Total messenger RNA was extracted from these cells using the RNeasy kit, and semiquantitative reverse transcription polymerase chain reaction was performed to detect the down-regulation of PIK3CA, AKT1, MYC, and CCND1 gene expression. Cells were harvested, proteins were resolved, and western blot was employed to detect the expression levels of PIK3CA, AKT1, MYC, and CCND1 gene. Cell proliferation was assessed by 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide assay and the inhibition rate was calculated. Soft agar colony formation assay was performed basing on colonies greater than 60 μm in diameter at ×100 magnification. The effect on cell cycle distribution and apoptosis was assessed by flow cytometry. All experiments were performed in triplicate.
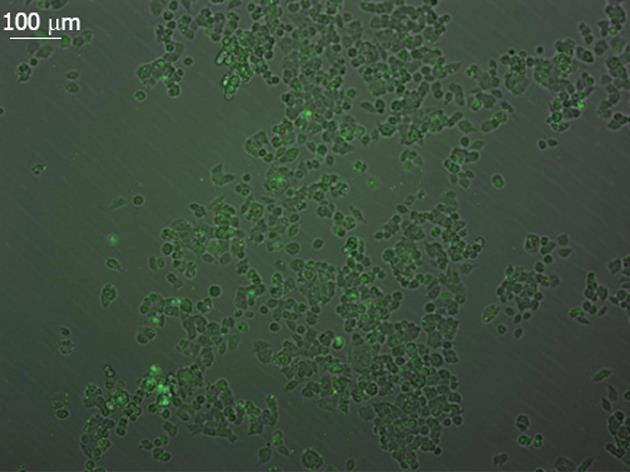
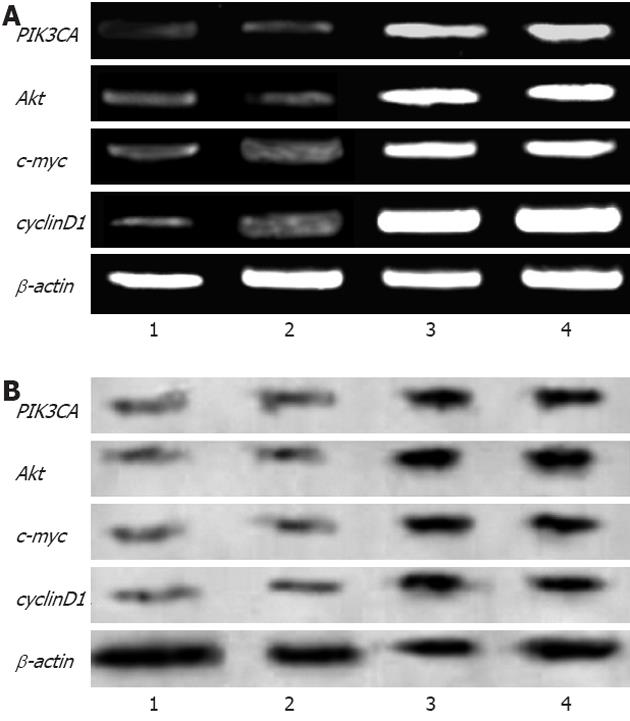
RESULTS: Green fluorescence was observed in SW948 cell transfected with plasmid Pgenesil-1, and the transfection effectiveness was about 65%. Forty-eight hours post-transfection, mRNA expression of PIK3CA in SW948 cells was 0.51 ± 0.04 vs 0.49 ± 0.03 vs 0.92 ± 0.01 vs 0.93 ± 0.03 (P = 0.001 ) in Pgenesil-CA1, Pgenesil-CA2, negative and blank group respectively. mRNA expression of AKT1 was 0.50 ± 0.03 vs 0.48 ± 0.01 vs 0.93 ± 0.04 vs 0.92 ± 0.02 (P = 0.000) in Pgenesil-CA1, Pgenesil-CA2, negative and blank group respectively. mRNA expression of MYC was 0.49 ± 0.01 vs 0.50 ± 0.04 vs 0.90 ± 0.02 vs 0.91 ± 0.03 (P = 0.001) in the four groups respectively. mRNA expression of CCND1 was 0.45 ± 0.02 vs 0.51 ± 0.01 vs 0.96 ± 0.03 vs 0.98 ± 0.01 (P = 0.001) in the four groups respectively. The protein level of PIK3CA was 0.53 ± 0.01 vs 0.54 ± 0.02 vs 0.92 ± 0.03 vs 0.91 ± 0.02 (P = 0.001) in Pgenesil-CA1, Pgenesil-CA2, negative and blank group respectively. The protein level of AKT1 in the four groups was 0.49 ± 0.02 vs 0.55 ± 0.03 vs 0.94 ± 0.03 vs 0.95 ± 0.04, P = 0.000). The protein level of MYC in the four groups was 0.51 ± 0.03 vs 0.52 ± 0.04 vs 0.92 ± 0.02 vs 0.95 ± 0.01 (P = 0.000). The protein level of CCND1 in the four groups was 0.54 ± 0.04 vs 0.56 ± 0.03 vs 0.93 ± 0.01 vs 0.93 ± 0.03 (P = 0.000). Both Pgenesil-CA1 and Pgenesil-CA2 plasmids significantly suppressed the growth of SW948 cells when compared with the negative or blank group at 48 h after transfection (29% vs 25% vs 17% vs 14%, P = 0.001), 60 h after transfection (38% vs 34% vs 19% vs 16%, P = 0.001), and 72 h after transfection (53% vs 48% vs 20% vs 17%, P = 0.000). Numbers of colonies in negative, blank, CA1, and CA2 groups were 42 ± 4, 45 ± 5, 8 ± 2, and 10 ± 3, respectively (P = 0.000). There were more than 4.5 times colonies in the blank and negative control groups as there were in the CA1 and CA2 groups. In addition, the colonies in blank and negative control groups were also larger than those in the CA1 and CA2 groups. The percentage of cells in the CA1 and CA2 groups was significantly higher in G0/G1 phase, but lower in S and G2/M phase when compared with the negative and control groups. Moreover, cell apoptosis rates in the CA1 and CA2 groups were 5.11 ± 0.32 and 4.73 ± 0.32, which were significantly higher than those in negative (0.95 ± 0.11, P = 0.000) and blank groups (0.86 ± 0.13, P = 0.001). No significant difference was found between CA1 and CA2 groups in cell cycle distribution and apoptosis.
CONCLUSION: PIK3CA-targeted short hairpin RNAs can block the phosphoinositide 3-kinase-Akt signaling pathway and inhibit cell growth, increase apoptosis, and induce cell cycle arrest in the PIK3CA-mutant colon cancer SW948 cells.
- Citation: Huang WS, Wang TB, He Y, Chen YJ, Zhong SL, Tan M. Phosphoinositide-3-kinase, catalytic, alpha polypeptide RNA interference inhibits growth of colon cancer cell SW948. World J Gastroenterol 2012; 18(26): 3458-3464
- URL: https://www.wjgnet.com/1007-9327/full/v18/i26/3458.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i26.3458
Colon cancer is the fifth most common cancer in the United States, and the third leading cause of cancer-related death in the Western world. The phosphoinositide 3-kinase (PI3K)/Akt signaling transduction pathway is believed to play an important role in carcinogenesis. PI3K is a major signaling component downstream of growth factor receptor tyrosine kinases (RTKs), which may serve as a potential target for colon cancer therapy[1,2]. PI3K is a heterodimer consisting of a regulatory subunit (p85) and a catalytic subunit (p110). The subtype p110a encoded by the gene phosphoinositide-3-kinase, catalytic, alpha polypeptide (PIK3CA) is very important in phosphorylating phosphatidylinositol (4,5)-bisphosphate (PIP2) to the lipid second messenger PIP3, and PIP3 in turn functions in the recruitment and activation of a wide range of downstream targets, including the serine-threonine protein kinase Akt. Gene reactions distal to the PI3K-Akt signaling pathway contribute to tumor cell proliferation, cell cycle progression, energy metabolism, and resistance to apoptosis[3-6]. Troxell et al[7] found PIK3CA mutations were identified in 13/24 breast columnar cell lesions (54%) and 3/8 invasive carcinomas (37%). The high prevalence of PIK3CA mutations in breast cancer is an emerging tumor marker which might become used in treatment-choosing process[8]. Higgins et al[9] detected the PIK3CA status in metastatic breast cancer using peripheral blood, and found plasma level of PIK3CA presented predictive biomarkers of response to targeted therapies. PIK3CA mutations were detected in 18% patients with breast and gynecologic malignancies, and cases with PIK3CA mutations treated with its inhibitors demonstrated a higher response rate than patients without mutations[10]. Studies demonstrated PIK3CA mutations were not common, but its amplification was very common in gastric cancer, Akt (p-Akt) was often functionally linked to tumor progression and metastasis in gastric cancer, and double-stranded RNA (dsRNA) mediated targeting of PIK3CA may specifically knockdown the expression of PIK3CA, providing a potential implication for therapy of cancer[11-13].
RNA interference (RNAi) is a potent gene-silencing tool, which is triggered by the introduction of dsRNA. Degradation of mRNA homologous to dsRNA can cause a post-transcriptional gene-silencing effect. Recently, the vector-based RNAi has been developed in order to achieve long-term and stable effects. Short hairpin RNA (shRNA) is formed by hairpin structures and stretches of dsRNA. After being cleaved by a ribonuclease dicer, it becomes mature microRNA (miRNA) inside the targeted cells[14-17].
In our study, two shRNA plasmid vectors were constructed against the gene PIK3CA, and were transfected into the colon cancer cell line SW948. Their effects on cell proliferation and cycle distribution in this cell line were investigated.
The human colon carcinoma cell line SW948 was routinely grown in Leibowitz’s L-15 medium supplemented with 10% fetal calf serum, penicillin (100 U/mL), and streptomycin (100 μg/mL) at 37 °C in a humidified 5% CO2/95% air atmosphere.
Two small interference RNA (siRNA) for PIK3CA were designed according to the consensus sequence of the PIK3CA gene (GeneBank NM_006218) obtained from the online database of the National Center for Biotechnology Information, and then were cloned separately into the vector plasmid Pgenesil-1. One sequence, 5’-GCTATCATCTGAACAATTA-3’ was desgnated Pgenesil-CA1, and the other 5’-GGATAGAGGCCAAATAATA-3’ was designated Pgenesil-CA2. Both were verified in a basic local alignment search tool search of the database. An siRNA scrambled sequence 5’-GACTTCATAAGGCGCATGC-3’, designated Pgenesil-Neg, was used as a negative control. All siRNA sequences were synthesized by Invitrogen (Carlsbad, CA). The multiclone sites of plasmid Pgenesil-1 containing enhanced green fluorescent protein gene were as follows: HindIII-ShRNA-BamHI-U6Promotor-EcoRI-SalI-XbaI-DraIII. All the above RNAi sequences were transcribed with DNA polymerase III U6 promoter. Cells at 80%-90% confluency were transfected with the three shRNA vectors CA1, CA2, and Neg group, described above. Transfections were performed using lipofectamine TM2000 (Invitrogen, United State) according to the manufacture’s instructions. The Blank group was also treated with only LipofectamineTM2000, but without vector. Six hours post-transfection, 500 μL of fetal bovine serum (FBS) was added per well. Twenty-six hours after transfection, the medium was replaced by normal medium containing 10% FBS and antibiotics up to 72 h post-transfection. The transfection effectiveness was calculated basing on the rate of fluorescence cell of SW948 at 6 h after transfection.
Cultured cells (described above) were harvested 48 h post-transfection. Total messenger RNA was extracted from these cells using the RNeasy kit. Reverse transcription polymerase chain reaction was performed using 500 ng of total RNA samples with oligo dT primers (Fermentas, United States). β-actin mRNA was included as an internal standard for quantitative analysis. The primer pairs used in this study are listed in Table 1. Samples in each group were run in triplicate. The photodensity of the goal gene product was normalized with respect to β-actin content.
| Goal gene | Upstream primer | Downstream primer | PCR frag (bp) |
| β-actin | 5’-TCCTGTGGATCCACGAAACT-3’ | 5’-GAAGCATTTGCGGTGGACGAT-3’ | 330-bp |
| PIK3CA | 5’-CCCTGCTCATCAACTAGGAAACC-3' | 5’-TTGCCGTAAATCATCCCCATT-3' | 160-bp |
| Akt | 5’-GGACAACGCCATCCAGACT-3’ | 5’-GCCAGGGACACCTCCATCTC-3’ | 121-bp |
| c-myc | 5’-TACCCTCTCAACGACAGCAG-3’ | 5’-TCTTGACA TTCTCCTCGGTG-3’ | 477-bp |
| cyclinD1 | 5’-GCCAACCTCCTCAACGACCGG-3' | 5’-GTCCATGTTCTGCTGGGCCTG-3’ | 744-bp |
Cells were harvested 48 h post-transfection, and were washed twice in ice-cold 1× phosphate-buffered saline (PBS). Cell pellets were treated with the lysis buffer, and whole cell extracts were isolated. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10%), then transferred onto a polyvinylidene fluoride membrane, and subjected to immunohybridization analysis using a monoclonal antibody against the targeted proteins (Santa Cruz Biotechnology, Inc., United States). Peroxidase-conjugated secondary antibody was added later (Bolster, China). Hybridized protein bands were displayed via chemiluminescence reagents (Santa Cruz Biotechnology, Inc., United State) according to the manufacturer’s instructions. β-actin staining was used as an internal standard. All experiments were performed in triplicate.
The transfected cells were seeded in 96-well plates (1×104/mL), and were allowed to attach for 24 h. Thiazolyl blue tetrazolium bromide (MTT, Sigma, United States) was dissolved in 1× PBS at a concentration of 5 mg/mL, and filtered as a stock solution. Ten microlitres of stock solution were added to 100 μL of medium in each well. The plates were incubated for 4 h at 37 °C. After loading of 3- (4,5-cimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), medium was replaced with 100 µL dimethyl sulfoxide, and was incubated for 30 min at room temperature for color development. Results were read by an enzyme-linked immunosorbent assay reader (570 nm, DG-3022A, United States) to determine absorbance values (A). In each group, time points for detection of absorbance values were 36, 48, 60 and 72 h after transfection. The inhibition rate was calculated as follows: Inhibition rate (IR) = [1 - A1/A2] × 100%, where A1 is the absorbance value of the observation group, and A2 is the absorbance value of the control group.
Different groups of cells were mixed with culture medium containing agar at a final concentration of 0.4% 24 h after transfection. One milliliter of cell suspension was seeded onto 6-well plates coated with 0.5% agar in culture medium. Colonies greater than 60 μm in diameter at ×100 magnification were counted in each plate to measure the colony efficiency after 10 d incubation. The assays were performed in triplicate.
Flow cytometry was used to assess cell cycle and apoptosis. Cells were harvested 72 h after incubation, and then were washed with cold 1 × PBS and fixed with 80% ethanol overnight at -20 °C. Next, cells were treated with RNase A (Sigma, United States), and were stained with propidium iodide (PI, Sigma, United States). All samples underwent analysis using flow cytometry (Becton Dickinson, United States). The experiments were performed three times.
Results of experimental data are reported as mean ± SE. Significance levels were determined by one-way analysis of variance and Student’s t test using SPSS 11.5 Statistics software. A statistically significant result is indicated by a P < 0.05.
SW948 transfected with plasmid Pgenesil-1 presented green fluorescence, and the transfection effectiveness was about 65% (Figure 1). Forty-eight hours post-transfection, the expression of mRNA and protein from targeted genes was tested in SW948 cells. The results are shown in Figure 2, Tables 1 and 2. All mRNA and protein expression from targeted genes in SW948 cells was down-regulated significantly after transfection with either plasmids Pgenesil-CA1 or Pgenesil-CA2 (P < 0.001, vs negative or blank group). No significant difference was found between the CA1 and CA2 groups. Pgenesil-Neg plasmid had no significant inhibitory effect on the expression of mRNA or protein.
| Goal gene | CA1 | CA2 | Negative | Blank |
| mRNA expression | ||||
| PIK3CA | 0.51 ± 0.04a | 0.49 ± 0.03a | 0.92 ± 0.01 | 0.93 ± 0.03 |
| Akt | 0.50 ± 0.03b | 0.48 ± 0.01b | 0.93 ± 0.04 | 0.92 ± 0.02 |
| c-myc | 0.49 ± 0.01a | 0.50 ± 0.04a | 0.90 ± 0.02 | 0.91 ± 0.03 |
| cyclinD1 | 0.45 ± 0.02a | 0.51 ± 0.01a | 0.96 ± 0.03 | 0.98 ± 0.01 |
| Protein expression | ||||
| PIK3CA | 0.53 ± 0.01a | 0.54 ± 0.02a | 0.92 ± 0.03 | 0.91 ± 0.02 |
| Akt | 0.49 ± 0.02b | 0.55 ± 0.03b | 0.94 ± 0.03 | 0.95 ± 0.04 |
| c-myc | 0.51 ± 0.03b | 0.52 ± 0.04b | 0.92 ± 0.02 | 0.95 ± 0.01 |
| cyclinD1 | 0.54 ± 0.04b | 0.56 ± 0.03b | 0.93 ± 0.01 | 0.93 ± 0.03 |
The effect of PIK3CA silencing on proliferation of SW948 cells was analyzed by the MTT assay. As demonstrated in Table 3, both Pgenesil-CA1 and Pgenesil-CA2 plasmids significantly suppressed the growth of SW948 cells when compared with the negative or blank group.
Soft-agar colony-formation assays were used to analyze the anchorage-independent proliferation of SW948 cells. Numbers of colonies in negative, blank, CA1, and CA2 group were 42 ± 4, 45 ± 5, 8 ± 2, and 10 ± 3, respectively. There were more than 4.5 times more than colonies in the blank and negative control groups as there were in the CA1 and CA2 groups. In addition, the colonies in blank and negative control groups were also larger than those in the CA1 and CA2 groups.
As shown in Table 4, the percentage of cells in the CA1 and CA2 groups was significantly higher in G0/G1 phase, but lower in S and G2/M phase when compared with the negative and control groups. Moreover, cell apoptosis rates in the CA1 and CA2 groups were around 5%, which was significantly higher than those in negative and blank groups. No significant difference was found between CA1 and CA2 groups in cell cycle distribution and apoptosis.
One of the areas of greatest interest in cancer research concerns the coordination of cancer cell proliferation and apoptosis. The balance between cell proliferation and apoptosis, and the distribution of cell cycles play very important roles in the process of carcinogenesis in colon cancer[18]. Abnormal activation of the PI3K signaling pathway can cause hyper-proliferation of intestinal crypt progenitors, and promote the transformation of a normal cell to a cancer cell. This process involves a series of complicated molecular activities. The PI3K signaling cascade begins with the phosphorylation of PIP2 to PIP3, which is largely regulated by the balance of activity of PTEN and PI3Ks. Many investigators have reported that mutations of PI3Ks contribute to the process of carcinogenesis in colon cancer[2,5]. Liao et al[19] reported that co-existence of PIK3CA exon 9 and 20 mutations, but not PIK3CA mutation in either exon 9 or 20 alone, was associated with poor prognosis of colorectal cancer patients. Thus, PI3K has become the focus of considerable attention for research into the treatment of colon cancer. Small-molecule inhibitors targeting PI3K are important to investigate, but their toxicity, drug resistance, and side-effects have prevented widespread use of these compounds[20]. Therefore, more efficient and safer new methods have been developed to solve those problems.
RNA interference is a ubiquitous mechanism of eukaryotic gene regulation, and an excellent strategy for specific gene silencing. Recently the vector-based approach of shRNA interference has been developed in order to achieve highly specific suppression of gene expression in mammalian cells. ShRNA is formed by hairpin structures and stretches of double-stranded RNA, which determine the specificity of RNA interference. Previous reports about ShRNA interference have demonstrated their easy and stable introduction into cells, and more importantly, their consistently efficacy, which has potential as a new method for cancer therapy[15-17,21].
PI3K is a heterodimer composed of a regulatory subunit (p85) and a catalytic subunit (p110), which has three isoforms. Among them, p110a (PIK3CA), is an important catalytic subunit that is implicated in a wide range of cancers including colon cancer. Beuers et al[22] reported strong associations were found between KRAS, PIK3CA mutations and colorectal cancers Dukes’ staging, and KRAS and PIK3CA bi-mutations were more likely to develop into liver metastasis. It naturally serves as a potent potential target for colon cancer therapy[2-6]; however, the use of an RNA interference technique for PIK3CA gene silencing in established colon cancer cell lines has rarely been reported. In a previous study, Wee et al[23] reported successful PIK3CA gene depletion by RNA interference and consequential inhibition of proliferation in the colon cancer cell lines HCT116 and DLD1; however, the down-regulation of PIK3CA gene expression by RNAi in other colon cancer cell line has not been reported, due to the limited availability of colorectal carcinoma cell lines with PIK3CA gene mutation. Herein we describe our experience with the effects of two PIK3CA-specific shRNAs on cell proliferation and apoptosis in the colon cancer cell line SW948, which harbors mutation in the PIK3CA gene.
In this study we used plasmid Pgenesil-1 as vector into which two PIK3CA-specific interference sequences were successfully inserted. The vector Pgenesil-1 could transcribe and generate interfering RNAs continually under the control of U6 promotor. Thus, a persistent gene knock-down effect could be achieved by successfully transfecting the vectors into SW948 cells. Our results demonstrated that these two PIK3C-specific shRNAs both showed evident effects on silencing the PIK3CA gene either at the mRNA or at the protein level. At the same time, the expression of downstream genes, namely AKT1, MYC, and CCND1 was also significantly suppressed. MTT assays demonstrated that cell proliferation in the PIK3CA knockdown groups was significantly inhibited from 48 to 72 h after transfection. Similar results were achieved in detecting anchorage-independent cell proliferation by soft agar colony formation assays, in which significant reduction in either the number or the size of colonies was found in both CA1 and CA2 groups. These results suggest that PIK3CA knockdown by specific shRNAs decreased the abilities of SW948 cells to form colonies in soft agar, which was consistent with Wee’s report[23]. Our results revealed that these two PIK3CA-specific shRNAs expressed an anti-proliferation function in the PIK3CA-mutant cancer cells, and suggest that they may have therapeutic potential for colon cancer.
Our flow cytometry results showed cell cycle arrest at G1/S, and an increase in apoptosis in these two PIK3CA-specific shRNAs-transfected SW948 cells, which may have caused the inhibition of cell growth. This finding is consistent with previously published results in the HCT116 cell line[23]. Akt, a major downstream effector of PI3K, is activated by phosphoinositide-dependent protein kinase 1, and plays an important role in cell proliferation, apoptosis, cell motility and invasion[12]. Lots of studies have shown that Akt is activated in a variety of malignances, and is often functionally linked to cancer progression and metastasis[13]. Another possible mechanism is that AKT down-regulation induced by PIK3CA gene silencing resulted in the degradation of cyclinD1 and down-regulation of c-myc expression via GSK3β activity. CyclinD1 is a regulatory kinase that is critical in modulating the cell cycle through G1 to S phase. C-myc is a positive regulator of G1-specific cyclin-dependent kinases. Over-expression of these two genes may stimulate cells to overcome the cell cycle checkpoints, and thus enhance cell proliferation[24,25]. Inhibition of these two genes may result in cell cycle arrest and increased apoptosis, and thus inhibit cell growth. Migliardi et al[26] reported that PI3K/mTor inhibitor BEZ235 could produce prevalent growth-suppressive effects in patient-derived xenografts of RAS-mutant colorectal carcinomas.
In conclusion, our experiment showed that the shRNA targeted against PIK3CA could specifically silence the PIK3CA gene and consequentially suppress the expression of its downstream genes including, AKT1, MYC, and CCND1. This silencing effect on these genes could efficiently inhibit the growth of PIK3CA-mutant colon cancer SW948 cells, and might be a potential approach for treating human PIK3CA-mutant colon cancer.
The phosphoinositide-3-kinase (PI3K)/Akt signaling transduction pathway is believed to play an important role in carcinogenesis. PI3K is a major signaling component, which is a heterodimer consisting of a regulatory subunit (p85) and a catalytic subunit (p110). The subtype p110a encoded by the gene phosphoinositide-3-kinase, catalytic, alpha polypeptide (PIK3CA) is very important in phosphorylating, and may serve as a potential target for colon cancer therapy.
The coordination of cell proliferation and apoptosis in cells is one of the hotspots in cancer research. The balance between cell proliferation and apoptosis, and the distribution of cell cycles play very important roles in the carcinogenesis process of many cancers. PIK3CA is the key component in this process. Thus PIK3CA might be a gene therapy target for lots of cancers include colon carcinoma.
In this study, two PIK3CA specific shRNAs showed their evident effect on silencing the PIK3CA gene. The results also indicated that PIK3CA shRNA inhibits cell growth and induces apoptosis of SW948 cell.
The shRNA interference targeted against PIK3CA may have potential therapeutic utility in human PIK3CA-mutant colon cancer.
The official name of PIK3CA is “phosphoinositide-3-kinase, catalytic, alpha polypeptide”. It is located on the long(q) arm of chromosome 3 at position 26.3. Phosphatidylinositol 3-kinase is composed of an 85 kDa regulatory subunit and a 110 kDa catalytic subunit. The protein encoded by PIK3CA gene represents the catalytic subunit. The PIK3CA gene has been found to be oncogenic and is mutated in a range of human cancers. Due to the association between PIK3CA and cancer, it is believed to be a promising drug target.
The PIK3CA knock-down effect on cells was investigated in the present study. The results showed that shRNAs transfection down-regulated PIK3CA and its downstream genes such as Akt1, MYC and CCND1, and inhibited cell growth. Further, it was demonstrated that cell cycle arrest and apoptosis were induced in PIK3CA shRNAs-transfected SW948 cells. The study appears to be interesting and may have therapeutic implication.
Peer reviewers: Ming Li, Associate Professor, Tulane Univer-sity Health Sciences Center, 1430 Tulane Ave Sl-83, New Orleans, LA 70112, United States; Dr. Lisardo Boscá, Professor, Instituto de Investigaciones Biomédicas Alberto Sols (CSIC-UAM), Arturo Duperier 4, 28029 Madrid, Spain
S- Editor Lv S L- Editor A E- Editor Li JY
| 1. | Hartmann W, Digon-Söntgerath B, Koch A, Waha A, Endl E, Dani I, Denkhaus D, Goodyer CG, Sörensen N, Wiestler OD. Phosphatidylinositol 3'-kinase/AKT signaling is activated in medulloblastoma cell proliferation and is associated with reduced expression of PTEN. Clin Cancer Res. 2006;12:3019-3027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 116] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 2. | Chaussade C, Rewcastle GW, Kendall JD, Denny WA, Cho K, Grønning LM, Chong ML, Anagnostou SH, Jackson SP, Daniele N. Evidence for functional redundancy of class IA PI3K isoforms in insulin signalling. Biochem J. 2007;404:449-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 183] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 3. | Condliffe AM, Davidson K, Anderson KE, Ellson CD, Crabbe T, Okkenhaug K, Vanhaesebroeck B, Turner M, Webb L, Wymann MP. Sequential activation of class IB and class IA PI3K is important for the primed respiratory burst of human but not murine neutrophils. Blood. 2005;106:1432-1440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 244] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 4. | Yu HG, Ai YW, Yu LL, Zhou XD, Liu J, Li JH, Xu XM, Liu S, Chen J, Liu F. Phosphoinositide 3-kinase/Akt pathway plays an important role in chemoresistance of gastric cancer cells against etoposide and doxorubicin induced cell death. Int J Cancer. 2008;122:433-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 172] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 5. | Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, Downward J. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 451] [Cited by in RCA: 465] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 6. | Zhao JJ, Cheng H, Jia S, Wang L, Gjoerup OV, Mikami A, Roberts TM. The p110alpha isoform of PI3K is essential for proper growth factor signaling and oncogenic transformation. Proc Natl Acad Sci USA. 2006;103:16296-16300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 187] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 7. | Troxell ML, Brunner AL, Neff T, Warrick A, Beadling C, Montgomery K, Zhu S, Corless CL, West RB. Phosphatidylinositol-3-kinase pathway mutations are common in breast columnar cell lesions. Mod Pathol. 2012;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 8. | Cizkova M, Susini A, Vacher S, Cizeron-Clairac G, Andrieu C, Driouch K, Fourme E, Lidereau R, Bièche I. PIK3CA mutation impact on survival in breast cancer patients and in ERα, PR and ERBB2-based subgroups. Breast Cancer Res. 2012;14:R28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 144] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 9. | Higgins MJ, Jelovac D, Barnathan E, Blair B, Slater S, Powers P, Zorzi J, Jeter SC, Oliver GR, Fetting J. Detection of Tumor PIK3CA Status in Metastatic Breast Cancer Using Peripheral Blood. Clin Cancer Res. 2012;18:3462-3469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 260] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 10. | Janku F, Wheler JJ, Westin SN, Moulder SL, Naing A, Tsimberidou AM, Fu S, Falchook GS, Hong DS, Garrido-Laguna I. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol. 2012;30:777-782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 11. | Shi J, Yao D, Liu W, Wang N, Lv H, Zhang G, Ji M, Xu L, He N, Shi B. Highly frequent PIK3CA amplification is associated with poor prognosis in gastric cancer. BMC Cancer. 2012;12:50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 12. | Liu JF, Zhou XK, Chen JH, Yi G, Chen HG, Ba MC, Lin SQ, Qi YC. Up-regulation of PIK3CA promotes metastasis in gastric carcinoma. World J Gastroenterol. 2010;16:4986-4991. [PubMed] |
| 13. | Zhou XK, Tang SS, Yi G, Hou M, Chen JH, Yang B, Liu JF, He ZM. RNAi knockdown of PIK3CA preferentially inhibits invasion of mutant PIK3CA cells. World J Gastroenterol. 2011;17:3700-3708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 12] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 14. | Guan HT, Xue XH, Dai ZJ, Wang XJ, Li A, Qin ZY. Down-regulation of survivin expression by small interfering RNA induces pancreatic cancer cell apoptosis and enhances its radiosensitivity. World J Gastroenterol. 2006;12:2901-2907. [PubMed] |
| 15. | Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci USA. 2002;99:6047-6052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 770] [Cited by in RCA: 776] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 16. | Liu F, He CW, Zhang YF, Zhou KY. RNA interference by expression of short hairpin RNAs suppresses bcl-xL gene expression in nasopharyngeal carcinoma cells. Acta Pharmacol Sin. 2005;26:228-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 17. | Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC, Shi Y. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci USA. 2002;99:5515-5520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 859] [Cited by in RCA: 882] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 18. | Huang WS, Wang JP, Wang T, Fang JY, Lan P, Ma JP. ShRNA-mediated gene silencing of beta-catenin inhibits growth of human colon cancer cells. World J Gastroenterol. 2007;13:6581-6587. [PubMed] |
| 19. | Liao X, Morikawa T, Lochhead P, Imamura Y, Kuchiba A, Yamauchi M, Nosho K, Qian ZR, Nishihara R, Meyerhardt JA. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18:2257-2268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 213] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 20. | Czauderna F, Fechtner M, Aygün H, Arnold W, Klippel A, Giese K, Kaufmann J. Functional studies of the PI(3)-kinase signalling pathway employing synthetic and expressed siRNA. Nucleic Acids Res. 2003;31:670-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 74] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 21. | Verma UN, Surabhi RM, Schmaltieg A, Becerra C, Gaynor RB. Small interfering RNAs directed against beta-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin Cancer Res. 2003;9:1291-1300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 74] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 22. | Beuers U, Ritter MM, Richter WO, Paumgartner G. Lipoprotein (a) serum levels in chronic cholestatic liver disease during treatment with ursodeoxycholic acid. Arch Intern Med. 1990;150:1542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 23. | Wee S, Wiederschain D, Maira SM, Loo A, Miller C, deBeaumont R, Stegmeier F, Yao YM, Lengauer C. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci USA. 2008;105:13057-13062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 460] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 24. | Niu ZS, Li BK, Wang M. Expression of p53 and C-myc genes and its clinical relevance in the hepatocellular carcinomatous and pericarcinomatous tissues. World J Gastroenterol. 2002;8:822-826. [PubMed] |
| 25. | Takahashi Y, Kawate S, Watanabe M, Fukushima J, Mori S, Fukusato T. Amplification of c-myc and cyclin D1 genes in primary and metastatic carcinomas of the liver. Pathol Int. 2007;57:437-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 26. | Migliardi G, Sassi F, Torti D, Galimi F, Zanella ER, Buscarino M, Ribero D, Muratore A, Massucco P, Pisacane A. Inhibition of MEK and PI3K/mTOR suppresses tumor growth but does not cause tumor regression in patient-derived xenografts of RAS-mutant colorectal carcinomas. Clin Cancer Res. 2012;18:2515-2525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 160] [Article Influence: 12.3] [Reference Citation Analysis (0)] |