Published online Jul 14, 2012. doi: 10.3748/wjg.v18.i26.3389
Revised: March 23, 2012
Accepted: March 29, 2012
Published online: July 14, 2012
AIM: To investigate the role of osteopontin (OPN) and its splice variants in the proliferation of hepatocellular carcinoma (HCC).
METHODS: The expression of OPN variants in HCC cell lines as well as HCC tissue samples and non-tumour tissue was studied using polymerase chain reaction. OPN variant cDNAs were cloned into a mammalian expression vector allowing both transient expression and the production of stable OPN expressing cell lines. OPN expression was studied in these cells using Western blotting, immunofluoresnce and enzyme linked immunosorbent assay. A CD44 blocking antibody and siRNA targeting of CD44 were used to examine the role of this receptor in the OPN stimulated cell growth observed in culture. Huh-7 cells stably expressing either OPN-A, -B or -C were injected subcutaneously into the flanks of nude mice to observe in vivo tumour growth. Expression of OPN mRNA and protein in these tumours was examined using reverse transcription-polymerase chain reaction and immunohistochemistry.
RESULTS: OPN is expressed in HCC in 3 forms, the full length OPN-A and 2 splice variants OPN-B and -C. OPN variant expression was noted in HCC tissue as well as cognate surrounding cirrhotic liver tissue. Expression of these OPN variants in the HCC derived cell line Huh-7 resulted in secretion of OPN into the culture medium. Transfer of OPN conditioned media to naïve Huh-7 and HepG2 cells resulted in significant cell growth suggesting that all OPN variants can modulate cell proliferation in a paracrine manner. Furthermore the OPN mediated increase in cellular proliferation was dependent on CD44 as only CD44 positive cell lines responded to OPN conditioned media while siRNA knockdown of CD44 blocked the proliferative effect. OPN expression also increased the proliferation of Huh-7 cells in a subcutaneous nude mouse tumour model, with Huh-7 cells expressing OPN-A showing the greatest proliferative effect.
CONCLUSION: This study demonstrates that OPN plays a significant role in the proliferation of HCC through interaction with the cell surface receptor CD44. Modulation of this interaction could represent a novel strategy for the control of HCC.
- Citation: Phillips RJ, Helbig KJ, Hoek KHVD, Seth D, Beard MR. Osteopontin increases hepatocellular carcinoma cell growth in a CD44 dependant manner. World J Gastroenterol 2012; 18(26): 3389-3399
- URL: https://www.wjgnet.com/1007-9327/full/v18/i26/3389.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i26.3389
Hepatocellular carcinoma (HCC) is a highly aggressive carcinoma of the liver, and is the fifth most common cancer worldwide, and the third leading cause of cancer related death[1,2]. Risk factors for HCC include; infection with either hepatitis B virus (HBV) or hepatitis C virus (HCV), alcoholic cirrhosis and exposure to environmental toxins such as aflatoxin. HCC is a global disease most prevalent in Southeast Asia and sub-Saharan Africa[3], however, in countries such as the United States and Japan, HCC incidence is on the increase, primarily as a result of infection with HCV[4,5]. Despite intense investigation, therapeutic intervention for HCC is extremely limited, with a poor prognosis due to high rates of recurrence and intrahepatic metastasis following surgical resection. Furthermore, there is a significant gap in our knowledge of the molecular mechanisms responsible for HCC development and progression.
Osteopontin (OPN) is a secreted multifunctional matrix-glycoprotein that is emerging as a significant protein in the biology of HCC. It is involved in normal tissue remodelling processes and is secreted to high levels in numerous tumours including HCC[6]. OPN is overexpressed in HBV-related metastatic HCC[7], while OPN antibodies can suppress pulmonary metastasis of HCC cells in a nude mouse model, suggesting that it plays a significant role in the metastatic potential of HCC[8]. However, its role in development and proliferation of HCC is relatively unexplored, although a recent study involving the silencing OPN mRNA expression in HCC cell lines, suggested it may also have proliferative effects[9]. OPN binds to the family of αvβ integrins, and the cell-surface adhesion molecule CD44, to initiate cellular signals that enable tumour progression[10-12]. However, the role, if any, of CD44 and OPN in modulating HCC cell growth, is unknown.
Osteopontin is expressed as a heterogenous protein, dependent on glycosylation patterns and the type of cell from which it is expressed. For example, OPN derived from osteosarcoma-derived cells is smaller than that from non-transformed bone cells, with expression of the smaller form correlating with ancorage independence, suggesting that different forms of OPN have different phenotypic effects[13]. In addition, the existence of two OPN splice variants have been described, with deletions in exon 4 (termed OPN-C) and 5 (termed OPN-B)[14]. These variants were originally described in glioma cells and more recently OPN-C has been implicated in the invasiveness of breast cancer cell lines[15] while in HCC derived cell lines OPN-C promotes the extracellular clea-vage by matrix metalloproteinase (MMP)-9, releasing a distinct 5 kDa OPN fragment that is essential for HCC cellular invasion[16]. However, the relative expression of the OPN variants in HCC has not been formally demonstrated, nor have their effects on HCC cell growth been studied. In this study we demonstrate that all splice variant forms of OPN are expressed in HCC at the mRNA level and that all have the ability to stimulate the growth of HCC derived cell lines in vitro and in an in vivo ectopic xenograft mouse model. Furthermore this growth promoting effect was mediated by interaction of OPN with CD44 and adds significantly to our understanding of the role of OPN in HCC.
The human hepatoma-derived cell lines used in this study were Huh-7, Hep G2 and Hep3B, while Hepa 1-6 cells are of mouse hepatoma origin. All cells were maintained in Dulbecco’s Modified Eagle Medium, containing 4.5 g/L D-Glucose, 25 mmol 2-[4-(2-Hydroxyethyl)-1-piperazinyl]ethanesulfonic acid and 2 mmol/L L-glutamine (Invitrogen, CA, United States). Media was supplemented with 10% fetal calf serum, 12 μg/mL penicillin and 16 μg/mL gentamycin. To monitor cell growth, cultured cells were seeded at a density of 7 × 104 cells per well in a 12-well plate and cell numbers monitored daily using trypan blue exclusion. All experiments were performed at least in triplicate. Human HCC tissue and cognate surrounding tissue were collected from patients undergoing HCC resection at the Royal Adelaide Hospital (collection was approved by the Hospital’s ethics committee).
Full-length OPN cDNA and splice variants were amplified from Huh-7 cells by reverse transcription polymerase chain reaction (RT-PCR). Total RNA and cDNA synthesis were performed as described elsewhere[17]. The coding sequence for OPN was amplified using the primers 5’-GTTGAAGCTTCTCACTACCATGAGAATTGCAGTG-3’ and 5-TAGTTCTAGACCTTTTAATTGACCTCAGAAGATG-3’ and cloned into the mammalian expression vector pRC-CMV using XbaIand Hind III to generate pRC-CMV-OPN-A. During sequence verification, two smaller cDNAs were isolated that corresponded to OPN-B and OPN-C. These vectors were used in transient expression assays and to generate stable OPN expressing clones in Huh-7 cells using the transfection reagent Fugene (Roche, Australia) and G418 selection.
Total cellular RNA was isolated from cells using Trizol (Invitrogen) and first strand cDNA was synthesized and standard PCR performed as mentioned previously[17]. Primers used for OPN cDNA detection were 5’- ATGAGA ATTGCAGTGATTTGCTTTTGCCT -3’ and 5’-CATGGTCATCATCATCTTCATCATC -3’; primers utilised for the detection of CD44 were 5’-GACACATATTGCTTCAATGCT TCAGC-3’ and 5’-GATGCCAAGATGATCAGCCATTCTGGAAT-3’; and glyceraldehyde-3-phosphate dehydrogenase primers were 5’-ACCACAGTCCATGCCATCAC-3’ and 5’-TCCACCACCCTGTTGCTGTA-3’. Amplicons were electrophoresed through a 1% agarose gel before visualisation on a ultraviolet transilluminator.
Huh-7 cells, either stably selected or transiently expressing OPN, were lysed using RIPA buffer and total protein separated via sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to nitrocellulose as previously described[18]. Membranes were blocked with 5% skim milk in 0.1% phosphate buffered saline Tween-20 (PBS-T) and incubated overnight at 4 °C with either 400 ng/mL of goat anti-human OPN antibody (K-20: SCBT, SantaCruz, CA) or mouse anti-human CD44 antibody (Labvision, Fremont, CA, United States) at 200 ng/mL followed by either 33 ng/mL of anti-goat or anti-mouse horseradish peroxidase (HRP) antibody (Rockland, Gilbertsville, PA, United States). Washes between antibody binding were with 0.1% PBS-T. Protein bound to antibody was visualised via chemiluminescence (ECL; Amersham Biosciences, Piscataway, NJ, United States).
Cellular localisation of transiently expressed OPN was performed via indirect immunofluorescence as previously described[19] with the exception that cells were incubated in 1 μg/mL of anti-OPN antibody followed by 10 mg/mL anti-goat Alexa 488-conjugated antibody (Molecular probes, Eugene, OR). CD44 expression was visualised using a mouse anti-human CD44 antibody at 4 μg/mL on cells that had been fixed in 5% formalin but not permeabilised for detection of surface CD44 only. Cells were visualised using a BioRad Radiance 2100 confocal microscope.
OPN concentration in cell culture supernatants was determined using an “in house” sandwich enzyme linked immunosorbent assay (ELISA) as described previously[17]; where plates were coated with a monoclonal anti-OPN antibody (3 μg/mL R and D Systems, Minneapolis, MN, United States) and detection performed with a polyclonal anti-OPN antibody (200 ng/mL R and D Systems). CD44 blocking antibody (sc-7946; Santa Cruz, CA, United States) for 30 min at room temperature. The blocking antibody is a polyclonal antiserum raised against amino acids 21-320 of CD44[20].
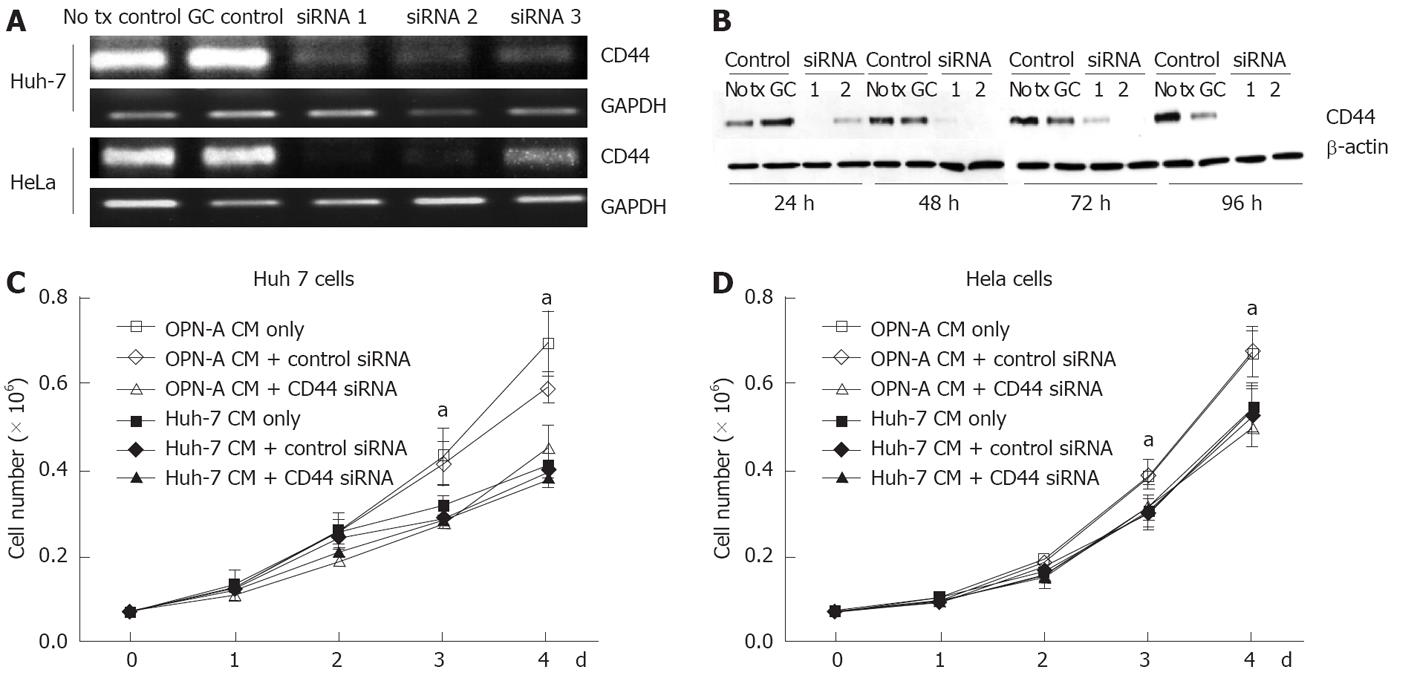
StealthTM siRNA double stranded RNA oligonucleotides (Invitrogen) designed to “knock down” or minimise expression of the OPN receptor CD44 were transfected into Huh-7 and HeLa cells using the reverse transcription method of LipofectamineTM 2000 (Invitrogen) as per the manufacturer’s instructions. Cells were assayed for CD44 expression (and growth rate when required) 24-96 h post transfection via immunoblot assay. Three different siRNA oligonucleotides, designed to target the conserved exons present in all CD44 isoforms, were tested for their ability to minimise CD44 expression in a transient system using both RT-PCR and immunoblotting as described above. Of the three, two demonstrated consistent knockdown of CD44 at the RNA level by RT-PCR and immunoblot analysis and were pooled for further experiments.
Five week-old female Balb/c nude mice were implanted subcutaneously with 5 × 106 cells from Huh-7 OPN-A expressing cell lines and the Huh-7 parent cell line as a control. Cells were diluted in 100 μL of PBS and injected into both dorsal flanks (n = 6). Tumour growth was monitored by calculation of tumour volume [V (mL) = (width2× length2)/2] and measured daily for approximately 25 d. The mice were sacrificed and tumours removed for histological analysis. These experiments were approved by the University of Adelaide and Institute of Medical and Veterinary Science animal ethics committees.
Paraffin embedded sections of liver and tumour tissue were processed as previously described[17]. Five micron sections were re-hydrated and incubated in either anti-OPN (LabVision, Fremont, CA, United States), or normal rabbit IgG (Cell signalling, Boston, MA, Unites States) (diluted to 1 μg/mL in PBS + 3% (v/v) normal horse serum) for 30 min, before being incubated in ADVANCETM HRP-Link (Dako, Glostrup, Denmark), and then in ADVANCETM HRP Enzyme (Dako) for 30 min each. Slides were then incubated in diluted DAB + chromagen (Dako; 20 μL DAB + chromagen in 1 mL DAB + substrate buffer) for 5 min and then following a 5 min wash in H2O, slides were counterstained with haematoxylin.
Student t tests were performed to analyse the distributions of 2 independent data sets and all statistical analysis were performed using SPSS 10.0.
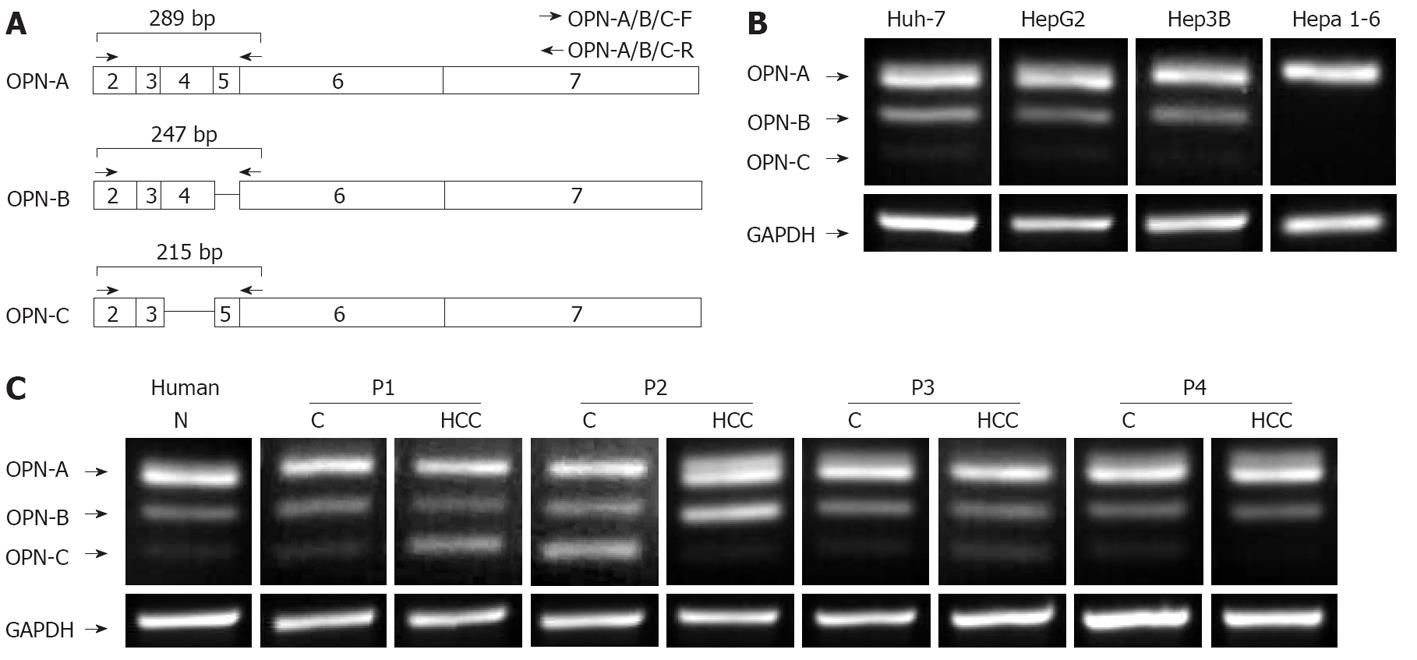
The existence of multiple forms of OPN, termed OPN-A, OPN-B and OPN-C, has been previously described[14], however, their expression and physiological roles in tumour biology, including HCC has not been well studied. We therefore investigated expression of these OPN variants in the HCC cell lines, Huh-7, HepG2, Hep3b and the mouse HCC line Hepa1-6, using PCR primers that spanned exons 2-6 (Figure 1A). As predicted, all HCC cell lines expressed the full-length OPN-A mRNA (Figure 1B). In addition, two smaller transcripts were identified in all cell lines (with the exception of Hepa 1-6 cells) with OPN-B consistently expressed to a higher level than OPN-C (OPN-C cDNA is readily visable on longer exposure). These smaller transcripts were identified by cloning and sequencing and shown to be specific for OPN-b and OPN-C that lack exons 5 and 4 respectively (Figure 1A). OPN-A is clearly the dominantly expressed OPN transcript followed by OPN-B and OPN-C respectively. Interestingly mouse tissue only expressed the full length OPN-A and this is consistent with a previous report indicating that OPN-A is exclusively expressed in mouse tissues[15].
We next investigated OPN mRNA expression in HCC tissue samples and cognate non-tumour tissue using the primers described above. Attempts to generate a quantitative real-time assay for detection of all OPN variant simultaneously was unreliable. Nevertheless, it is clear that normal human liver tissue expressed the 3 OPN transcripts to variable degrees, while OPN variant expression was variable between HCC samples. For example, OPN-C expression was increased in HCC P1 compared to cognate liver tissue, while in P2, expression of OPN transcripts in non-tumour tissue was significantly different from that of the corresponding tumour (Figure 1C). This suggests that OPN transcript expression is variable and that individual OPN variants may have specific physiological roles. The detection of all OPN splice variants in normal liver tissue (OPN-C visible on long exposure) is in contrast to previous reports in which it was suggested that OPN-C expression is specific to tumour cells[14,15].
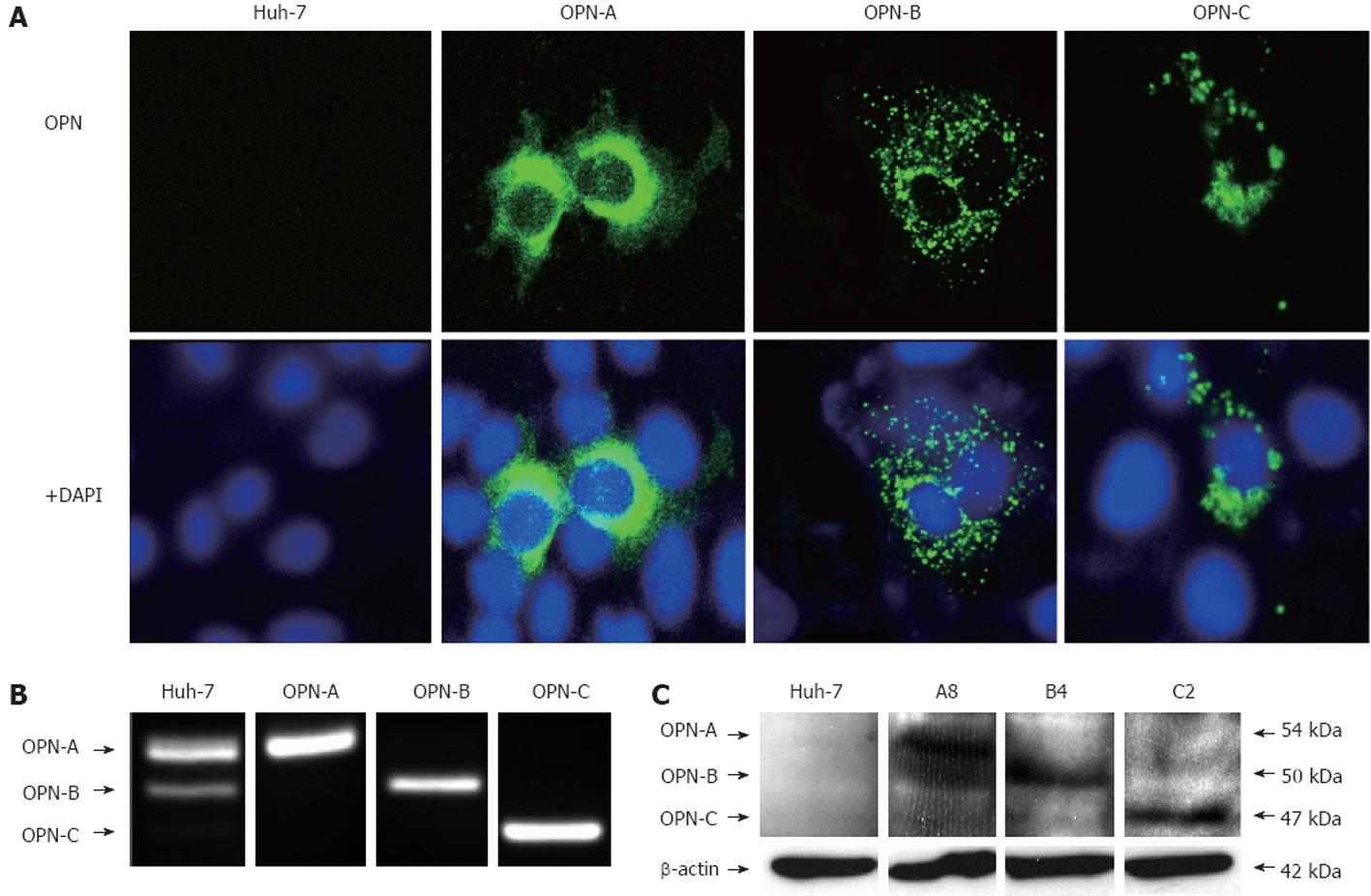
In order to study the physiological roles of the distinct OPN splice variants, cDNAs representing OPN-A, OPN-B and OPN-C, were cloned into a mammalian expression vector to allow both transient expression and the production of stable OPN expressing cell lines. Following transfection of cDNA constructs representing the OPN variants, detection of OPN expression by confocal microscopy was unsuccessful, suggesting efficient secretion of OPN into the media. However, following treatment of transiently transfected cells with brefeldin A, which inhibits cellular secretion pathways and causes retention of OPN within the cell, we could detect intracellular expression of OPN variants (Figure 2A). OPN is predominantly thought to act at the extracellular level, however, OPN can be detected in tumourogenic hepatocytes by immunohistochemistry suggesting it may, at least in part, have a role at the intracellular level[6]. Following brefeldin-A treatment, OPN variants exhibited different cytoplasmic distribution in Huh-7 cells, suggesting that each variant may have different effects on tumourogenic hepatocytes, however, this requires further investigation.
To investigate the role of the OPN variants on cell growth, stable clones were produced in a Huh-7 cell line that has low OPN mRNA expression and undetectable protein expression by ELISA (results not shown) and immunoblot. Multiple clones were isolated after G418 selection with no adverse effect on cell morphology or growth. Intracellular OPN was not detected in any clones (results not shown) consistent with efficient expression of OPN, however, OPN mRNA, specific for the OPN variant of interest, was detected by RT-PCR (Figure 2B), while OPN protein was detected by immunoblot (Figure 2C). Furthermore, OPN ELISA performed on media from stable cell lines revealed secretion of OPN in parent Huh 7 and vector tranfected cells to be below the limit of detection (< 7.81 ng/mL). In contrast OPN-A was detected at 160.4 ng/mL and OPN-B at 127.1 ng/mL. We could not detect OPN-C by ELISA for reasons that are not entirely clear, however the OPN detection ELISA Ab was raised against OPN purified from human milk in which OPN-C is not present[21]. However, OPN-C is clearly expressed in our system as detected by RT-PCR, immunofluorescence and immunoblotting (Figure 2).
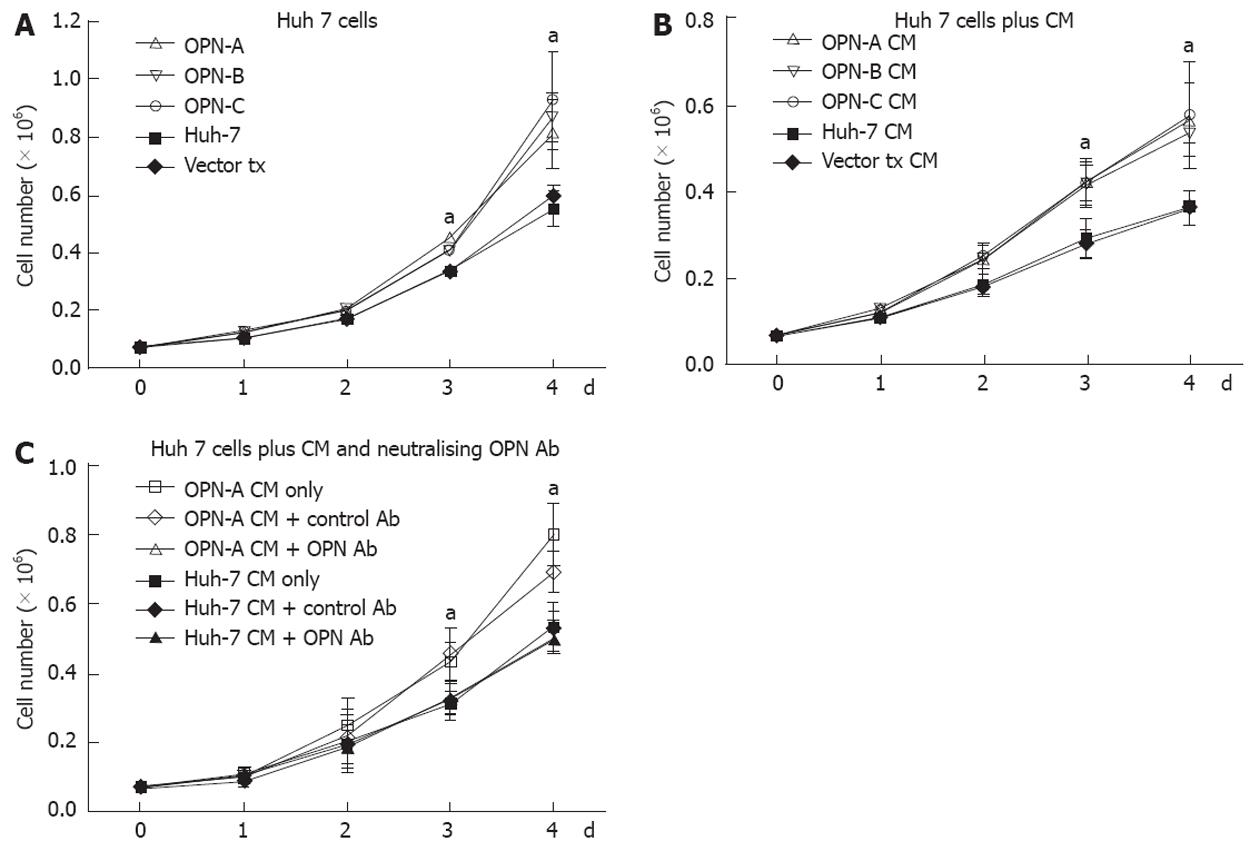
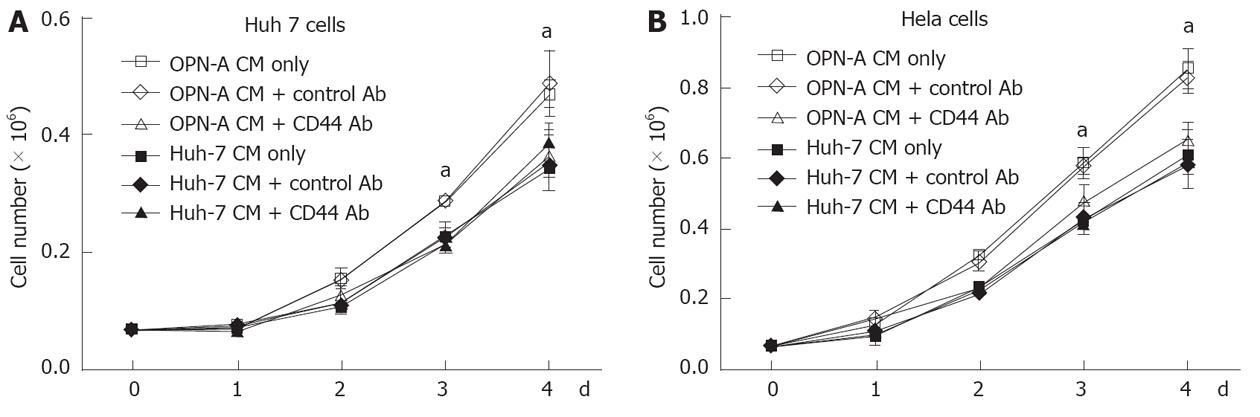
Little work has been done regarding the role of OPN and associated splice variants on cell growth. However recent work from Sun et al[9] suggests that OPN can enhance cell growth of liver-derived cell lines, although the role of OPN splice variants was not investigated. Therefore, using our stably transduced OPN Huh-7 cells, we determined the impact of the individual variants on Huh-7 cell growth. Huh-7 cells expressing either OPN-A, -B or -C grew significantly faster after 4 d of culture compared to cells transfected with the vector only and the parent cell line (Figure 3A). This increase in growth rate was seen in multiple selected OPN transformants, removing the possibility of clonal selection bias. OPN is readily secreted from these Huh-7 cells and to determine if OPN was exerting its effect by either an autocrine or paracrine mechanism, we incubated Huh-7 cells with conditioned media from OPN-A, -B or -C stable transformants. As can be seen in Figure 3B, conditioned media from all stable transformants increased Huh-7 cell growth, suggesting OPN was exerting its effect in a paracrine manner. This increase in Huh-7 cell growth was directly attributable to the action of OPN because the addition of an anti-OPN antibody, but not a control immunoglobulin, to the cultures, suppressed the increase in cell growth when OPN-A conditioned media was added to Huh-7 cells (Figure 3C). Similar results were obtained for OPN-B and -C conditioned media (results not shown). These results suggest that all variants of OPN can enhance Huh-7 cell proliferation in vitro.
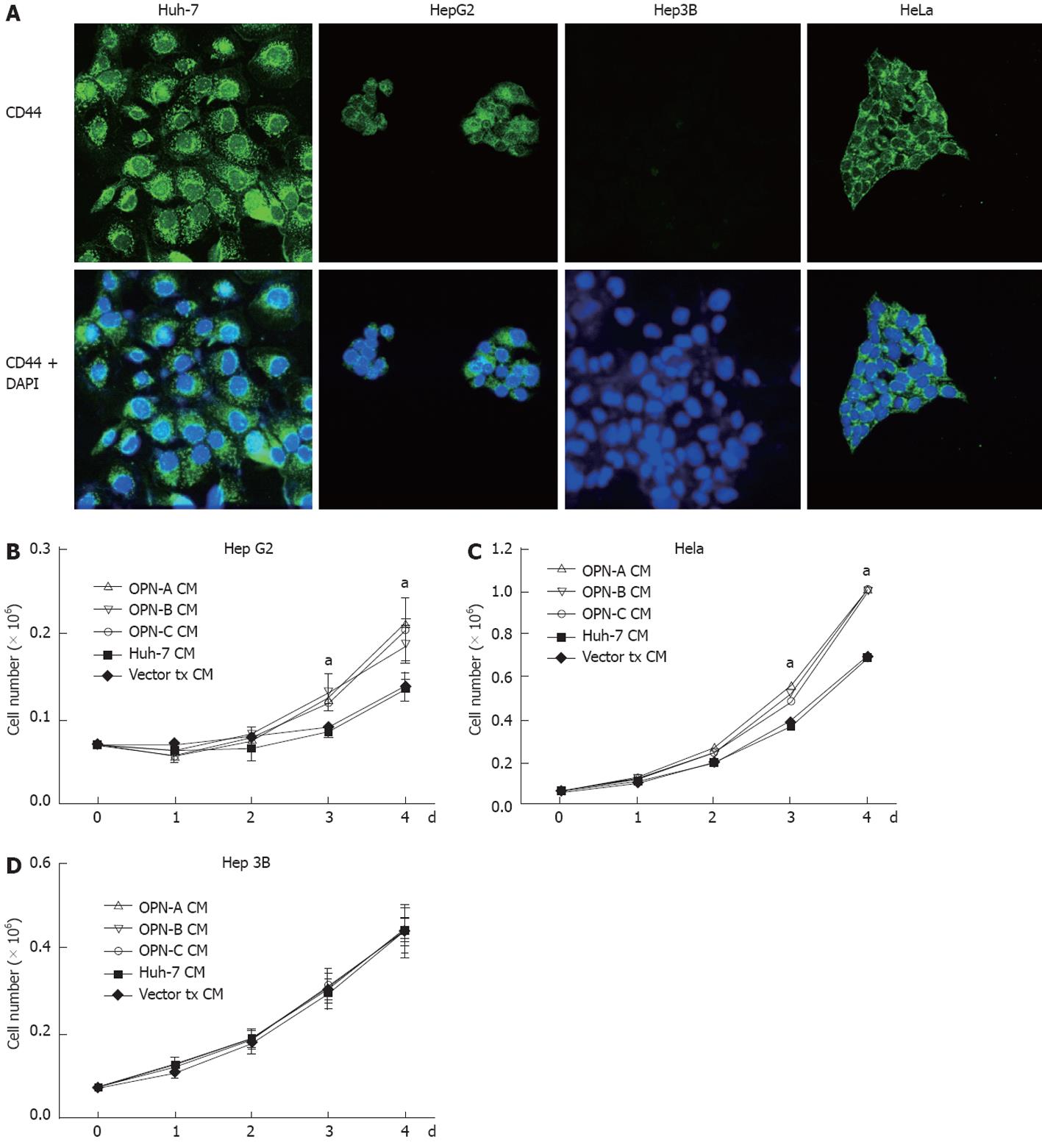
The C-terminal region of OPN binds the CD44 variant v6/7 to induce cellular signals responsible for tumour progression[22]. To investigate if OPN was exerting its growth effect in a CD44-dependent manner, we stimulated a number of cell lines possessing different levels of CD44 with OPN conditioned media. As can be seen in Figure 4A, Huh-7, HepG2 and HeLa cells all express cell surface CD44 as determined by immunofluorescence microscopy of unpermeabilised cells, while Hep3b cells do not express CD44. Consistent with this observation, Huh-7, HepG2 and HeLa cells, when stimulated with OPN-A conditioned media, all showed a significant increase in cell growth (Figures 3B, 4B and C). However no cell growth was noted in Hep3Bs consistent with a lack of CD44 expression (Figure 4D). These observations were confirmed following the addition of CD44 blocking antibody to the conditioned media that completely suppressed the OPN growth effect (Figure 5A and B). Addition of control immunoglobulin to the cultures had no effect on OPN induced cell growth. To further confirm the role of CD44, we suppressed CD44 expression using an siRNA approach. Transfection of an siRNA targeting CD44 resulted in a significant decrease in CD44 mRNA and CD44 protein expression as determined by PCR and immunoblot in Huh-7 and HeLa cells (Figure 6A and B). This knockdown, in turn, correlated with a decrease in OPN-mediated dependent cell growth of both cell lines (Figure 6C and D). Collectively, these experiments indicate that the increase in OPN induced cell growth results from an interaction between OPN and CD44.
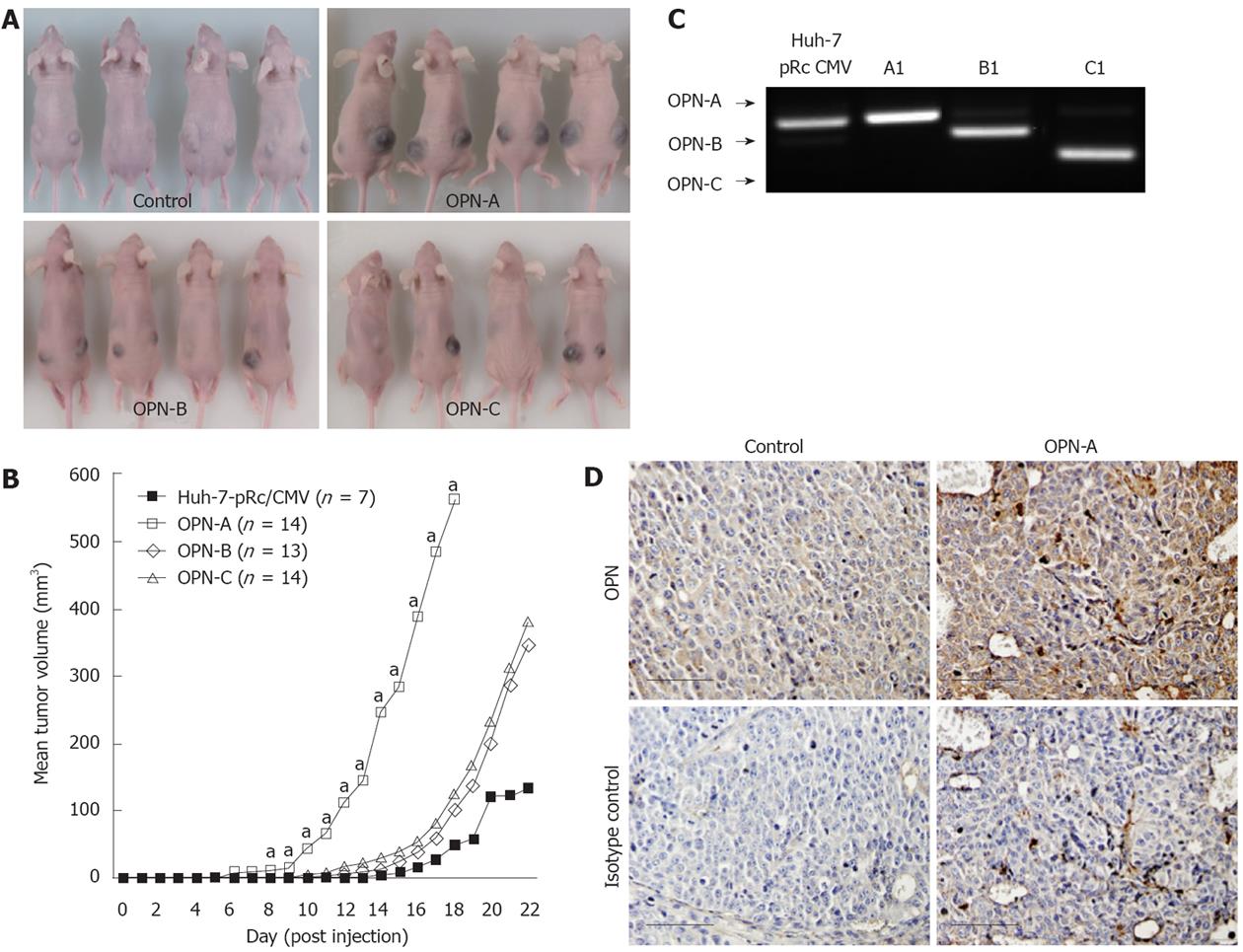
Our in vitro data prompted us to examine whether the different OPN splice variants regulate tumour growth in an ectopic xenograft nude mouse model. Accordingly, Huh-7 cells stably expressing either OPN-A, -B or -C were injected subcutaneously into the flanks of nude mice. Mice injected with OPN-A expressing Huh-7 cells were sacrificed approximately 18 d post injection as the tumour size was causing mice discomfort, while all other mice were sacrificed at approximately 22 d post injection. The typical gross appearance of the tumours is shown in Figure 7A, while the mean tumour volume throughout the experiment is shown in Figure 7B. Compared to parent control Huh-7 cells, mice injected with OPN expressing cell lines showed an increase in tumour growth. This increase in cell growth was most noticeable in mice injected with Huh-7 cells expressing full-length OPN-A (Figure 7B). These mice developed tumours at day 10, after which the growth rate was faster than that of the control tumours. To confirm OPN expression in isolated tumours, RNA was isolated and mRNA expression of the different OPN splice variants was confirmed by RT-PCR (Figure 7C). Detection of OPN in situ was performed by immunohistochemisty in which OPN-A expressing tumours expressed significantly more OPN-A than control Huh-7 cells (Figure 7D). OPN-B and -C were similarly detected by immunohistochemistry (results not shown). No differences were noted histologically between the tumours, with the exception that OPN-A tumours contained a greater number of cells undergoing various stages of the mitotic pathway. These data clearly suggest a role for OPN in promoting the growth of Huh-7 cells in an ectopic xenograft mouse model.
OPN is a secreted phosphoglycoprotein that binds to the family of cellular receptors integrin αvβ and CD44 to exert its biological effects, and of late has gained attention as its expression is associated with invasion and metastasis of a number of malignant tumours, including HCC[8,23-26]. In many of these tumours, increased plasma levels of OPN have shown promise as potential prognostic biomarkers as OPN expression correlates with metastasic potential, early recurrence and decreased patient survival. While our understanding of the role of OPN in HCC biology is expanding, there is still a paucity of information regarding the role of OPN (and its splice variants) in proliferation of HCC and the molecular mechanisms responsible.
OPN undergoes post-translational modifications via phosphorylation, glycosylation and cleavage by thrombin and MMPs to produce differentially active forms[10]. For example, the MMP-9 cleaves OPN into specific fragments of which a 5 kDa fragment induces HCC cellular invasion via the CD44 receptor[16]. Furthermore, stromelysin (MMP-3) and matrilysin (MMP-7) have been reported to cleave OPN at residues 166 and 210, resulting in tumour cell adhesion, while thrombin cleavage of OPN mediates increased adhesion and migration of tumour cells, via binding of the OPN RGD motif to cell surface integrins[27,28]. Clearly the role of OPN and its derivatives in tumour biology is complex, which is further complicated by the expression of OPN splice variants. In humans, the existence of two OPN splice variants with deletion of exon 4 (OPN-C) or exon 5 (OPN-B) were first described in glioma cells[29] and more recently in breast cancer cell lines[15], however, in HCC, the roles of the OPN splice variants is relatively undefined, although a recent report suggests that expression of OPN-A and -B in HCC-derived cell lines confers a robust migratory phenotype through activation of the p42/44 Map kinase pathway[30].
There has been little investigation of the expression of OPN splice variants and the mechanisms by which OPN regulates cellular proliferation. Our initial investigations focused on OPN splice variant expression in various HCC cell lines, and in HCC and its cognate surrounding normal liver. At the mRNA level, we observed expression of all three OPN splice variants in human HCC cell lines, although OPN-A was clearly the dominant species followed by OPN-B and to a lesser extent-c. This observation is consistent with the work of Takafuji et al[16] who showed that greater levels OPN-C were expressed in HCC cell lines with high metastatic potential (the cell lines studied here are not of high metastatic potential). However, the expression of OPN-B and other splice variants was not investigated in primary HCC. Thus this study represents a comprehensive description of complete OPN variant expression in vivo and in vitro. Transcripts for all OPN variants were expressed in both primary HCC and cognate surrounding tissue, however, levels of expression varied between paired samples and between different patient HCCs. While our RT-PCR is only semi-quantitative (development of a quantitative real time assay for all OPN variants was not reliable) these results in-part confirm the work of He et al[15] and show that expression of OPN splice variants can be detected in surrounding non-cancerous tissue. It has been reported that the OPN splice variants are specific to tumour cells and are rarely, if ever, expressed in normal tissue[15]. However, we noted expression of all OPN variants in non-diseased liver tissue, suggesting that OPN splice variant expression is not restricted to tumour cells, and different forms of OPN may play different roles depending on the cell type that expresses OPN and the context in which it is expressed.
A significant finding of this study was the role of OPN and splice variants in in vitro proliferation and in vivo tumour growth of HCC. It is becoming increasingly clear that in addition to the role of OPN in the metastatic process it also has a role in cellular proliferation of a number of tumours. However, its role in HCC is not well studied, although, recently, an siRNA OPN knock-down approach suggested that OPN is important for HCC proliferation. Nevertheless, the relative roles of OPN variants or the cellular receptor responsible were not investigated[9]. In contrast to the above OPN RNA interference study, we exogenously expressed OPN variants in the HCC Huh-7 cell line, and investigated cellular proliferation in vitro and in an ectopic xenograft nude mouse model. Stable ectopic expression of OPN-A, -B and -C resulted in secretion of all OPN forms into the culture media, and significant proliferation of Huh-7 cells, either directly or through the addition of conditioned media to naive Huh-7 cells. This suggests that at least in vitro OPN exerts its proliferative effect in either an autocrine or paracrine manner, which is consistent with the secreted nature of this protein, and was confirmed through the use of a neutralizing OPN antibody that abolished the proliferative effect of conditioned media. This proliferation effect was also seen in an in vivo HCC proliferation model in which Huh-7 cell lines, expressing either OPN-A, -B or -C, all proliferated faster than the control Huh-7 line. Interestingly compared to the in vitro proliferation assays, OPN-A showed the most significant proliferation advantage over OPN-B and -C. The reason for this discrepancy is unclear as all cell lines expressed similar amounts of OPN but may be related to stability or processing of OPN in an in vitro environment. To our knowledge, this is the first report that OPN and its splice variants play a significant role in cellular proliferation and growth of HCC, in addition to its role in invasiveness. Our collective studies strongly suggest that OPN plays a role in cellular proliferation however we cannot exclude the possibility that OPN expression also plays a role in increased cell survival given the recent observations from Zhao et al[31].
The CD44 family are cell surface adhesion molecules that mediate cell-matrix interactions, and in addition to the standard form, multiple isoforms arise from differential splicing of ten variant exons, and are designated CD44v6-15[32]. CD44 variants, especially CD44v6, have been identified as protein markers for metastatic behaviour in a number of cancers, including HCC[33-36]. A C-terminal region of OPN can specifically interact with CD44v6 and or v7 to mediate cellular chemotaxis[37] and a thrombin-cleaved C-terminal OPN fragment induces macrophage migration via CD44[38]. While OPN CD44v6-7 binding has been associated with metastasis and invasion, little work has been done in regard to the CD44 OPN interaction and tumour growth, although one study revealed that knockdown of CD44 expression reduces tumour growth in colon carcinoma cells[39]. In this study, we show that the increased growth rate of Huh-7 cells in response to OPN conditioned media was as a result of CD44 expression because blocking CD44 with a neutralising antibody completely abolished this growth effect. Furthermore, HepG2 and HeLa cells, which both express CD44v6/7, also showed this growth effect while the Hep3B cell line, which is negative for CD44v6/7, was unresponsive to OPN in the conditioned media. To conclusively demonstrate a role for CD44, a siRNA approach was employed that successfully reduced CD44 expression in both Huh-7 and HeLa cells. Consistent with the above, this reduction in CD44 in both cell lines resulted in a decrease in cellular proliferation in response to OPN conditioned media. To our knowledge this is the first report that the interaction between OPN and CD44 results in signals that drive cellular proliferation in HCC. This is not surprising considering the recent work describing a role for OPN in regulating MKK3/6 and p38 dependent nuclear factor kappa B activation in cervical cancer leading to furin expression, a proprotein convertase that plays crucial roles in regulation of tumour progression metastasis and angiogenesis[40]. Cervical cancer cells over expressing OPN showed enhanced cell growth, while shRNA-mediated silencing of OPN suppressed cell growth in a CD44-mediated MKK3/6 dependent p38 activation mechanism[40]. While similar mechanisms may be at play in HCC, the signalling pathways driven by OPN and CD44 activation in HCC require further investigation. Thus, understanding the molecular mechanisms that underpin increased HCC cell proliferation through the OPN/CD44 interaction and activation of downstream signalling pathways could provide potential targets for therapeutic strategies for the treatment of HCC.
Hepatocellular carcinoma (HCC) is a highly aggressive carcinoma of the liver, and is the fifth most common cancer worldwide. It is the third leading cause of cancer related death. Osteopontin (OPN) is a secreted multifunctional matrix-glycoprotein involved in normal tissue remodelling processes but is secreted at high levels in numerous tumours, including HCC. OPN is expressed as 3 isoforms and their role in HCC biology is not entirely known.
Human OPN is known to be expressed in three different isoforms with potentially differing functions. Despite evidence of elevated OPN in several cancers and correlation of levels to malignant invasivness, the expression of OPN isoforms in the context of HCC has not previously been investigated. Similarly the affect of OPN and its isoforms on HCC growth is unexplored.
This study demonstrates that all isoforms of OPN are expressed in HCC tissues and cell lines, at the mRNA level, and that each isoform can stimulate the growth of HCC derived cell lines both in vitro and in vivo, in an ectopic xenograft mouse model. Furthermore, this growth promoting effect has been shown to occur through a mechanism requiring functional CD44. These cellular proliferation signalling pathways, driven by OPN and CD44 activation, require further investigation.
Understanding the molecular mechanisms that underpin increased HCC cell proliferation could provide potential targets for therapeutic strategies for the treatment of HCC.
Authors elegantly and convincingly demonstrated that OPN plays a relevant role in the expansion of HCC cells through interaction with the cell surface receptor CD44; this information is potentially interesting for novel anti HCC therapeutic approaches.
Peer reviewers: Dr. C Bart Rountree, Pediatrics and Phar-macology, Penn State College of Medicine, 500 University Drive, H085, Hershey, PA 17033, United States; Dr. Luca Valenti, Internal Medicine, Università degli Studi di Milano, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, via Francesco Sforza 35, 20122 Milano, Italy; Gabriele Grassi, Associate Professor, Department of Medical, Technological and Tran, University Hospital of Cattinara, Strada di Fiume 447, 34100 Trieste, Italy
S- Editor Gou SX L- Editor A E- Editor Li JY
| 1. | Okuda K. Hepatocellular carcinoma. J Hepatol. 2000;32:225-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 397] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 2. | Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2651] [Cited by in RCA: 2597] [Article Influence: 108.2] [Reference Citation Analysis (0)] |
| 3. | Wong CM, Ng IO. Molecular pathogenesis of hepatocellular carcinoma. Liver Int. 2008;28:160-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 108] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 4. | Bosch FX, Ribes J, Cléries R, Díaz M. Epidemiology of hepatocellular carcinoma. Clin Liver Dis. 2005;9:191-211, v. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 625] [Cited by in RCA: 604] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 5. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3846] [Cited by in RCA: 4259] [Article Influence: 236.6] [Reference Citation Analysis (2)] |
| 6. | Gotoh M, Sakamoto M, Kanetaka K, Chuuma M, Hirohashi S. Overexpression of osteopontin in hepatocellular carcinoma. Pathol Int. 2002;52:19-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 114] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 7. | Xie H, Song J, Du R, Liu K, Wang J, Tang H, Bai F, Liang J, Lin T, Liu J. Prognostic significance of osteopontin in hepatitis B virus-related hepatocellular carcinoma. Dig Liver Dis. 2007;39:167-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 52] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 8. | Ye QH, Qin LX, Forgues M, He P, Kim JW, Peng AC, Simon R, Li Y, Robles AI, Chen Y. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 642] [Cited by in RCA: 643] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 9. | Sun BS, Dong QZ, Ye QH, Sun HJ, Jia HL, Zhu XQ, Liu DY, Chen J, Xue Q, Zhou HJ. Lentiviral-mediated miRNA against osteopontin suppresses tumor growth and metastasis of human hepatocellular carcinoma. Hepatology. 2008;48:1834-1842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 141] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 10. | Rangaswami H, Bulbule A, Kundu GC. Osteopontin: role in cell signaling and cancer progression. Trends Cell Biol. 2006;16:79-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 502] [Cited by in RCA: 533] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 11. | Rittling SR, Chambers AF. Role of osteopontin in tumour progression. Br J Cancer. 2004;90:1877-1881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 310] [Cited by in RCA: 334] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 12. | Weber GF. The metastasis gene osteopontin: a candidate target for cancer therapy. Biochim Biophys Acta. 2001;1552:61-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 114] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 13. | Chackalaparampil I, Banerjee D, Poirier Y, Mukherjee BB. Altered processing of a major secreted phosphoprotein correlates with tumorigenicity in Rous sarcoma virus-transformed mammalian cells. J Virol. 1985;53:841-850. [PubMed] |
| 14. | Saitoh Y, Kuratsu J, Takeshima H, Yamamoto S, Ushio Y. Expression of osteopontin in human glioma. Its correlation with the malignancy. Lab Invest. 1995;72:55-63. [PubMed] |
| 15. | He B, Mirza M, Weber GF. An osteopontin splice variant induces anchorage independence in human breast cancer cells. Oncogene. 2006;25:2192-2202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 146] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 16. | Takafuji V, Forgues M, Unsworth E, Goldsmith P, Wang XW. An osteopontin fragment is essential for tumor cell invasion in hepatocellular carcinoma. Oncogene. 2007;26:6361-6371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 141] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 17. | Helbig KJ, Ruszkiewicz A, Semendric L, Harley HA, McColl SR, Beard MR. Expression of the CXCR3 ligand I-TAC by hepatocytes in chronic hepatitis C and its correlation with hepatic inflammation. Hepatology. 2004;39:1220-1229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 98] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 18. | Li K, Prow T, Lemon SM, Beard MR. Cellular response to conditional expression of hepatitis C virus core protein in Huh7 cultured human hepatoma cells. Hepatology. 2002;35:1237-1246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 70] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Helbig KJ, Lau DT, Semendric L, Harley HA, Beard MR. Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology. 2005;42:702-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 193] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 20. | Teramoto H, Castellone MD, Malek RL, Letwin N, Frank B, Gutkind JS, Lee NH. Autocrine activation of an osteopontin-CD44-Rac pathway enhances invasion and transformation by H-RasV12. Oncogene. 2005;24:489-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 72] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 21. | Nagatomo T, Ohga S, Takada H, Nomura A, Hikino S, Imura M, Ohshima K, Hara T. Microarray analysis of human milk cells: persistent high expression of osteopontin during the lactation period. Clin Exp Immunol. 2004;138:47-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 57] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 22. | Weber GF, Ashkar S, Glimcher MJ, Cantor H. Receptor-ligand interaction between CD44 and osteopontin (Eta-1). Science. 1996;271:509-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 675] [Cited by in RCA: 710] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 23. | Boldrini L, Donati V, Dell'Omodarme M, Prati MC, Faviana P, Camacci T, Lucchi M, Mussi A, Santoro M, Basolo F. Prognostic significance of osteopontin expression in early-stage non-small-cell lung cancer. Br J Cancer. 2005;93:453-457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 63] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 24. | Hotte SJ, Winquist EW, Stitt L, Wilson SM, Chambers AF. Plasma osteopontin: associations with survival and metastasis to bone in men with hormone-refractory prostate carcinoma. Cancer. 2002;95:506-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 119] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 25. | Irby RB, McCarthy SM, Yeatman TJ. Osteopontin regulates multiple functions contributing to human colon cancer development and progression. Clin Exp Metastasis. 2004;21:515-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 26. | Rudland PS, Platt-Higgins A, El-Tanani M, De Silva Rudland S, Barraclough R, Winstanley JH, Howitt R, West CR. Prognostic significance of the metastasis-associated protein osteopontin in human breast cancer. Cancer Res. 2002;62:3417-3427. [PubMed] |
| 27. | Agnihotri R, Crawford HC, Haro H, Matrisian LM, Havrda MC, Liaw L. Osteopontin, a novel substrate for matrix metalloproteinase-3 (stromelysin-1) and matrix metalloproteinase-7 (matrilysin). J Biol Chem. 2001;276:28261-28267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 304] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 28. | Senger DR, Brown LF, Perruzzi CA, Papadopoulos-Sergiou A, Van de Water L. Osteopontin at the tumor/host interface. Functional regulation by thrombin-cleavage and consequences for cell adhesion. Ann N Y Acad Sci. 1995;760:83-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 29. | Young MF, Kerr JM, Termine JD, Wewer UM, Wang MG, McBride OW, Fisher LW. cDNA cloning, mRNA distribution and heterogeneity, chromosomal location, and RFLP analysis of human osteopontin (OPN). Genomics. 1990;7:491-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 284] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 30. | Chae S, Jun HO, Lee EG, Yang SJ, Lee DC, Jung JK, Park KC, Yeom YI, Kim KW. Osteopontin splice variants differentially modulate the migratory activity of hepatocellular carcinoma cell lines. Int J Oncol. 2009;35:1409-1416. [PubMed] |
| 31. | Zhao J, Dong L, Lu B, Wu G, Xu D, Chen J, Li K, Tong X, Dai J, Yao S. Down-regulation of osteopontin suppresses growth and metastasis of hepatocellular carcinoma via induction of apoptosis. Gastroenterology. 2008;135:956-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 109] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 32. | Goodison S, Urquidi V, Tarin D. CD44 cell adhesion molecules. Mol Pathol. 1999;52:189-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 434] [Cited by in RCA: 501] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 33. | Asosingh K, Günthert U, De Raeve H, Van Riet I, Van Camp B, Vanderkerken K. A unique pathway in the homing of murine multiple myeloma cells: CD44v10 mediates binding to bone marrow endothelium. Cancer Res. 2001;61:2862-2865. [PubMed] |
| 34. | Ponta H, Sleeman J, Dall P, Moll J, Sherman L, Herrlich P. CD44 isoforms in metastatic cancer. Invasion Metastasis. 1984;14:82-86. [PubMed] |
| 35. | Rudzki Z, LeDuy L, Jothy S. Changes in CD44 expression during carcinogenesis of the mouse colon. Exp Mol Pathol. 1997;64:114-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 36. | Takahashi K, Stamenkovic I, Cutler M, Saya H, Tanabe KK. CD44 hyaluronate binding influences growth kinetics and tumorigenicity of human colon carcinomas. Oncogene. 1995;11:2223-2232. [PubMed] |
| 37. | Weber GF, Ashkar S, Cantor H. Interaction between CD44 and osteopontin as a potential basis for metastasis formation. Proc Assoc Am Physicians. 1997;109:1-9. [PubMed] |
| 38. | Weber GF, Zawaideh S, Hikita S, Kumar VA, Cantor H, Ashkar S. Phosphorylation-dependent interaction of osteopontin with its receptors regulates macrophage migration and activation. J Leukoc Biol. 2002;72:752-761. [PubMed] |
| 39. | Harada N, Mizoi T, Kinouchi M, Hoshi K, Ishii S, Shiiba K, Sasaki I, Matsuno S. Introduction of antisense CD44S CDNA down-regulates expression of overall CD44 isoforms and inhibits tumor growth and metastasis in highly metastatic colon carcinoma cells. Int J Cancer. 2001;91:67-75. [PubMed] |
| 40. | Kumar V, Behera R, Lohite K, Karnik S, Kundu GC. p38 kinase is crucial for osteopontin-induced furin expression that supports cervical cancer progression. Cancer Res. 2010;70:10381-10391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |