Published online Jun 28, 2012. doi: 10.3748/wjg.v18.i24.3129
Revised: May 6, 2011
Accepted: May 12, 2012
Published online: June 28, 2012
AIM: To assess BGC823 gastric cancer (GC) cell metastasis after knockdown of liver-intestine cadherin (CDH17) and the therapeutic value of CDH17-RNAi-lentivirus in vivo.
METHODS: We evaluated primary tumor growth and assessed local infiltration and systemic tumor dissemination using an orthotopic implantation technique. The therapeutic value of CDH17 knockdown was examined by intratumoral administration of CDH17-RNA interference (RNAi)-lentivirus in an established GC tumor xenograft mouse model. Furthermore, a comparative proteomic approach was utilized to identify differentially expressed proteins in BGC823 and lenti-CDH17-miR-neg cells following CDH17 knockdown.
RESULTS: Metastases in the liver and lung appeared earlier and more frequently in animals with tumors derived from BGC823 or lenti-CDH17-miR-neg cells than in tumors derived from lenti-CDH17-miR-B cells. Average tumor weight and volume in the CDH17-RNAi-lentivirus-treated group were significantly lower than those in the control group (tumor volume: 0.89 ± 0.04 cm3vs 1.16 ± 0.06 cm3, P < 0.05; tumor weight: 1.15 ± 0.58 g vs 2.09 ± 0.08 g, P < 0.05). Fifteen differentially expressed proteins were identified after CDH17 silencing in BGC823 cells, including a variety of cytoskeletal and chaperone proteins as well as proteins involved in metabolism, immunity/defense, cell proliferation and differentiation, cell cycle, and signal transduction.
CONCLUSION: Our data establish a foundation for future studies of the comprehensive protein expression patterns and effects of CDH17 in GC.
-
Citation: Xu Y, Zhang J, Liu QS, Dong WG. Knockdown of liver-intestine cadherin decreases BGC823 cell invasiveness and metastasis
in vivo . World J Gastroenterol 2012; 18(24): 3129-3137 - URL: https://www.wjgnet.com/1007-9327/full/v18/i24/3129.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i24.3129
Gastric cancer (GC) is one of the most prevalent and lethal malignancies worldwide owing to difficulties in early detection and high postsurgical recurrence rates[1]. Currently, there are no effective drugs for curing GC. Gastrocarcinogenesis is a complex process in which the altered proliferation and migration properties of cancer cells play important roles in tumor growth and metastasis[2]. Identification of specific markers for predicting recurrence will permit detection of early dissemination and perhaps even eradication.
Cadherins are transmembrane glycoproteins that mediate calcium-dependent cell adhesion and that are strongly implicated in tumorigenesis[3-5]. Liver-intestine cadherin (CDH17) has been identified as a novel member of the cadherin superfamily, which is distinguished from classic cadherins by unique structural and functional features[6,7]. Previous studies have demonstrated the clinicopathological significance and prognostic influence of CDH17 expression in GC. For example, CDH17 has been proposed as a marker of gastric metaplasia and neoplasia[8-11]. In our earlier study, we found that downregulation of CDH17 inhibits proliferation and adherence of the poorly differentiated GC cells, BGC823, in vitro and induces cell cycle arrest[12]. Using an in vivo tumor growth assay with human tumor xenografts grown subcutaneously in nude mice, we also confirmed that CDH17 silencing slows the growth of GC derived from BGC823 cells. Although human tumor xenografts, which grow subcutaneously in nude mice, closely resemble the original tumors morphologically, biologically, and biochemically, these tumors do not metastasize[13]. Orthotopic tumor models are regarded as more suitable for mimicking human tumor disease because they encompass the entire process of primary tumor growth, local tumor infiltration, and subsequent distant metastasis[14,15]. Thus, in our current study, we evaluated primary tumor growth and assessed local infiltration and tumor dissemination using an orthotopic implantation technique.
The therapeutic value of lentivirus and adeno-associated virus has been identified using RNA interference (RNAi) technology[16-19]. Gene therapy for GC is a rationalized strategy because various genes are associated with this disease[20]. Previous research has proposed CDH17 as an in vivo target for liver cancer therapy[21]. In our current research, we further examined the therapeutic value of CDH17-RNAi-lentivirus in established GC xenografts in nude mice.
Proteomic technologies have been used to identify proteins involved in various pathways altered in disease[22,23]. Global analysis of protein expression complements and in some cases has certain advantages over genomic analyses. Given the cumulative data indicating possible roles for CDH17 in cancer development and/or outcomes[24-26], the important function of CDH17 in GC has elicited the need to further investigate the underlying mechanism. Thus, we also used 2-dimensional polyacrylamide gel electrophoresis (2-DE) followed by tandem mass spectrometry (MS) to perform proteome-wide profiling of GC cells in which CDH17 expression had been knocked down to identify proteins whose cellular levels are affected by with CDH17 expression.
Dithiothreitol, urea, agarose, glycerol, bromophenol blue, 3-[(3-cholamidopropyl) dimethylammonio] propanesulfonic acid (CHAPS), Immobiline DryStrips (4-7 L), DryStrip cover fluid, and PhastGel Blue R (Coomassie Brilliant Blue R-350 stain) were purchased from Amersham Pharmacia Biotech AB (Uppsala, Sweden). Acrylamide, Bis, tri(hydroxymethyl)aminomethane (Tris), glycine, sodium dodecyl sulfate (SDS), ammonium persulfate, and tetramethylethylenediamine were from Bio-Rad (Hercules, CA, United States). Iodoacetamide, ammonium bicarbonate, and acetic acid were from Sigma (St. Louis, MO, United States). Acetonitrile and methanol were from Fisher (Fair Lawn, NJ, United States). Trifluoroacetic acid was from Merck (Darmstadt, Germany). Other chemicals are domestic products (analytical grade). All buffers were prepared with Milli-Q water.
The poorly differentiated human GC cell line BGC823 was obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI 1640 medium supplemented with 10% (v/v) fetal bovine serum (Gibco, Carlsbad, CA, United States) at 37 °C in a humidified incubator containing 5% CO2.
The stably transfected cell line in which CDH17 had been knocked down by RNAi (lenti-CDH17-miR-B), the empty vector-transfected control cells (lenti-CDH17-miR-neg), and the CDH17-RNAi-lentivirus and green fluorescent protein (GFP)-lentivirus were obtained as described[12]. Stably transfected cells were cultured at 37 °C in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and 3 mg/mL blasticidin (Invitrogen, Carlsbad, CA, United States) in a 5% CO2 environment.
All nude mice in this study were treated following the experimental animal ethics rules of Wuhan University (Wuhan, China). BALB/c-nu mice (5-6 wk old) weighing 18-20 g were obtained from the Central Laboratory of Animal Science at Wuhan University. The animals were housed in laminar-flow cabinets and kept at a constant humidity (50%-70%) and temperature (25-28 °C) according to standard guidelines under an approved protocol of Wuhan University. Donor nude mice were anesthetized with isoflurane (Abbott, Wiesbaden, Germany) inhalation. Then, 1 × 106 BGC823, lenti-CDH17-miR-neg, or lenti-CDH17-miR-B cells were injected subcutaneously at a single site each animal’s flank. The mice were killed at the same time after several weeks when the largest diameter of the subcutaneous tumors had reached a size of 1 cm. The donor tumors were harvested and minced with a scalpel (No. 11) into small (1 mm3) pieces. To avoid necrotic tissue from the central tumor areas, only macroscopically viable tumor tissue from the outer part of the donor tumors was used for orthotopic implantation.
Thirty-six tumor recipient nude mice were anesthetized with an intraperitoneal injection of xylazine hydrochloride (Rompun, 12 mg/kg BW; Bayer, Leverkusen, Germany). Each animal’s abdomen was aseptically opened with a midline incision, and the stomach was gently exteriorized. One small tissue pocket was prepared in the submucosa of the distal stomach. One donor tumor fragment was placed into the gastric tissue pocket and fixed with one drop of tissue adhesive (Histoacryl, B Braun, Tuttlingen, Germany). The stomach was relocated into the abdominal cavity, which was then closed with two layers of 4-0 absorbable sutures (Ethicon, Norderstedt, Germany).
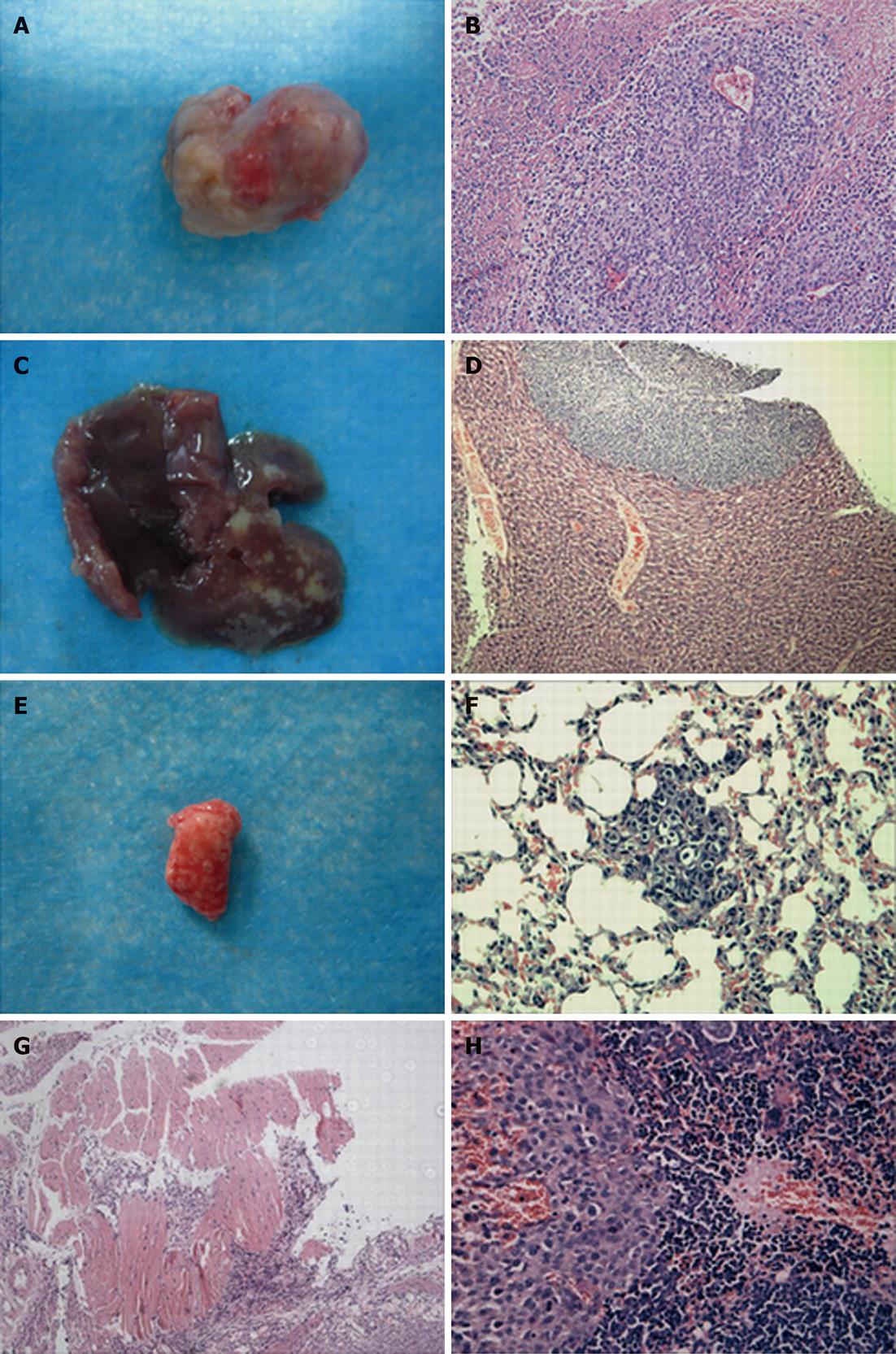
At 4 and 6 wk after transplantation, animals with non transfected BGC823 cells, empty vector-transfected cells (lenti-CDH17-miR-neg), and CDH17 microRNA-transfected cells (lenti-CDH17-miR-B) (n = 6 per cell line) were sacrificed, and an autopsy was done to examine tumor growth. The perpendicular diameters of the primary orthotopic tumors were measured with calipers, and the volume was calculated using the following formula: volume = (shortest diameter)2× (longest diameter)/2. Local infiltration was determined in the liver, lung, spleen, and diaphragmatic muscle. Parts of the tumor tissue and the organ were fixed in 4% paraformaldehyde in PBS and embedded in paraffin. Serial sections were cut at 3 μm thickness, stained with hematoxylin and eosin, and reviewed to confirm the findings of the macroscopic dissemination as described previously[11].
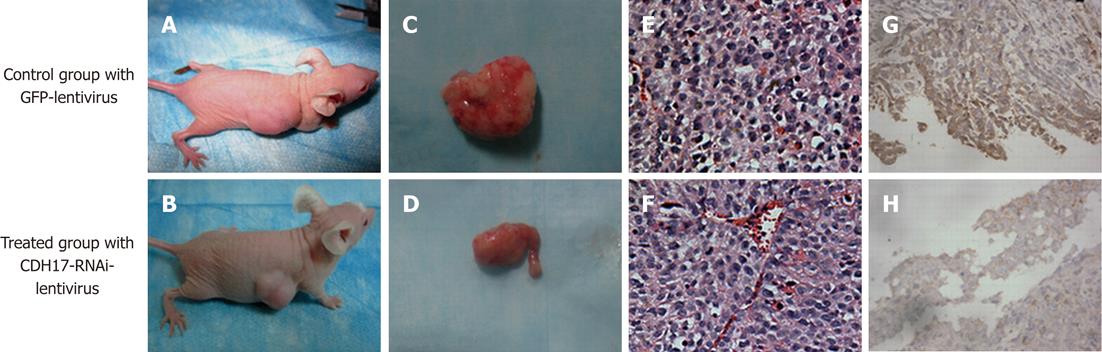
To investigate whether CDH17-RNAi-lentivirus could serve as a therapeutic agent against GC, subcutaneous tumors were induced in nude mice using BGC823 cells as above. Once tumors reached approximately 0.5-0.6 cm3, animals in the treated group were intratumorally injected with 1 × 109 copies of CDH17-RNAi-lentivirus, and those in the control group were treated with 1 × 109 copies of GFP-lentivirus, twice weekly for 2 wk. Tumor formation and growth were monitored daily. Tumors were harvested from mice 1 wk after the end of treatment and evaluated with hematoxylin and eosin. CDH17 expression was examined by immunohistochemistry[11].
In agreement with our previous research[12], we found no differences between BGC823 and lenti-CDH17-miR-neg cells with respect to proliferation, adherence, or invasive ability in our current study. Therefore, we compared the protein expression between BGC823 and lenti-CDH17-miR-B cells in our current study. Briefly, cells were suspended in 1 mL lysis buffer containing 7 mol/L urea, 2 mol/L thiourea, 4% CHAPS, 65 mmol/L dithiothreitol, 2% IPG buffer (pH 4-7), and 1 mmol/L phenylmethylsulfonyl fluoride. After extraction, the protein concentration was quantified using Bradford’s method. Five samples per cell strain were prepared from individual cell cultures. All samples were stored at -80 °C prior to electrophoresis.
For each sample, 1 mg protein was mixed with a rehydration solution containing 8 mol/L urea, 2% CHAPS, 0.5% IPG buffer, pH 4-7 (nonlinear), 18 mmol/L dithiothreitol, and a trace of bromophenol blue, to a total volume of 250 μL, and applied to IPG dry strips. After rehydration for 12 h, isoelectric focusing was performed using ReadyStripTM IPG strips (17 cm, PH 3-10 NL, Bio-Rad) on a PROTEAN IEF system (Bio-Rad) until a total of 80 kVh was reached. Following isoelectric focusing separation, the gel strips were equilibrated two times for 15 min each in equilibration buffer (50 mmol/L Tris-HCl, pH 8.0, 6 mol/L urea, 30% glycerol, 2% SDS, and a trace of bromophenol blue) and directly applied to a 13% sodium dodecyl sulfate-polyacrylamide gel electrophoresi (SDS-PAGE) vertical slab gel for electrophoresis using a PROTEAN II xi Cell system (Bio-Rad) until the bromophenol blue dye marker reached the bottom of the gel. Dithiothreitol (1%) was added to the first equilibration buffer and was replaced with 2.5% Iodoacetamide in the second equilibration buffer. The gels were visualized with Coomassie Blue G250 (Bio-Rad) staining after electrophoresis. Spot detection, quantification, and matching were performed using PDQuest Advanced 8.0.1 software (Bio-Rad). Five analytical gels for each cell strain, resulting from triplicate runs of individual samples, were completed. Quantitative analysis was performed using the Student’s t-test between BGC823 and lenti-CDH17-miR-B gels. The confidence level was 95%.
The separated proteins in SDS-PAGE gels were excised, in-gel digested, and extracted according to the manufacturer’s instructions (Promega, Madison, WI, United States). Briefly, protein spots of interest were destained with 100 μL 50% v/v acetonitrile in 25 mmol/L ammonium bicarbonate for 1 h. In-gel digestion was performed with 10 ng/μL trypsin (Promega) in 40 mmol/L ammonium bicarbonate for 15 h at 37 °C. The resulting peptides were extracted with 5% trifluoroacetic acid in 50% acetonitrile for 1 h at 37 °C and dried completely with centrifugal lyophilization.
Peptide mixtures were redissolved in 0.5% trifluoroacetic acid, and 1.5 μL of the peptide elute was mixed with an identical volume of α-cyano-4-hydroxycinnamic acid matrix on the stainless matrix-assisted laser desorption ionization (MALDI) target until dry (i.e., air dried). All mass spectra were obtained on an Autoflex MALDI-time of flight (TOF) mass spectrometer (Bruker, Bremen, Germany) to generate peptide mass fingerprints. The analyses of peptides were performed in the reflectron mode with a 337-nm nitrogen laser with an acceleration voltage of 20 kV and a reflected voltage of 23 kV. Spectra were accumulated until a satisfactory signal/noise ratio had been obtained from a range of 800 MHz to 4000 MHz. After MS acquisition, 10 ions of maximum intensity were selected automatically for MS/MS analysis. Trypsin autolysis products and keratin-derived precursor ions were automatically excluded. The collision voltage was varied depending on the mass of the precursor. MS/MS data were processed with the search algorithm MASCOT (Version 2.2; Matrix Science, London, United Kingdom) against the SWISS-PROT protein database (UniProt_SP sprot_84; 230,133 sequences). Confident identification had a statistically significant (P≤ 0.05) protein score (based on combined MS spectra) and best ion score (based on MS/MS spectra).
All quantitative data were recorded as the mean ± SD. Comparisons between two groups were performed with the Student’s t-test. Comparisons among multiple groups were performed with one-way analysis of variance. P < 0.05 was considered statistically significant. Computations were performed using the SPSS version 13.0 software package (Chicago, IL, United States).
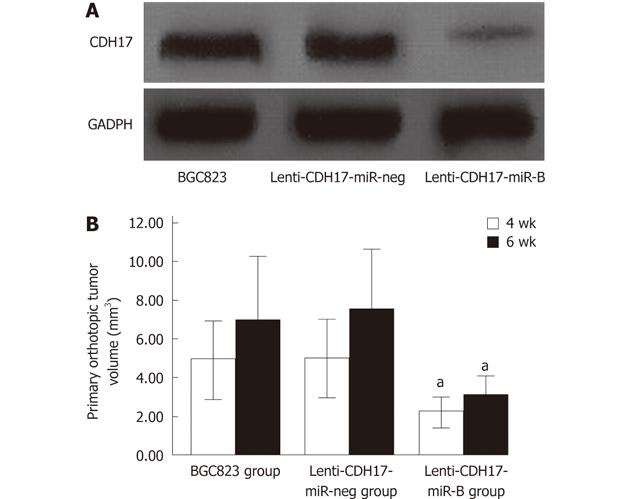
Downregulation of CDH17 in stably transfected lenti-CDH17-miR-B cells was confirmed by Western blotting (Figure 1A). We found that subcutaneous tumors reached a size of 1 cm after approximately 4 wk in BGC823- and lenti-CDH17-miR-neg-treated groups, whereas 5 wk was required after subcutaneous injection with lenti-CDH17-miR-B cells, indicating that proliferation of GC cells was inhibited after CDH17 knockdown. These results were consistent with our previous report[12].
To demonstrate the progress of tumor growth, the animals were randomized into two groups and killed at 4 or 6 wk after tumor implantation. We found that the primary orthotopic tumor volume in the lenti-CDH17-miR-B group was much smaller than that of the BGC823 and lenti-CDH17-miR-neg groups (Figures 1B, 2A and B) (P < 0.05). In addition, nude mice in the lenti-CDH17-miR-neg and BGC823 groups developed liver and lung metastases after 4 wk (lung 1/6, liver 1/6, in the lenti-CDH17-miR-neg group; lung 1/6, liver 1/6, in the BGC823 group), whereas neither type of metastasis was found in the lenti-CDH17-miR-B group at that time. Moreover, 6 wk after transplantation, we found that mice transplanted with lenti-CDH17-miR-B cells developed fewer metastases (lung 1/6, liver 1/6) than animals in the BGC823 group (lung 2/6, liver 3/6) and animals in the lenti-CDH17-miR-neg group (lung 2/6, liver 3/6) (Figure 2C-F). In addition, nude mice injected with BGC823 or lenti-CDH17-miR-neg cells also developed diaphragmatic muscle (1/6) (Figure 2G) and spleen (1/6) (Figure 2H) infiltration, neither of which was found in the lenti-CDH17-miR-B group. These results indicated that the cell line lenti-CDH17-miR-B displayed a less aggressive phenotype regarding tumor dissemination than the other two cell lines and suggested that CDH17 knockdown in GC cells downregulated the invasive and metastatic ability in vivo.
All 12 mice that were injected subcutaneously with 1 × 106 BGC823 cells developed detectable tumors at the termination of the experiment. The mice treated with CDH17-RNAi-lentivirus showed significantly suppressed tumor growth compared with those treated with GFP-lentivirus (Figure 3A-F). The average tumor volume (0.89 ± 0.04 cm3) in the former group was significantly lower (P < 0.05) than that (1.16 ± 0.06 cm3) in the latter group. The average tumor weight (1.15 ± 0.58 g) was also much lower (P < 0.05) than that in the control group (2.09 ± 0.08 g). Furthermore, we found that intratumoral injection of CDH17-RNAi-lentivirus reduced CDH17 levels (Figure 3G-H). These results indicated that CDH17 knockdown may be an effective means of slowing GC tumor growth in vivo.
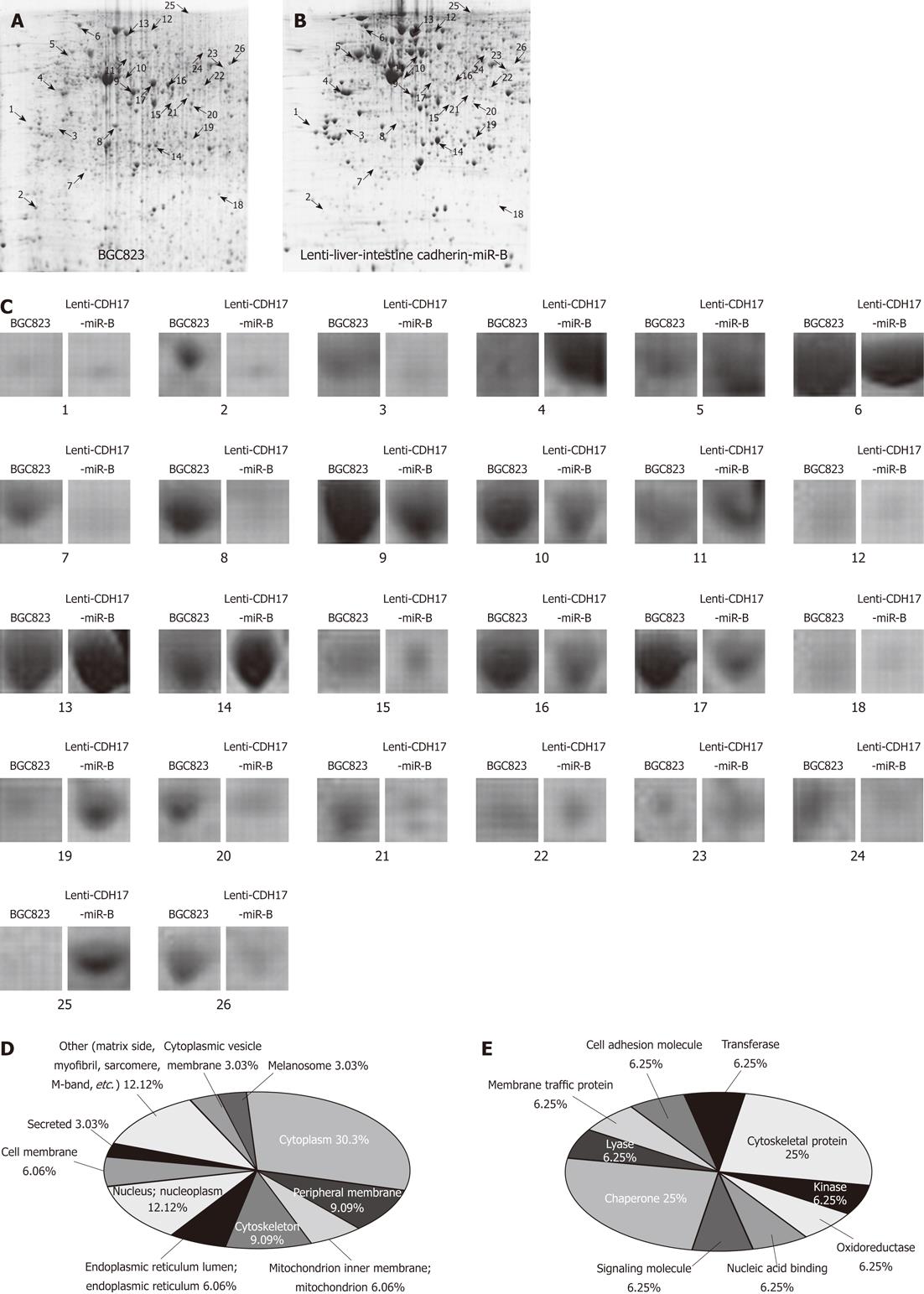
Proteins extracted from lenti-CDH17-miR-B or BGC823 cells were subjected to 2-DE and stained with Coomassie, which revealed a match rate of 80.5% and showed an average of 1032 ± 31 and 1279 ± 2 spots, respectively (Figure 4A and B). A total of 26 differentially expressed protein spots were defined as statistically significant based on two criteria: (1) intensity alterations > 3-fold; and (2) recurrence four or more times in the five pairs examined (Figure 4C). To eliminate the redundancy of proteins appearing in the database under different names or accession numbers, the protein member belonging to Homo sapiens with the highest MASCOT score was selected[27]. Fifteen different proteins from 2-DE gels were successfully identified using MALDI-TOF tandem MS (Table 1). Several protein spots could not be identified, possibly owing to a lower amount of protein as revealed by a retrospective analysis of the spot intensities. Differences between the experimental MW/pI and the theoretical value were observed for some of the proteins, which may have been a consequence of post-translational modifications such as truncation and/or phosphorylation[28].
| Protein description | Gene name | Accessionno.1 | MW/pI(theo)1 | Sequencecoverage2(%) | Mascotscore3 | MS/MS peptide sequence4 | Cellular location |
| Clathrin light chain A | CLCA | P09496 | 27 174/4.43 | 15 | 59 | K.AIKELEEWYAR.Q | Cytoplasmic vesicle membrane; peripheral membrane; cytoplasm |
| Nucleophosmin | NPM | Q96AT6 | 32 726/4.64 | 23 | 56 | K.MSVQPTVSLGGFEITPPVVLR.L | Nucleus; nucleoplasm |
| Tropomyosin α-3 chain | TPM3 | P06753 | 32 856/4.68 | 7 | 56 | R.KLVIIEGDLER.T | Cytoplasm; cytoskeleton |
| Tubulin beta-2C chain | TBB2C | P68371 | 50 255/4.79 | 19 | 61 | R.AVLVDLEPGTMDSVR.S | Cytoplasm |
| Glutathione S-transferase P | GSTP1 | P09211 | 23 569/5.43 | 43 | 298 | M.PPYTVVYFPVR.G | Nucleus; cytoplasm |
| Activator of 90 kDa heat shock protein ATPase homolog 1 | AHSA1 | O95433 | 38 421/5.41 | 30 | 143 | K.ETFLTSPEELYR.V | Cytoplasm; endoplasmic reticulum |
| Interleukin-1 α | IL1A | P01583 | 30 758/5.04 | 23 | 64 | K.LTFKESMVVVATNGK.V | Secreted |
| Heat shock 70 kDa protein 1A/1B | HSP71 | P08107 | 70 294/5.48 | 27 | 239 | K.DAGVIAGLNVLR.I | Cytoplasm |
| 78 kDa glucose-regulated protein | GRP78 | P11021 | 72 402/5.07 | 18 | 100 | R.ITPSYVAFTPEGER.L | Endoplasmic reticulum lumen; melanosome |
| Actin, cytoplasmic 1 | ACTB | Q96HG5 | 42 052/5.29 | 36 | 210 | R.AVFPSIVGRPR.H | Cytoplasm; cytoskeleton |
| Pyruvate kinase isozymes M1/M2 | KPYM | Q9BWB5 | 58 470/7.96 | 30 | 161 | R.LDIDSPPITAR.N | Cytoplasm; nucleus |
| Mitochondrial 28S ribosomal protein S22 | RT22 | P82650 | 41 425/7.70 | 21 | 131 | K.YVFTDISYSIPHR.E | Mitochondria |
| NADH dehydrogenase (ubiquinone) iron-sulfur protein 2, mitochondrial | NDUS2 | O75306 | 52 911/7.21 | 25 | 86 | K.TYLQALPYFDR.L | Mitochondrial inner membrane; peripheral membrane; matrix side |
| Vinculin | VINC | P18206 | 124 292/5.50 | 26 | 183 | R.EAFQPQEPDFPPPPPDLEQLR.L | Cytoplasm; cytoskeleton; cell membrane; peripheral membrane |
| α-enolase | ENOA | Q6GMP2 | 47 481/7.01 | 29 | 134 | R.AAVPSGASTGIYEALELR.D | Cytoplasm; cell membrane; myofibril; sarcomere; m-band |
As listed in Table 1, we found that the altered proteins after CDH17 knockdown were mainly located in the cytoplasm (30.3%), nucleus (12.12%), or peripheral membrane (9.09%) (Figure 4D). In brief, these differentially expressed proteins between lenti-CDH17-miR-B and BGC823 cells play important and fundamental roles in cellular physiology, e.g., as chaperones (25%), cytoskeletal proteins (25%), transferases (6.25%), and oxidoreductases (6.25%), among others (Figure 4E).
Using an orthotopic implantation technique, we examined the growth and metastatic potential of BGC823 cells in which CDH17 had been knocked down. Our results showed that BGC823 or lenti-CDH17-miR-neg cells produced more extensive local tumor growth during the observation period. Moreover, local infiltration into the liver and lung appeared much earlier and more frequently in animals with tumors derived from BGC823 or lenti-CDH17-miR-neg cells than in those derived from lenti-CDH17-miR-B cells, suggesting the less aggressive behavior of GC after CDH17 knockdown. However, the effects of CDH17 in gastric carcinogenesis should be examined in other human GC cell lines.
To investigate whether CDH17-RNAi-lentivirus could serve as a therapeutic agent against GC, BALB/c-nu mice with induced tumors of BGC823 cells were treated with CDH17-RNAi-lentivirus. The results showed that both the average tumor weight and volume were significantly lower in the CDH17-RNAi-lentivirus-treated group compared with the control group treated with GFP-lentivirus. Thus, intratumoral administration of CDH17-RNAi-lentivirus, which reduced CDH17 levels, significantly slowed tumor growth. However, our results also demonstrated that CDH17 knockdown did not eliminate the tumor completely, suggesting that CDH17 is not the only protease involved in invasion and growth of GC.
Although the CDH17 gene was identified as a biomarker or a potential diagnostic and therapeutic target for GC using gene expression profiling[29,30], the molecular mechanisms of CDH17 remain largely undefined due to limited knowledge of CDH17 target recognition. Although 2-DE and MS-based proteomic strategies have certain limitations, these strategies provide high-throughput simultaneous identification of hundreds of proteins and are considered powerful approaches for analyzing alterations in protein expression in complex biological systems[31-34]. Using the 2-DE/MS approach, we identified 15 proteins in BGC823 cells whose expression levels are affected by CDH17.
Among these proteins, six were upregulated, and nine were downregulated after CDH17 knockdown. We found that these 15 proteins play important roles in metabolism, immunity/defense, cell structure and motility, intracellular protein trafficking, protein modification, cell cycle, signal transduction, cell proliferation and differentiation, electron transport, and muscle contraction, suggesting extensive roles for CDH17 in biological processes. These altered proteins may be potentially involved in the effect(s) of CDH17 in BGC823 cells and represent candidate proteins that may be regulated by CDH17. These diverse proteins may thus serve as focused targets for future studies to determine their potential genetic and/or physical interactions with CDH17 and their functional relevance in GC. Although previous studies have demonstrated possible interactions between CDH17 and galectin-3, metal-responsive transcription factor-1, and placental growth factor in ductal adenocarcinoma of the pancreas and GC[11,24,26], these proteins were not identified in our current study. Thus, further validation experiments should be carried out to confirm our results. In-depth and detailed investigation of the interactions between CDH17 and other proteins may enhance our understanding of the mechanisms by which CDH17 affects carcinogenesis in humans. The mechanisms by which downregulation of CDH17 inhibits proliferation and invasion of BGC823 cells need to be explored in further studies.
Gastrocarcinogenesis is a complex process in which alterations in proliferation and migration properties of cancer cells have an important role in cancer infiltrative growth and metastasis. If specific markers related to cancer invasion were to be identified, dissemination could be detected early and perhaps even eradicated.
Cadherins are transmembrane glycoproteins that mediate calcium-dependent cell adhesion and have strong implications in tumorigenesis. Liver-intestine cadherin (CDH17), also known as LI-cadherin, has been identified as a novel member of the cadherin superfamily, which is distinguished from classic cadherins by its unique structural and functional features. Therefore, the invasive and metastasis ability of gastric cancer (GC) cells after CDH17 silencing were investigated.
Their results presented further evidence that CDH17 was closely correlated to GC invasion. The invasive and metastasis ability of GC cells was found to be significantly reduced in vivo after the downregulation of CDH17. Furthermore, protein profiles in GC cells were also significantly altered after CDH17 knockdown, which suggested the interactions of CDH17 with many other important proteins.
CDH17 could be recognized as an important marker of cancer invasion. Knockdown of CDH17 may become a way to downregulate the invasive ability of GC cells.
The authors investigated the metastatic ability of the GC cells BGC823 with CDH17 silencing and the therapeutic value of CDH17-RNA interference-lentivirus. They demonstrated that knockdown of CDH17 in BGC823 cells down-regulated invasive and metastatic ability in vivo. The work is well written. The methods are adequate and the results obtained justify the conclusions drawn.
Peer reviewer: Mario M D’Elios, Professor, University of Florence, Viale Morgagni 85, 50134 Florence, Italy
S- Editor Cheng JX L- Editor Cant MR E- Editor Zheng XM
| 1. | Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137-2150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2591] [Cited by in RCA: 2639] [Article Influence: 138.9] [Reference Citation Analysis (0)] |
| 2. | Sawada K, Mitra AK, Radjabi AR, Bhaskar V, Kistner EO, Tretiakova M, Jagadeeswaran S, Montag A, Becker A, Kenny HA. Loss of E-cadherin promotes ovarian cancer metastasis via alpha 5-integrin, which is a therapeutic target. Cancer Res. 2008;68:2329-2339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 301] [Cited by in RCA: 304] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 3. | Birchmeier W, Behrens J. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim Biophys Acta. 1994;1198:11-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 361] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 4. | Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol. 1993;5:806-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 643] [Cited by in RCA: 674] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 5. | Hirohashi S, Kanai Y. Cell adhesion system and human cancer morphogenesis. Cancer Sci. 2003;94:575-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 312] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 6. | Gessner R, Tauber R. Intestinal cell adhesion molecules. Liver-intestine cadherin. Ann N Y Acad Sci. 2000;915:136-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 7. | Kreft B, Berndorff D, Böttinger A, Finnemann S, Wedlich D, Hortsch M, Tauber R, Gessner R. LI-cadherin-mediated cell-cell adhesion does not require cytoplasmic interactions. J Cell Biol. 1997;136:1109-1121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 74] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 8. | Grötzinger C, Kneifel J, Patschan D, Schnoy N, Anagnostopoulos I, Faiss S, Tauber R, Wiedenmann B, Gessner R. LI-cadherin: a marker of gastric metaplasia and neoplasia. Gut. 2001;49:73-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 77] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | Ito R, Oue N, Yoshida K, Kunimitsu K, Nakayama H, Nakachi K, Yasui W. Clinicopathological significant and prognostic influence of cadherin-17 expression in gastric cancer. Virchows Arch. 2005;447:717-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 10. | Dong W, Yu Q, Xu Y. Altered expression of a Li-cadherin in gastric cancer and intestinal metaplasia. Dig Dis Sci. 2007;52:536-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 11. | Dong WG, Yu QF, Xu Y, Fan LF. Li-cadherin is inversely correlated with galectin-3 expression in gastric cancer. Dig Dis Sci. 2008;53:1811-1817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 12. | Liu QS, Zhang J, Liu M, Dong WG. Lentiviral-mediated miRNA against liver-intestine cadherin suppresses tumor growth and invasiveness of human gastric cancer. Cancer Sci. 2010;101:1807-1812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 13. | Kerbel RS. Human tumor xenografts as predictive preclinical models for anticancer drug activity in humans: better than commonly perceived-but they can be improved. Cancer Biol Ther. 2003;2:S134-S139. [PubMed] |
| 14. | Onn A, Isobe T, Itasaka S, Wu W, O'Reilly MS, Ki Hong W, Fidler IJ, Herbst RS. Development of an orthotopic model to study the biology and therapy of primary human lung cancer in nude mice. Clin Cancer Res. 2003;9:5532-5539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 15. | Takahashi T, Morotomi M, Nomoto K. A novel mouse model of rectal cancer established by orthotopic implantation of colon cancer cells. Cancer Sci. 2004;95:514-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | Morris KV, Rossi JJ. Lentiviral-mediated delivery of siRNAs for antiviral therapy. Gene Ther. 2006;13:553-558. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 89] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 17. | Orlacchio A, Bernardi G, Orlacchio A, Martino S. RNA interference as a tool for Alzheimer's disease therapy. Mini Rev Med Chem. 2007;7:1166-1176. [PubMed] |
| 18. | Lundberg C, Björklund T, Carlsson T, Jakobsson J, Hantraye P, Déglon N, Kirik D. Applications of lentiviral vectors for biology and gene therapy of neurological disorders. Curr Gene Ther. 2008;8:461-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 100] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 19. | Maeda Y, Sheffield AM, Smith RJ. Therapeutic regulation of gene expression in the inner ear using RNA interference. Adv Otorhinolaryngol. 2009;66:13-36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 20. | Fumoto S, Nishi J, Nakamura J, Nishida K. Gene therapy for gastric diseases. Curr Gene Ther. 2008;8:187-200. [PubMed] |
| 21. | Liu LX, Lee NP, Chan VW, Xue W, Zender L, Zhang C, Mao M, Dai H, Wang XL, Xu MZ. Targeting cadherin-17 inactivates Wnt signaling and inhibits tumor growth in liver carcinoma. Hepatology. 2009;50:1453-1463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 96] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 22. | Nishigaki R, Osaki M, Hiratsuka M, Toda T, Murakami K, Jeang KT, Ito H, Inoue T, Oshimura M. Proteomic identification of differentially-expressed genes in human gastric carcinomas. Proteomics. 2005;5:3205-3213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 91] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 23. | Sickmann A, Mreyen M, Meyer HE. Identification of modified proteins by mass spectrometry. IUBMB Life. 2002;54:51-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 24. | Takamura M, Sakamoto M, Ino Y, Shimamura T, Ichida T, Asakura H, Hirohashi S. Expression of liver-intestine cadherin and its possible interaction with galectin-3 in ductal adenocarcinoma of the pancreas. Cancer Sci. 2003;94:425-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 25. | Su MC, Yuan RH, Lin CY, Jeng YM. Cadherin-17 is a useful diagnostic marker for adenocarcinomas of the digestive system. Mod Pathol. 2008;21:1379-1386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 85] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 26. | Takamura M, Yamagiwa S, Wakai T, Tamura Y, Kamimura H, Kato T, Tsuchiya A, Matsuda Y, Shirai Y, Ichida T. Loss of liver-intestine cadherin in human intrahepatic cholangiocarcinoma promotes angiogenesis by up-regulating metal-responsive transcription factor-1 and placental growth factor. Int J Oncol. 2010;36:245-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 27. | Zheng X, Hong L, Shi L, Guo J, Sun Z, Zhou J. Proteomics analysis of host cells infected with infectious bursal disease virus. Mol Cell Proteomics. 2008;7:612-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 104] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 28. | Zhu K, Zhao J, Lubman DM, Miller FR, Barder TJ. Protein pI shifts due to posttranslational modifications in the separation and characterization of proteins. Anal Chem. 2005;77:2745-2755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 137] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 29. | Yasui W, Oue N, Sentani K, Sakamoto N, Motoshita J. Transcriptome dissection of gastric cancer: identification of novel diagnostic and therapeutic targets from pathology specimens. Pathol Int. 2009;59:121-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 30. | Lee HJ, Nam KT, Park HS, Kim MA, Lafleur BJ, Aburatani H, Yang HK, Kim WH, Goldenring JR. Gene expression profiling of metaplastic lineages identifies CDH17 as a prognostic marker in early stage gastric cancer. Gastroenterology. 2010;139:213-25.e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 130] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 31. | Celis JE, Gromov P. Proteomics in translational cancer research: toward an integrated approach. Cancer Cell. 2003;3:9-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 109] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 32. | Rabilloud T. Two-dimensional gel electrophoresis in proteomics: old, old fashioned, but it still climbs up the mountains. Proteomics. 2002;2:3-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 33. | Sickmann A, Mreyen M, Meyer HE. Mass spectrometry--a key technology in proteome research. Adv Biochem Eng Biotechnol. 2003;83:141-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |