Published online Jun 28, 2012. doi: 10.3748/wjg.v18.i24.3119
Revised: November 26, 2011
Accepted: December 3, 2011
Published online: June 28, 2012
AIM: To assess the rigorous relationship between human leukocyte antigens (HLA)-DR alleles and outcomes of hepatitis B virus (HBV) infections by means of meta-analysis.
METHODS: Medline/PubMed, EMBASE, CNKI and VIP were searched to identify relevant studies. Study quality was evaluated using the Newcastle-Ottawa Scale. Odds ratios (OR) and 95% confidence interval (95% CI) were pooled using Stata 11.0. Subgroup analyses were performed by ethnicity. Heterogeneity and publication bias analyses were performed to validate the credibility.
RESULTS: A total of 2609 patients with chronic hepatitis B and 2606 controls spontaneously recovering from prior HBV infection were included. Meta-analysis showed that HLA-DR*04 (OR = 0.72, 95% CI: 0.60-0.85) and DR*13 (OR = 0.27, 95% CI: 0.19-0.37) alleles were significantly associated with HBV clearance while patients carrying HLA-DR*03 (OR = 1.47, 95% CI: 1.16-1.87) or DR*07 (OR = 1.59, 95% CI: 1.24-2.03) alleles had a significantly increased risk of chronic HBV persistence. For the HLA-DR*01 polymorphism, a significantly association with HBV clearance was found in Chinese Han group (OR = 0.48, 95% CI: 0.26-0.86), but not found in other ethnic groups (P = 0.191). For other polymorphisms, no association with the HBV infection outcome was found.
CONCLUSION: HLA-DR*04 and DR*13 alleles may be the protective factors for HBV clearance and HLA-DR*03, and DR*07 alleles may be the risk factors for HBV persistence.
-
Citation: Yan ZH, Fan Y, Wang XH, Mao Q, Deng GH, Wang YM. Relationship between
HLA-DR gene polymorphisms and outcomes of hepatitis B viral infections: A meta-analysis. World J Gastroenterol 2012; 18(24): 3119-3128 - URL: https://www.wjgnet.com/1007-9327/full/v18/i24/3119.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i24.3119
Hepatitis B virus (HBV) infection is a serious public health problem affecting more than 400 million people worldwide. The clinical features of HBV infection vary from clearance of the virus to fulminate hepatitis. Although some HBV carriers spontaneously eliminate the virus, 2%-10% of the individuals with chronic hepatitis B (CHB) are estimated to develop liver cirrhosis, and a subset of these individuals eventually suffer from liver failure or hepatocellular carcinoma[1]. A complex combination of environmental, pathogenic and host genetic factors plays a role in determining both susceptibility to HBV and the course of the infection[2]. Several epidemiological factors, such as age at infection, sex, chronic alcohol abuse and co-infection with other hepatitis viruses, were suspected to affect viral persistence. In addition, host immunological factors and genetic background were also considered to influence the susceptibility to and outcome of persistent HBV infection[3]. Family studies in China provide some evidences that the host genetic factors affect viral persistence, as concordance rates of HBeAg persistence were higher in identical twins than in non-identical twins[4].
Although genetic variants in IFNG[5], TNF[6], VDR[7], MBP[8], ESR1[9], CXCL10[10], IL-10[11] and several human leukocyte antigen (HLA) loci[12,13] have been shown to associate with CHB, none of the associations has been proven to be conclusive. The mechanism of susceptibility to chronic persistent HBV infection is not well clarified. Since the outcome of HBV infection mainly depends on the host immune response, and HLA, an integral component of the immune response, plays an important role in immunological reaction to HBV infection[14], the highly polymorphic HLA gene has been considered as an appropriated biological candidate susceptibility gene that is associated with the development and the progression of chronic HBV infection. Indeed, previous studies have highlighted that HLA-DR polymorphisms influence individual immune responses, thus affecting the outcome of diseases, and that many different HLA alleles play a role in HBV infection[15]; however, this relationship between HLA-DR polymorphisms and HBV infection is not universal for all investigated populations. As for Caucasians[16,17] and Koreans[18], HLA-DRB1*1301-02 has been found to be associated with acute self-limited hepatitis B. For Taiwanese people, HLA-DRB1*0406 is associated with recovery from HBV infection in the Han Chinese, as is HLA-DRB1*4001 in indigenous Taiwanese people[19]. Han Chinese with HLA-DR12 (especially one of its alleles, DRB1*1201) or HLA-DRB1*1101/1104 are able to resist HBV infection, while those with HLA-DR9, DQ9, HLA-DRB1*0301, HLA-DQB1*0301[20,21], and DRB1*10 are susceptible to chronic HBV infection[22].
In spite of the pivotal role that the polymorphic HLA antigens play in immune surveillance and immune response and the multitude of studies performed, there is still a lack of conclusive evidence of the association between polymorphisms and the outcomes of HBV infection; the relationship between them is not universal for all the investigated populations. In this meta-analysis, the identification of common HLA-DR alleles causing CHB susceptibility was examined through a systematic review of the literature followed by a meta-analysis of all case-control studies. Meta-analysis is a powerful method for quantitatively summarizing the results from different studies. One of the advantages is to enhance the statistical power of outcomes in ethnically- and ancestrally-rich populations and to enlarge the sample sizes, which may reduce the possibility of producing false-positive or false-negative association by random error[23].
A systematic search was conducted by two investigators independently (Yan ZH and Fan Y). All articles were retrieved from four main databases: Medline/PubMed, EMBASE, CNKI (China National Knowledge Infrastructure) and VIP database (Chinese Journal of Science and Technology of VIP). The latest search was updated on April 20, 2011, using the search terms: “hepatitis B” or “HBV”, “polymorphism”, “human leukocyte antigen” or “HLA” or “Major Histocompatibility Complex” or “MHC”. The search was not confined to articles written in a certain language, but focused on studies conducted on human subjects. All searched studies were retrieved and the relevant publications in the bibliographies were checked simultaneously. Abstracts and unpublished reports were not considered; only published studies with full-text articles were included. When the same patient population was included in several publications, only the most recent or complete study was used in this meta-analysis. When a study reported results of different subpopulations, we treated each subpopulation as a separate comparison in the meta-analysis. This work has been carried out in accordance with the Declaration of Helsinki (2000) of the World Medical Association. This study was approved ethically by University Hospital Birmingham Trust (LREC 2002/166). All patients provided informed written consent.
The inclusion and exclusion criteria were drawn up on the basis of discussion. The articles to be included in this meta-analysis should meet these inclusion criteria: (1) the full text was available for the published study; (2) the study was designed as an unrelated case-control study; (3) the study provided the number of persistent chronic HBV infection cases and controls; (4) the study provided the number of individual HLA-DR genotypes in cases and controls; (5) the study provided enough information for odd ratio (OR) and 95% confidence interval (95% CI) calculation; and (6) the diagnosis for CHB was based on the following criteria: seropositive for HBsAg for at least 6 mo; serum HBV DNA > 1 × 105 copies/mL for HBeAg-positive and > 1 × 104 copies/mL for HBeAg-negative patients; and persistent or intermittent elevation in alanine aminotransferase (ALT) level, or liver biopsy showing chronic hepatitis with moderate or severe necroinflammation. The criteria for spontaneous recovery from infection were: positive for both anti-HBs and anti-HBc antibodies, negative for HBsAg, normal liver function tests and no history of dominant hepatitis B and HBV vaccination.
The exclusion criteria were: (1) the study was not a case-control study; (2) the control group was only with healthy patients and without those who recovered from prior HBV infection; (3) the study did not fit the diagnosis criteria; (4) the individuals included in the case-control study had serologic evidence for co-infection with hepatitis C virus, hepatitis D virus, or human immunodeficiency virus (HIV); and (5) the studies were duplicated (only the most recent or complete study was used in this meta-analysis when the same patient population was included in several publications[24]). In addition, interim analyses and comparisons of laboratory methods were all excluded.
In order to retrieve articles as completely and correctly as possible, two investigators extracted data independently using a standardized form and they reached a consensus on all items. Disagreement was resolved by discussion among the whole study groups. For each study, the following data were extracted: name of the first author, year of publication, ethnicity, ethnic origin of the studied population, number of patients with persistent HBV infection (including asymptomatic carriers and patients with chronic liver diseases), number of spontaneously recovered controls, number of individual genotypes, source of controls, genotype frequency of cases and controls, genotyping methods, statistical methods and the results of the study.
The quality of the primary studies was evaluated using the Newcastle-Ottawa Scale (NOS)[25]. This scale judges the study on three broad perspectives: the selection of the study groups; the comparability of the groups; and the ascertainment of either the exposure or outcome of interest for case-control.
We compared persistent HBV infection cases with spontaneously recovered controls to assess the relationship between HLA-DR polymorphisms and HBV infection clearance. Subgroup analyses were mainly performed by ethnicity. Ethnic groups were mainly defined as Chinese Han and other ethnic group. Statistical analysis was conducted using the Stata 11.0 (StataCorp, College Station, TX, United States). The association between HLA-DR polymorphisms and the CHB risk was measured by OR with 95% CI. The pooled OR was determined by the Z test and statistical significance was set at P < 0.05. All P values were two-sided. To ensure the reliability and the accuracy of the results, two investigators uploaded the data into the statistic software programs independently and obtained the same results. In our study, two models of meta-analysis for dichotomous outcomes were constructed: the random-effects model and the fixed-effects model. The random-effects model was constructed using the DerSimonian and Laird’s method[26], which assumed that studies were conducted in populations with varying effect sizes and calculated the study weights from both in-study and between-study variances. The fixed-effects model was constructed using the Mantel-Haenszel’s method[27], which assumed that studies were conducted in populations with the same effect sizes and made an adjustment to the study weights according to the in-study variances.
To assess the between-study heterogeneity more precisely, both the χ2-based Cochran’s Q-statistic[28] (to test the heterogeneity) and the Higgins (I2) test[29] (to quantify the proportion of the total variation due to heterogeneity) were calculated. Because of the low power of Cochran’s Q statistic, heterogeneity was considered significant at Ph < 0.10, and the random-effects model was used to pool the results. When the P value of Cochran’s Q statistic was greater than 0.10, the fixed-effects model was used to pool the results. Besides, the Galbraith plot was used to spot the outliers as the possible major sources of heterogeneity[30]. To better investigate the possible sources of between-study heterogeneity, meta-regression analysis was also applied to both general analyses and subgroup analyses, when heterogeneity was observed. To confirm the effect of clinical heterogeneity between studies on the conclusions of the meta-analysis, subgroup analysis was conducted based on races. To validate the credibility of outcomes in this meta-analysis, a sensitivity analysis was performed by sequential omission of individual studies or by omitting studies plotted by the Galbraith plot method as the possible major source of heterogeneity. The potential publication bias was estimated by funnel plot, in which the standard error of logOR of each study was plotted against its logOR. An asymmetric plot suggests a possible publication bias. Visual inspection of asymmetry in funnel plots was conducted to assess the potential for publication bias. In addition, Begg’s rank correlation method and the Egger weighted regression method were also used to statistically assess the publication bias (publication bias with a P≤ 0.05 was considered statistically significant)[31].
A flow diagram of the study selection process is shown in Figure 1. We identified a total of 313 potentially relevant articles to our search criteria. After carefully reviewing the titles, abstracts and full text of the literature, we identified 72 articles with analysis on the association between HLA-DR polymorphisms and persistent HBV infection. However, according to the exclusion criteria, 54 publications were excluded including 21 articles involving the same study subjects as other included articles, nine papers not about case-control study, 22 publications without spontaneously recovered patients in the control group, and two papers in which the individuals included in the case-control study had serologic evidence for co-infection with HIV. Finally, 18 articles evaluating HLA-DR polymorphisms met the inclusion criteria and were chosen for this systematic review and meta-analysis. All the selected studies presented original data on independent samples.
The characteristics of each study are presented in Table 1. These 18 articles were published from 1995 to 2011 with 15 in English[16,18-21,32-41] and three in Chinese[42-44]. Of them, nine studies were on Han Chinese populations[19-21,34-36,42-44] and 10 studies on other race (Gambia[32], Caucasian[16], Korean[18,37,39], Iran[33,40], Thai[38] and Turkish[41]). Besides, one publication contained two individual case-control studies from two different ethnic populations (Chinese Han and Taiwanese aborigines[19]). All studies used the individuals who spontaneous recovered from prior HBV infection as control subjects. The sample size in the individual case-control studies ranged from 60 to 638. A total of 2609 patients with CHB and 606 spontaneously recovered controls prior to HBV infection, were included from 19 studies (cases and controls were counted only once for each study). Genotyping methods used in the studies included PCR-restriction fragment length polymorphism, PCR-sequence specific primer, PCR-sequence specific oligonucleotide probe and sequence-specific oligonucleotide probe hybridization.
| No. | Study | Yr | Language | Ethnicity | Genotyping | Spontaneously recovered control | Chronic hepatitis B case | NOS score | ||
| n | Characteristics | n | Characteristics | |||||||
| 1 | Thursz et al[32] | 1995 | English | Gambia | PCR-RFLP | 413 | 218 children; 195 adults | 225 | 185 children; 40 adults | 7 |
| 2 | Hohler et al[16] | 1997 | English | Caucasian | PCR-SSP | 24 | NA | 70 | Outpatients | 7 |
| 3 | Ahn et al[18] | 2000 | English | Korean | PCR-SSP | 243 | 156 males, mean age: 41 yr | 83 | 53 males, mean age: 35 yr | 8 |
| 4 | Chen et al[42] | 2002 | Chinese | Chinese Han | PCR-SSP | 56 | 43 males, mean age: 47.6 yr | 30 | 22 males, mean age: 38.7 yr | 7 |
| 5 | Akcam et al[33] | 2002 | English | Iran | PCR-SSP | 30 | 20 males, mean age: 35.9 yr | 30 | 7 males, mean age: 31.0 yr | 7 |
| 6 | Jiang et al[21] | 2003 | English | Chinese Han | PCR-SSP | 30 | 24 males, mean age: 33.2 yr | 52 | 43 males, mean age: 33.46 yr | 7 |
| 7 | Meng et al[20] | 2003 | English | Chinese Han | PCR-SSP | 56 | 22 males, mean age: 38.7 yr | 30 | 43 males, mean age: 47.6 yr | 7 |
| 8 | Wu et al[19] | 2004 | English | Chinese Han | SSOPH | 324 | 169 males, mean age: 39.1 yr | 98 | 66 males, mean age: 50.9 yr | 8 |
| 9 | Wu et al[19] | 2004 | English | Taiwanese aborigines | SSOPH | 229 | 90 males, mean age: 52.1 yr | 138 | 60 males, mean age: 45.7 yr | 8 |
| 10 | Lu et al[43] | 2006 | Chinese | Chinese Han | PCR-SSP | 148 | 70 males, mean age: 37.75 yr | 417 | 289 males; 207 CHB, 212ASC ASC | 6 |
| 11 | Zhang et al[34] | 2006 | English | Chinese Han | PCR-SSP | 32 | 20 males, mean age: 38.4 yr | 61 | 40 males, mean age: 38.4 yr | 7 |
| 14 | Yang et al[35] | 2007 | English | Chinese Han | PCR-SSP | 108 | Age and sex matched | 108 | 90 males, 58 LC, 24 HCC | 7 |
| 15 | Song et al[44] | 2007 | Chinese | Chinese Han | PCR-SSP | 102 | 63 males, mean age: 33.9 yr | 276 | 168 males, 77 LC, 106 ASC | 5 |
| 14 | Zhu et al[36] | 2007 | English | Chinese Han | PCR-SSP | 133 | 63 males, mean age: 37 yr | 151 | 120 males, mean age: 40 yr | 7 |
| 15 | Hwang et al[37] | 2007 | English | Korean | PCR-SSP | 438 | 286 man, mean age: 39 yr | 198 | 148 males, 76 LC, 28 HCC | 7 |
| 16 | Kummee et al[38] | 2007 | English | Thai | PCR-SSP | 100 | 48 males, mean age: 51.0 yr | 150 | 80 males, mean age: 30.9 yr | 8 |
| 17 | Cho et al[39] | 2008 | English | Korean | PCR-SSP | 80 | 60 males, mean age: 47.9 yr | 384 | 283 males, mean age: 41.0 yr | 8 |
| 18 | Remezani et al[40] | 2008 | English | Iran | PCR-SSP | 30 | 17 males, mean age: 32.2 yr | 33 | 20 males, mean age: 38 yr | 8 |
| 19 | Albayrak et al[41] | 2011 | English | Turkish | PCR-SSP | 30 | 15 males, mean age: 33.9 yr | 75 | 48 males, mean age: 33.0 yr | 8 |
The NOS results showed that the median overall score was 7 (range, 5-8), which indicated that the methodological quality was generally good. We defined studies that scored a 7 or above as having high methodological quality, and judged that 2 out of the 19 studies to be of low quality (one study[44] scored 5 and another study[43] scored 6) primarily due to either no description of case selection, no definition of control, a lack of adjusted analysis, or no description of the method of ascertainments for case-controls.
Table 2 lists the main results of meta-analysis of the relationship between HLA-DR polymorphisms and the clearance of persistent HBV infection. We found no evidence of associations between the clearance of persistent HBV infection and HLA-DR*08, DR*09, DR*10, DR*11, DR*12, DR*14, DR*15, DR*16 alleles (POR > 0.05). For these negatively associated alleles, the results of subgroup analyses by ethnicity also showed no associations in either Chinese Han group or other ethnic group (POR > 0.05).
| Allele | Present vs null | StudiesNo. | SRC (n/N) | CHB (n/N) | Heterogeneity | M | OR | Publication bias | |||
| I2 (%) | PH | OR (95% CI) | POR | Pbeggr’s | PEgger’s | ||||||
| DR*01 | Total studies | 13 | 109/1500 | 132/1673 | 0 | 0.826 | F | 0.69 (0.52-0.93) | 0.016 | 0.143 | 0.221 |
| Chinese Han group | 6 | 18/432 | 34/415 | 0 | 0.928 | F | 0.48 (0.26-0.86) | 0.014 | 0.348 | 0.219 | |
| Other ethnic groups | 7 | 91/1068 | 98/1258 | 0 | 0.643 | F | 0.79 (0.56-1.12) | 0.191 | 0.133 | 0.104 | |
| DR*03 | Total studies | 12 | 180/1489 | 177/1789 | 16.0 | 0.287 | F | 1.47 (1.16-1.87) | 0.002 | 0.945 | 0.572 |
| Chinese Han group | 5 | 72/470 | 74/627 | 37.0 | 0.174 | F | 1.57 (1.07-2.30) | 0.020 | 0.806 | 0.838 | |
| Other ethnic groups | 7 | 108/1019 | 103/1162 | 8.6 | 0.363 | F | 1.41 (1.03-1.91) | 0.029 | 0.548 | 0.444 | |
| DR*04 | Total studies | 16 | 358/1766 | 549/2256 | 0 | 0.757 | F | 0.72 (0.60-0.85) | 0.000 | 0.207 | 0.255 |
| Chinese Han group | 7 | 104/530 | 215/739 | 0 | 0.920 | F | 0.63 (0.48-0.84) | 0.002 | 0.453 | 0.299 | |
| Other ethnic groups | 9 | 254/1236 | 334/1517 | 0 | 0.459 | F | 0.77 (0.62-0.95) | 0.016 | 0.175 | 0.178 | |
| DR*07 | Total studies | 13 | 239/1373 | 146/1584 | 14 | 0.304 | F | 1.59 (1.24-2.03) | 0.000 | 0.625 | 0.634 |
| Chinese Han group | 7 | 111/530 | 82/739 | 13.6 | 0.326 | F | 1.50 (1.07-2.10) | 0.017 | 0.881 | 0.601 | |
| Other ethnic groups | 6 | 128/843 | 64/845 | 27.7 | 0.227 | F | 1.69 (1.17-2.44) | 0.005 | 0.452 | 0.924 | |
| DR*08 | Total studies | 15 | 290/1736 | 301/2226 | 0 | 0.541 | F | 1.19 (0.98-1.44) | 0.083 | 0.255 | 0.250 |
| Chinese Han group | 7 | 105/530 | 124/739 | 0 | 0.850 | F | 1.30 (0.96-1.77) | 0.086 | 0.293 | 0.188 | |
| Other ethnic groups | 8 | 185/1206 | 177/1487 | 27.7 | 0.207 | F | 1.11 (0.86-1.43) | 0.409 | 0.711 | 0.447 | |
| DR*09 | Total studies | 14 | 344/1703 | 385/2196 | 33.3 | 0.108 | F | 1.17 (0.98-1.40) | 0.075 | 0.661 | 0.740 |
| Chinese Han group | 7 | 157/530 | 206/739 | 32.3 | 0.182 | F | 1.10 (0.85-1.42) | 0.467 | 0.230 | 0.241 | |
| Other ethnic groups | 7 | 187/1173 | 179/1457 | 40.7 | 0.120 | F | 1.25 (0.98-1.59) | 0.077 | 0.764 | 0.786 | |
| DR*10 | Total studies | 11 | 62/955 | 96/1447 | 0 | 0.921 | F | 1.08 (0.76-1.53) | 0.682 | 0.815 | 0.343 |
| Chinese Han group | 6 | 9/469 | 24/707 | 0 | 0.871 | F | 0.68 (0.29-1.49) | 0.314 | 0.039 | 0.040 | |
| Other ethnic groups | 5 | 51/1206 | 70/1487 | 0 | 0.860 | F | 1.22 (0.82-1.81) | 0.325 | 0.806 | 0.687 | |
| DR*11 | Total studies | 14 | 221/1459 | 229/1630 | 41.3 | 0.053 | R | 1.03 (0.75-1.42) | 0.839 | 0.477 | 0.915 |
| Chinese Han group | 7 | 73/530 | 107/739 | 59.1 | 0.023 | R | 1.08 (0.59-1.97) | 0.806 | 0.652 | 0.848 | |
| Other ethnic groups | 7 | 148/929 | 122/891 | 20.0 | 0.277 | F | 1.00 (0.74-1.35) | 0.993 | 0.548 | 0.953 | |
| DR*12 | Total studies | 12 | 336/1478 | 360/1590 | 30.1 | 0.152 | F | 1.08 (0.90-1.31) | 0.406 | 0.945 | 0.948 |
| Chinese Han group | 6 | 142/500 | 173/683 | 33.7 | 0.183 | F | 1.23 (0.93-1.62) | 0.149 | 0.707 | 0.642 | |
| Other ethnic groups | 6 | 194/978 | 187/978 | 27.2 | 0.231 | F | 0.98 (0.76-1.26) | 0.845 | 1.000 | 0.709 | |
| DR*13 | Total studies | 16 | 69/1779 | 211/2127 | 0 | 0.699 | F | 0.27 (0.19-0.37) | 0.000 | 0.964 | 0.473 |
| Chinese Han group | 7 | 12/531 | 48/739 | 0 | 0.867 | F | 0.36 (0.18-0.70) | 0.003 | 0.368 | 0.510 | |
| Other ethnic groups | 9 | 57/1248 | 163/1388 | 1.3 | 0.424 | F | 0.24 (0.17-0.35) | 0.000 | 0.175 | 0.017 | |
| DR*14 | Total studies | 14 | 281/1511 | 335/739 | 12.2 | 0.319 | F | 1.17 (0.94-1.46) | 0.151 | 0.071 | 0.073 |
| Chinese Han group | 7 | 73/530 | 97/739 | 0.2 | 0.422 | F | 1.11 (0.79-1.57) | 0.552 | 0.176 | 0.108 | |
| Other ethnic groups | 7 | 208/981 | 238/1074 | 29.7 | 0.201 | F | 1.22 (0.92-1.62) | 0.172 | 0.022 | 0.076 | |
| DR*15 | Total studies | 12 | 248/1230 | 236/1132 | 0 | 0.525 | F | 0.84 (0.67-1.05) | 0.127 | 0.075 | 0.052 |
| Chinese Han group | 7 | 113/530 | 161/739 | 13.2 | 0.329 | F | 0.92 (0.69-1.22) | 0.556 | 0.293 | 0.155 | |
| Other ethnic groups | 5 | 135/700 | 75/393 | 0 | 0.717 | F | 0.73 (0.51-1.04) | 0.084 | 0.221 | 0.133 | |
| DR*16 | Total studies | 12 | 41/876 | 78/1082 | 22.4 | 0.224 | F | 0.71 (0.47-1.06) | 0.097 | 0.170 | 0.140 |
| Chinese Han group | 7 | 24/530 | 59/739 | 12.7 | 0.333 | F | 0.60 (0.42-1.13) | 0.137 | 0.652 | 0.042 | |
| Other ethnic groups | 5 | 17/346 | 19/343 | 44.8 | 0.124 | F | 0.76 (0.37-1.54) | 0.447 | 0.221 | 0.764 | |
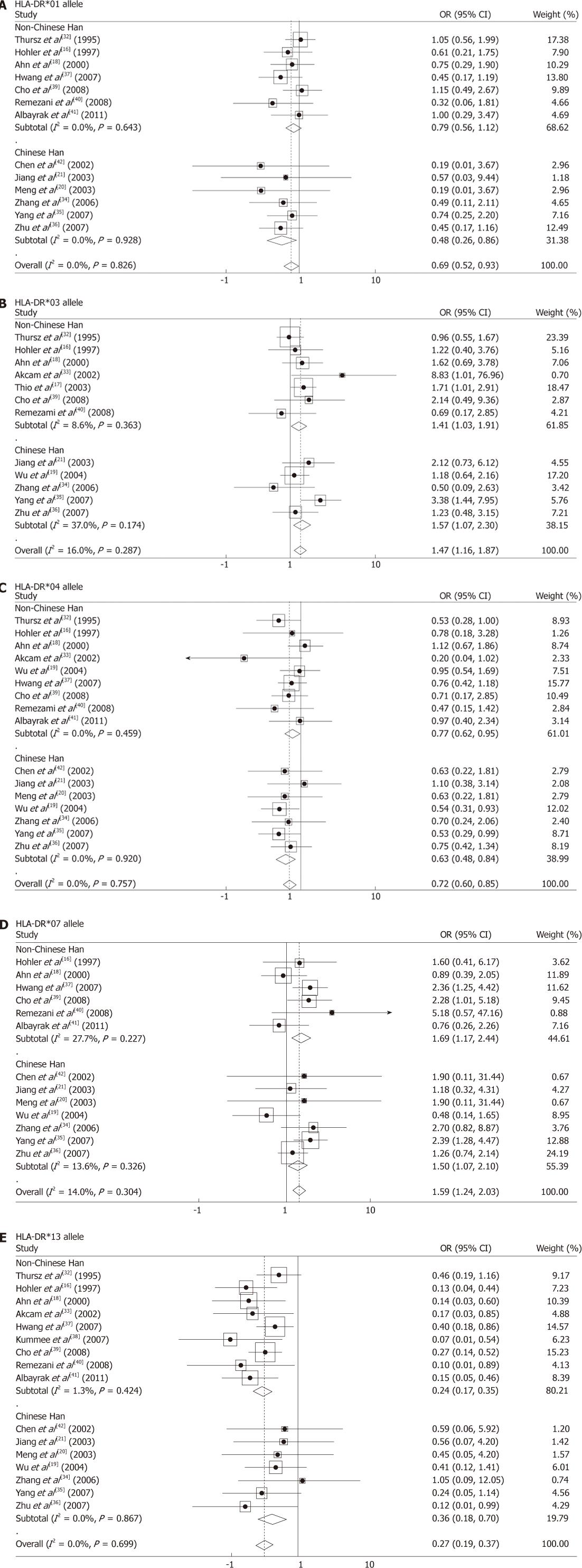
For the HLA-DR*01 polymorphism, the fixed-effects model was used to pool the meta-analysis result, as the between-study heterogeneity was not significant (I2 = 0%, PH = 0.826) when all eligible studies were pooled into meta-analysis. The results of pooling all 13 eligible studies showed that HLA-DR*01 allele was associated with an increased clearance rate of HBV infection (POR = 0.016, ORfixed-effects = 0.69, 95% CI: 0.52-0.93). In the subgroup analyses by ethnicity, the association between the DR*01 allele and HBV clearance was found in Chinese Han group (POR = 0.014, ORfixed-effects = 0.48, 95% CI: 0.26-0.886), but not found in other ethnic groups (POR = 0.191) (Figure 2A).
For the HLA-DR*03 polymorphism, the between-study heterogeneity was also not significant when all 12 eligible studies were pooled into meta-analysis (I2 = 16.0%, PH = 0.287). The results of the fixed-effects model meta-analysis which pools all studies showed that the HLA-DR*03 allele was associated with a significantly increased risk of chronic HBV persistence (POR = 0.002, ORfixed-effects = 1.47, 95% CI: 1.16-1.87). The subgroup analyses by ethnicity showed that the DR*03 allele was associated with a significantly increased risk of chronic HBV persistence in both Chinese Han group (POR = 0.020, ORfixed-effects = 1.57, 95% CI: 1.07-2.30) and other ethnic groups (POR = 0.029, ORfixed-effectss = 1.41, 95% CI: 1.03-1.91) (Figure 2B).
For the HLA-DR*04 polymorphism, the between-study heterogeneity was also not significant when all 16 studies were pooled into meta-analysis (I2 = 0%, PH = 0.757). The results of pooling all studies using the fixed-effects model meta-analysis showed that the HLA-DR*04 allele was associated with the clearance of HBV infection (POR = 0.000, ORfixed-effects = 0.72, 95% CI: 0.60-0.85). The subgroup analyses by ethnicity showed that the DR*04 allele was associated with an increased clearance rate of HBV infection in both Chinese Han group (POR = 0.002, ORfixed-effects = 0.63, 95% CI: 0.48-0.84) and other ethnic groups (POR = 0.016, ORfixed-effectss = 0.77, 95% CI: 0.62-0.95) (Figure 2C).
For the HLA-DR*07 polymorphism, the fixed-effects model was used to pool the meta-analysis result, since the between-study heterogeneity was not significant (I2 = 14.0%, PH = 0.304) when all 13 studies were pooled into meta-analysis. The results showed that HLA-DR*07 allele was associated with a significantly increased risk of chronic HBV persistence (POR = 0.000, ORfixed-effects = 1.59, 95% CI: 1.24-2.03). In the subgroup analyses by ethnicity, the results showed that the DR*07 allele was associated with a significantly increased risk of chronic HBV persistence among Chinese Han group (POR = 0.017, ORfixed-effects = 1.50, 95% CI: 1.07-2.10) and other ethnic groups (POR = 0.005, ORfixed-effectss = 1.69, 95% CI: 1.17-2.44) (Figure 2D).
For the HLA-DR*13 polymorphism, the fixed-effects model was used to pool the meta-analysis result, since the between-study heterogeneity was not significant (I2 = 0.00%, PH = 0.699). The results showed that HLA-DR*13 allele was associated with an increased clearance rate of HBV infection (POR = 0.000, ORfixed-effects = 0.27, 95% CI: 0.19-0.37). In the subgroup analyses by ethnicity, the results showed that the DR*13 allele was associated with HBV clearance among Chinese Han group (POR = 0.003, ORfixed-effects =0.36, 95% CI: 0.18-0.70) and other ethnic groups (POR = 0.000, ORfixed-effectss = 0.24, 95% CI: 0.17-0.35) (Figure 2E).
The heterogeneity was calculated among all studies using the Q-statistic and the I2 test. As shown in Table 2, for the HLA-DR*11 polymorphism, the between-study heterogeneity was significant (I2 = 41.3%, PH = 0.053) when all 14 studies were pooled into meta-analysis. In the subgroup analyses by ethnicity for this allele, the between-study heterogeneity was also significant in Chinese Han subgroups (I2 = 59.1%, PH = 0.023), while heterogeneity was not found in other ethnic groups (I2 = 20.0%, PH = 0.277). Except for the meta-analysis for DR*13 allele, heterogeneity in the all meta-analysis for other HLA-DR alleles was not found (PH > 0.10).
Univariate analysis of meta-regression suggested that the publishing year and language were not important sources of between-study heterogeneity in both generate analyses and subgroup analyses. We carried out a sensitivity analysis for each HLA-DR allele interaction analysis by sequential omission of individual studies. A single study involved in the meta-analysis was deleted each time to investigate the influence of the individual dataset on the pooled ORs. The corresponding pooled ORs were not materially altered, indicating that our results were statistically robust.
Begg’s funnel plot and Egger’s test were performed to assess the publication bias of the literature in this meta-analysis. All the P values of Begg’s test (Pbeggr’s) and Egger’s tests (PEgger’s) are shown in Table 2. The PEgger’s with more than 0.05 was considered as statistical evidence of the funnel plots’ asymmetry. The Egger’s test results suggested that publication bias in our meta-analyses of HLA-DR alleles was not remarkable except for the three subgroup meta-analyses. As shown in Table 2, the publication bias was borderline significant in the analysis of HLA-DR*10 allele (PEgger’s = 0.040) and Chinese Han subgroups analysis of HLA-DR*16 (PEgger’s = 0.042). The publication bias was evident in other ethnic group analysis of HLA-DR*13 allele (PEgger’s = 0.017). Then, the Duval and Tweedie non-parametric “trim and fill” method was used to adjust publication bias[45]. Meta-analysis with and without using the “trim and fill” method did not draw different conclusions, indicating that our results were statistically robust.
It is believed that the host genetic factors involved in genetic polymorphisms are responsible for the susceptibility to and clinical outcomes of infectious diseases. In recent years, genetic susceptibility to chronic HBV infection has been a research focus, and it has been identified that the polymorphisms of a number of immune-response-associated genes, including HLA loci, affected the susceptibility to and clearance of persistent chronic HBV infection among different populations. HLA plays an essential role in the pathogenesis of virus-associated hepatitis. The HLA class II molecules are expressed as cell surface glycol-proteins that bind short peptide epitope to CD4+ T cells. HLA-DR, a subtype of HLA class II molecule, has a particular binding motif that dictates a specific range of peptides that can physically bind in a groove on the surface of the HLA molecule[14].
Wide variations have been documented in the frequencies of HLA-DR gene polymorphisms which have been most widely investigated in healthy populations and been demonstrated to influence TNF-α expression. The association between HLA-DR polymorphisms and outcome of HBV infection has been investigated by several research groups. However, the previous studies have yielded conflicting results, and included no more than a few hundred CHB cases, which are too few to assess the genetic effects reliably. Meta-analysis has been recognized as an important tool to precisely assess the effect of the selected genetic polymorphisms on the risk of diseases and to identify potentially important sources of between-study heterogeneity. In our present meta-analysis, a total of 2609 patients with CHB and 2606 controls spontaneously recovering prior to HBV infection were included from 19 case-control studies which were evaluated using the NOS. It could provide the most comprehensive assessment to draw reliable conclusions.
Heterogeneity is a potential problem when interpreting the results of all meta-analyses, and finding the sources of heterogeneity is one of the most important goals of meta-analysis[46]. In this present meta-analysis, we assessed the between-study heterogeneity by different methods, including the χ2 based Q statistic test (Cochran’s Q statistic)[28] (to test for heterogeneity) and the I2 statistic (to quantify the between-study heterogeneity)[29]. For the meta-analyses comparing persistent HBV infection cases with spontaneously recovering controls, there was significant between-study heterogeneity in pooled meta-analyses of total eligible studies on HLA-DR*11 allele, which suggested obvious consistency of effects across those included studies. Subgroup analyses by ethnicity showed that the heterogeneity was still significant in subgroup analyses in Chinese Han populations for HLA-DR*11 polymorphism. Univariate analysis of meta-regression suggested that the publishing year and language were not important causes of between-study heterogeneity in both general analyses and subgroup analyses. We presumed that the quality of the primary studies would be the main cause of heterogeneity. Interesting, we found no significant between-study heterogeneity in pooled meta-analyses of total eligible studies and subgroup analyses by ethnicity for other alleles.
Some limitations still exist in this meta-analysis. First, meta-analysis essentially remains with observational study that was subject to the methodological deficiencies of the included studies. Since only published studies written in English and Chinese were included in the meta-analysis, publication bias may occur, even though it was not found by statistical tests. Second, the associations were investigated in all kinds of cases (asymptomatic carriers, patients with CHB, patients with liver cirrhosis), and there may be specific genetic effects among these cases, but we could not obtain enough information to further estimate these effects. It is necessary to conduct large trials using standardized unbiased methods on homogeneous CHB patients and well matched controls, with the assessors blinded to the data. Third, our results were based on unadjusted estimates. A more precise analysis should be conducted with individual data, which would allow the adjustment by other co-varieties including age, ethnicity, family history, environmental factors and lifestyle. Finally, gene-gene and gene-environment interactions were not addressed in this meta-analysis due to the lack of sufficient data. For instance, the major genotypes of HBV in Chinese are B and C, but most of the studies did not analyze them separately, which could not be solved because of the methodological limitations of the meta-analysis.
Despite these limitations, this meta-analysis suggests that HLA-DR*04 and DR*13 alleles may be the protective factors for HBV clearance, and HLA-DR*03 and DR*07 alleles may be the risk factors for HBV persistence. For the HLA-DR*01 polymorphism, a significantly association with HBV clearance was found in Chinese Han group, but not found in other ethnic groups. In summary, ethnicity may play an important role in HBV infection outcome, leading to conflicting results. More studies on individuals from various ethnic groups and large and carefully designed case-control studies will be necessary to determine the role of HLA-DR polymorphisms in the outcome of HBV infection.
Chronic hepatitis B virus (HBV) infection is a serious public health problem worldwide. Host genetic factors play a role in determining both susceptibility to HBV and the outcome of the infection. A large number of studies on the association between human leukocyte antigens (HLA)-DR gene polymorphisms and the risk of chronic hepatitis B (CHB) have been conducted, but their conclusions are different or even contradictory.
The polymorphic HLA antigens play an important role in immune surveillance and immune response. However, the relationships HLA-DR polymorphisms and the outcomes of HBV infection are not universal for all the investigated populations and no meta-analysis has been conducted.
This meta-analysis systemically assessed the associations of HLA-DR gene polymorphisms with the outcomes of HBV infections, and concluded that HLA-DR*03, DR*04, DR*07 and DR*13 alleles have significant associations with HBV clearance in both Chinese Han population and other ethnic groups.
The results of meta-analysis in this study show that HLA-DR*04 and DR*13 alleles may be the protective factors for HBV clearance, while HLA-DR*03 and DR*07 alleles may be the risk factors for HBV persistence, which may benefit early prevention and treatment of CHB.
This meta-analysis of HLA-DR gene polymorphisms and HBV infection outcomes is well written, addressed an important in the field. However, there are still some problems.
Peer reviewer: Dr. Guang-Wen Cao, MD, PhD, Professor and Chairman, Department of Epidemiology, The Second Military Medical University, 800 Xiangyin Road, Shanghai 200433, China
S- Editor Yang XC L- Editor Ma JY E- Editor Zheng XM
| 1. | Lai CL, Ratziu V, Yuen MF, Poynard T. Viral hepatitis B. Lancet. 2003;362:2089-2094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 595] [Cited by in RCA: 590] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 2. | Segal S, Hill AV. Genetic susceptibility to infectious disease. Trends Microbiol. 2003;11:445-448. [PubMed] |
| 3. | Frodsham AJ. Host genetics and the outcome of hepatitis B viral infection. Transpl Immunol. 2005;14:183-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 43] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 4. | Lin TM, Chen CJ, Wu MM, Yang CS, Chen JS, Lin CC, Kwang TY, Hsu ST, Lin SY, Hsu LC. Hepatitis B virus markers in Chinese twins. Anticancer Res. 1989;9:737-741. [PubMed] |
| 5. | Ben-Ari Z, Mor E, Papo O, Kfir B, Sulkes J, Tambur AR, Tur-Kaspa R, Klein T. Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol. 2003;98:144-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 180] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 6. | Höhler T, Kruger A, Gerken G, Schneider PM, Meyer zum Büschenefelde KH, Rittner C. A tumor necrosis factor-alpha (TNF-alpha) promoter polymorphism is associated with chronic hepatitis B infection. Clin Exp Immunol. 1998;111:579-582. [PubMed] |
| 7. | Suneetha PV, Sarin SK, Goyal A, Kumar GT, Shukla DK, Hissar S. Association between vitamin D receptor, CCR5, TNF-alpha and TNF-beta gene polymorphisms and HBV infection and severity of liver disease. J Hepatol. 2006;44:856-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 93] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 8. | Chong WP, To YF, Ip WK, Yuen MF, Poon TP, Wong WH, Lai CL, Lau YL. Mannose-binding lectin in chronic hepatitis B virus infection. Hepatology. 2005;42:1037-1045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 76] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 9. | Deng G, Zhou G, Zhai Y, Li S, Li X, Li Y, Zhang R, Yao Z, Shen Y, Qiang B. Association of estrogen receptor alpha polymorphisms with susceptibility to chronic hepatitis B virus infection. Hepatology. 2004;40:318-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 83] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 10. | Deng G, Zhou G, Zhang R, Zhai Y, Zhao W, Yan Z, Deng C, Yuan X, Xu B, Dong X. Regulatory polymorphisms in the promoter of CXCL10 gene and disease progression in male hepatitis B virus carriers. Gastroenterology. 2008;134:716-726. [PubMed] |
| 11. | Yan Z, Tan W, Zhao W, Dan Y, Wang X, Mao Q, Wang Y, Deng G. Regulatory polymorphisms in the IL-10 gene promoter and HBV-related acute liver failure in the Chinese population. J Viral Hepat. 2009;16:775-783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 12. | Guo X, Zhang Y, Li J, Ma J, Wei Z, Tan W, O'Brien SJ. Strong influence of human leukocyte antigen (HLA)-DP gene variants on development of persistent chronic hepatitis B virus carriers in the Han Chinese population. Hepatology. 2011;53:422-428. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 117] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 13. | Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, Kubo M, Tsunoda T, Kamatani N, Kumada H. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet. 2009;41:591-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 430] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 14. | Godkin A, Davenport M, Hill AV. Molecular analysis of HLA class II associations with hepatitis B virus clearance and vaccine nonresponsiveness. Hepatology. 2005;41:1383-1390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 104] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 15. | Singh R, Kaul R, Kaul A, Khan K. A comparative review of HLA associations with hepatitis B and C viral infections across global populations. World J Gastroenterol. 2007;13:1770-1787. [PubMed] |
| 16. | Höhler T, Gerken G, Notghi A, Lubjuhn R, Taheri H, Protzer U, Löhr HF, Schneider PM, Meyer zum Büschenfelde KH, Rittner C. HLA-DRB1*1301 and *1302 protect against chronic hepatitis B. J Hepatol. 1997;26:503-507. [PubMed] |
| 17. | Thio CL, Thomas DL, Karacki P, Gao X, Marti D, Kaslow RA, Goedert JJ, Hilgartner M, Strathdee SA, Duggal P. Comprehensive analysis of class I and class II HLA antigens and chronic hepatitis B virus infection. J Virol. 2003;77:12083-12087. [PubMed] |
| 18. | Ahn SH, Han KH, Park JY, Lee CK, Kang SW, Chon CY, Kim YS, Park K, Kim DK, Moon YM. Association between hepatitis B virus infection and HLA-DR type in Korea. Hepatology. 2000;31:1371-1373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 104] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 19. | Wu YF, Wang LY, Lee TD, Lin HH, Hu CT, Cheng ML, Lo SY. HLA phenotypes and outcomes of hepatitis B virus infection in Taiwan. J Med Virol. 2004;72:17-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 20. | Meng XQ, Chen HG, Ma YL, Liu KZ. Influence of HLA class II molecules on the outcome of hepatitis B virus infection in population of Zhejiang Province in China. Hepatobiliary Pancreat Dis Int. 2003;2:230-233. [PubMed] |
| 21. | Jiang YG, Wang YM, Liu TH, Liu J. Association between HLA class II gene and susceptibility or resistance to chronic hepatitis B. World J Gastroenterol. 2003;9:2221-2225. [PubMed] |
| 22. | Shen JJ, Ji Y, Guan XL, Huang RJ, Sun YP. [The association of HLA-DRB110 with chronic hepatitis B in Chinese patients]. Zhonghua Weishengwuxue He Mianyixue Zazhi. 1999;19:58-59. |
| 23. | Blettner M, Sauerbrei W, Schlehofer B, Scheuchenpflug T, Friedenreich C. Traditional reviews, meta-analyses and pooled analyses in epidemiology. Int J Epidemiol. 1999;28:1-9. [PubMed] |
| 24. | Mays N, Pope C, Popay J. Systematically reviewing qualitative and quantitative evidence to inform management and policy-making in the health field. J Health Serv Res Policy. 2005;10 Suppl 1:6-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 820] [Cited by in RCA: 723] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 25. | Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8858] [Cited by in RCA: 12634] [Article Influence: 842.3] [Reference Citation Analysis (0)] |
| 26. | DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177-188. [PubMed] |
| 27. | Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719-748. [PubMed] |
| 28. | Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101-129. |
| 29. | Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39087] [Cited by in RCA: 46470] [Article Influence: 2112.3] [Reference Citation Analysis (3)] |
| 30. | Galbraith RF. A note on graphical presentation of estimated odds ratios from several clinical trials. Stat Med. 1988;7:889-894. [PubMed] |
| 31. | Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629-634. [PubMed] |
| 32. | Thursz MR, Kwiatkowski D, Allsopp CE, Greenwood BM, Thomas HC, Hill AV. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med. 1995;332:1065-1069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 317] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 33. | Akcam Z, Sunbul M, Durupinar B, Eroglu C, Esen S, Leblebicioglu H. Tissue types as prognostic risk factor in hepatitis B virus infection. Indian J Gastroenterol. 2002;21:139-141. [PubMed] |
| 34. | Zhang SY, Gu HX, Li D, Yang SF, Zhong ZH, Li XK, Jin X. Association of human leukocyte antigen polymorphism with hepatitis B virus infection and genotypes. Jpn J Infect Dis. 2006;59:353-357. [PubMed] |
| 35. | Yang G, Liu J, Han S, Xie H, Du R, Yan Y, Xu D, Fan D. Association between hepatitis B virus infection and HLA-DRB1 genotyping in Shaanxi Han patients in northwestern China. Tissue Antigens. 2007;69:170-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 36. | Zhu XL, Du T, Li JH, Lu LP, Guo XH, Gao JR, Gou CY, Li Z, Liu Y, Li H. Association of HLA-DQB1 gene polymorphisms with outcomes of HBV infection in Chinese Han population. Swiss Med Wkly. 2007;137:114-120. [PubMed] |
| 37. | Hwang SH, Sohn YH, Oh HB, Hwang CY, Lee SH, Shin ES, Lee KJ. Human leukocyte antigen alleles and haplotypes associated with chronicity of hepatitis B virus infection in Koreans. Arch Pathol Lab Med. 2007;131:117-121. [PubMed] |
| 38. | Kummee P, Tangkijvanich P, Poovorawan Y, Hirankarn N. Association of HLA-DRB1*13 and TNF-alpha gene polymorphisms with clearance of chronic hepatitis B infection and risk of hepatocellular carcinoma in Thai population. J Viral Hepat. 2007;14:841-848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 44] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 39. | Cho SW, Cheong JY, Ju YS, Oh do H, Suh YJ, Lee KW. Human leukocyte antigen class II association with spontaneous recovery from hepatitis B virus infection in Koreans: analysis at the haplotype level. J Korean Med Sci. 2008;23:838-844. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 40. | Ramezani A, Hasanjani Roshan MR, Kalantar E, Eslamifar A, Banifazl M, Taeb J, Aghakhani A, Gachkar L, Velayati AA. Association of human leukocyte antigen polymorphism with outcomes of hepatitis B virus infection. J Gastroenterol Hepatol. 2008;23:1716-1721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 41. | Albayrak A, Ertek M, Tasyaran MA, Pirim I. Role of HLA allele polymorphism in chronic hepatitis B virus infection and HBV vaccine sensitivity in patients from eastern Turkey. Biochem Genet. 2011;49:258-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 42. | hen HG, Jiang GF, Meng XQ, Ma YL, Liu KZ. [Preliminary report on the influence of HLA class II molecules on outcome of hepatitis B virus infection in Zhejiang district]. Zhonghua Chuanranbing Zazhi. 2002;20:164-167. |
| 43. | Lu LP, Li XW, Liu Y, Sun GC, Chen ZH, Zhu XL, Hu QY, Li H. [Association of haplotype formed on HLA-DRB1 and HLA-DQA1 alleles with outcomes of hepatitis B virus infection]. Zhonghua Yixue Yichuanxue Zazhi. 2006;23:427-430. [PubMed] |
| 44. | Song MS, Li HW, Peng HY, Duan BN, Chen H, Xu LQ. [Association of polymorphism on HLA-DRB1*04 alleles with outcome of hepatitis B virus infection]. Zhonghua Yixue Yichuanxue Zazhi. 2007;24:467-469. [PubMed] |
| 45. | Duval S, Tweedie R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455-463. [PubMed] |
| 46. | Ioannidis JP, Patsopoulos NA, Evangelou E. Uncertainty in heterogeneity estimates in meta-analyses. BMJ. 2007;335:914-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 790] [Cited by in RCA: 948] [Article Influence: 52.7] [Reference Citation Analysis (0)] |