Published online May 7, 2012. doi: 10.3748/wjg.v18.i17.2035
Revised: November 12, 2011
Accepted: February 26, 2012
Published online: May 7, 2012
AIM: To investigate the role of transforming growth factor (TGF)-β-inducible early gene 1 (TIEG1) in TGF-β-induced growth inhibition in hepatocellular carcinoma (HCC) cells.
METHODS: Human hepatocyte and HCC cell lines with varied susceptibilities to TGF-β1 were tested by methylthiazoletetrazolium (MTT) assay. The expression changes of Smad2, Smad3, Smad4, Smad7, TIEG1 and TIEG2 gene following treatment with TGF-β1 in a TGF-β-sensitive hepatocyte cell line (MIHA), a TGF-β-sensitive hepatoma cell line (Hep3B) and two TGF-β-insensitive hepatoma cell lines (HepG2 and Bel7404) were examined. SiRNA targeting TIEG1 was transfected into Hep3B cells and the sensitivity of cells to TGF-β1 was examined. Overexpression of TIEG1 was induced by lentiviral-mediated transduction in TGF-β1-resistant hepatoma cell lines (Bel7404 and HepG2). MTT assay and 4’,6-Diamidino-2-phenylindole staining were used to identify cell viability and apoptosis, respectively. The expression level of stathmin was measured by reverse transcriptase polymerase chain reaction and Western-blotting analysis, and stathmin promoter activity by TIEG1 was monitored by a luciferase reporter gene system.
RESULTS: TIEG1 was significantly upregulated by TGF-β1 in the TGF-β1-sensitive HCC cell line, Hep3B, but not in the resistant cell lines. The suppression of TIEG1 by siRNAs decreased the sensitivity of Hep3B cells to TGF-β1, whereas the overexpression of TIEG1 mediated growth inhibition and apoptosis in TGF-β1-resistant HCC cell lines, which resembled those of TGF-β1-sensitive HCC cells treated with TGF-β1. Our data further suggested that stathmin was a direct target of TIEG1, as stathmin was significantly downregulated by TIEG1 overexpression, and stathmin promoter activity was inhibited by TIEG1 in a dose-dependent manner.
CONCLUSION: Our data suggest that transactivation of TIEG1 conferred growth inhibition of TGF-β-susceptible human HCC cells.
- Citation: Jiang L, Lai YK, Zhang JF, Chan CY, Lu G, Lin MC, He ML, Li JC, Kung HF. Transactivation of the TIEG1 confers growth inhibition of transforming growth factor-β-susceptible hepatocellular carcinoma cells. World J Gastroenterol 2012; 18(17): 2035-2042
- URL: https://www.wjgnet.com/1007-9327/full/v18/i17/2035.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i17.2035
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third most common cause of cancer death worldwide, and there are few effective therapeutic options available for those suffering from advanced disease[1]. HCC poses a major challenge because of its clinical heterogeneity and lack of good diagnostic markers and treatment strategies[2]. Transforming growth factor-β (TGF-β) is a multifunctional cytokine which regulates cell proliferation, migration and differentiation[3]. TGF-β has been shown to inhibit cell proliferation and to induce apoptosis to control excessive growth of hepatocytes and maintain liver size, and is considered a liver tumor suppressor[4]. However, many HCC cells are thought to have lost their sensitivity to TGF-β, and thus escape the antiproliferative effect of TGF-β[4,5]. The loss of TGF-β activities resulting in hyperproliferative disorders and cancer in the liver and derangement of TGF-β signaling are associated with an increased incidence of HCC[6]. TGF-β has shown dual effects on tumors, in that it can either be pro- or anti-tumorigenetic, depending on the stage of tumorigenesis and the responsiveness of the tumor cells[7]. Various studies have shown that TGF-β signaling may suppress human hepatocarcinogenesis, possibly via cyclin D1 deregulation[8], and that TGF-β could serve as a potential senescence inducer in HCC cells and thereby inhibit tumor growth in vivo[9]. However, in advanced cancers, TGF-β has been found to be a tumor enhancer because it can promote tumor progression by facilitating tumor invasion, neoangiogenesis, and immunosuppression[10].
The TGF-β-inducible early gene 1 (TIEG1) can be activated at the initial stage of the TGF-β pathway. Previous studies have shown that TIEG1 has an important role in regulating cell growth[11,12]. This gene is classified as a member of the Krüppel-like family of transcription factors (KLF10), all of which bind to GC-rich Sp1-like binding sites to regulate gene transcription[12]. In addition, TIEG1 is also regarded as a potent transcriptional repressor. The overexpression of TIEG1 has been found to induce apoptosis in pancreatic cancer cells, and this indicates that it has a pivotal role in mediating TGF-β-induced apoptosis[13,14]. In addition, TIEG1 was found to induce apoptosis via a mechanism which involves the formation of reactive oxygen species[15]. The transcription level of TIEG1 was found to be predominantly expressed in various tissues, and the associated levels were regulated by cytokines and growth factors[12]. It is well known that different HCC cell lines respond differently to TGF-β treatment, and that some HCC cell lines are sensitive to TGF-β, whereas others are resistant. However, the molecular mechanism underlying the differential responses has never been elucidated. In the present study, we investigated the role of TIEG1 in TGF-β-induced growth inhibition in HCC cells. We deduced that transactivation of the TIEG1 conferred growth inhibition of TGF-β-susceptible human HCC cells.
Immortalized human hepatocyte (MIHA), HCC cell lines (HepG2, Hep3B, Bel7404, Huh-7, and PLC), and human HEK293T cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and 100 U/mL penicillin/streptomycin (Invitrogen, Carlsbad, CA, United States). All cultures were maintained in a humidified 37 °C incubator with 5% CO2.
Prior to treatment with TGF-β1, the cells were seeded, allowed to attach for 24 h and then starved in a serum-free medium for another 24 h. The cells were then treated with 5 ng/mL TGF-β1 (R&D System, Minneapolis, MN, United States) for the indicated time periods. Cell survival and changes in nuclei morphology were respectively monitored using methylthiazoletetrazolium (MTT) assays and 4’,6-Diamidino-2-phenylindole (DAPI) staining[13] after 96 h of treatment.
For gene response studies, the total RNA was extracted using TRIZOL (Invitrogen), and then cDNA synthesis was performed using the Superscript First-Strand Synthesis Kit (Promega, United States)[13]. The mRNA levels of genes were determined by quantitative real-time polymerase chain reaction (PCR) or semi-quantitative reverse transcriptase (RT)-PCR[13]. The following forward and reverse primers were used respectively: TIEG1: 5’-GTCACATCTGTAGCCACCCA-3’ and 5’-CCTCCTTTCACAACCTTTCC-3’; TIEG2: 5’-TCTGACTCTGGGGATGTCAC-3’ and 5’-CGGCAATCTGGAGTCTGGA-3’; Smad2: 5’-GCCACGGTAGAAATGACAAG-3’ and 5’-CAGACTGAGCCAGAAGAGCA-3’; Smad3: 5’-GAACGGGCAGGAGGAGAAAT-3’and 5’-ACAGGCGGCAGTAGATGACA-3’; Smad4: 5’-CCATTTCCAATCATCCTGCT-3’ and 5’-ACCTTTGCCTATGTGCAACC-3’; Smad7: 5’-CTTAGCCGACTCTGCGAACT-3’ and 5’-CCCAGGCTCCAGAAGAAGTT-3’; Stathmin (STMN): 5’-TTTTCAATCCCAATTCTGTC-3’ and 5’-GAAAGTAACAGCTGACCTGG-3’; glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (loading control): 5’-CCAGCCGAGCCACATCGCTC-3’ and 5’-ATGAGCCCCAGCCTTCTCCAT-3’. Quantitative real-time PCR was performed using SYBRs GREEN PCR Master Mix (Applied Biosystems, Warrington, United Kingdom) and an ABI 7500 Real-time PCR system. The relative amount of mRNA expression was normalized based on the expression of human GAPDH. Each of the normalized gene expression values was calibrated to the normalized gene expression of cells with TGF-β1 treatment at time zero. The experiments were repeated thrice to build a geometric mean. Alternatively, RT-PCR was employed for semi-quantitative analysis of gene expression levels. GAPDH acted as the internal control[13].
For siRNA transfection, Hep3B cells were transfected with 50 pmol control siRNA or TIEG1 siRNA (Santa Cruz) using oligofectamime (Invitrogen). A second identical transfection was carried out 24 h later. Seventy-two hours after the first transfection, the total RNA was extracted and real-time RT-PCR was performed to evaluate the downregulatory effects. Moreover, 5 × 103 cells were seeded onto 96-well plates and transfected with siRNA twice and then treated with 5 ng/mL TGF-β1 for 72 h. MTT assay was then performed to determine the changes in cell growth.
Lentiviral vectors expressing TIEG1 were constructed, as previously described[13]. The VSV-G pseudotyped lentiviruses were produced by cotransfecting 293T cells with the transfer vector and three packaging vectors. The cells were transduced with lentivirus, as described[13].
The SDS-PAGE and Western blotting analysis were performed, as previously described[13]. The primary antibodies used were polyclonal antibodies against TIEG1 (sc-23159; Santa Cruz) and Actin (80-50; Abcam).
The promoter of STMN was constructed into the pGL3-basic luciferase reporter vector (Promega, Madison, WI, United States) using primer: F-Kpn I: 5’-CCGGTACCTCTAAGGCACGGTCAGACCA-3’, R-Bgl II: 5’-CGCAGATCTCCTGACCACACTCTGAGC-3’. For the luciferase assay, 293T cells were co-transfected with pGL3-promoter-STMN vector, a renilla plasmid, along with different amount of TIEG1-expressed construct or pEGFP-N1 vector. Cell extracts were lysed 48 h post-transfection and assayed for luciferase activities using the Dual-Luciferase Reporter Assay System (Promega).
Data are expressed as the mean ± SD error of the mean. Statistical differences between groups were compared using the Student’s t test. P values less than 0.05 were considered significant.
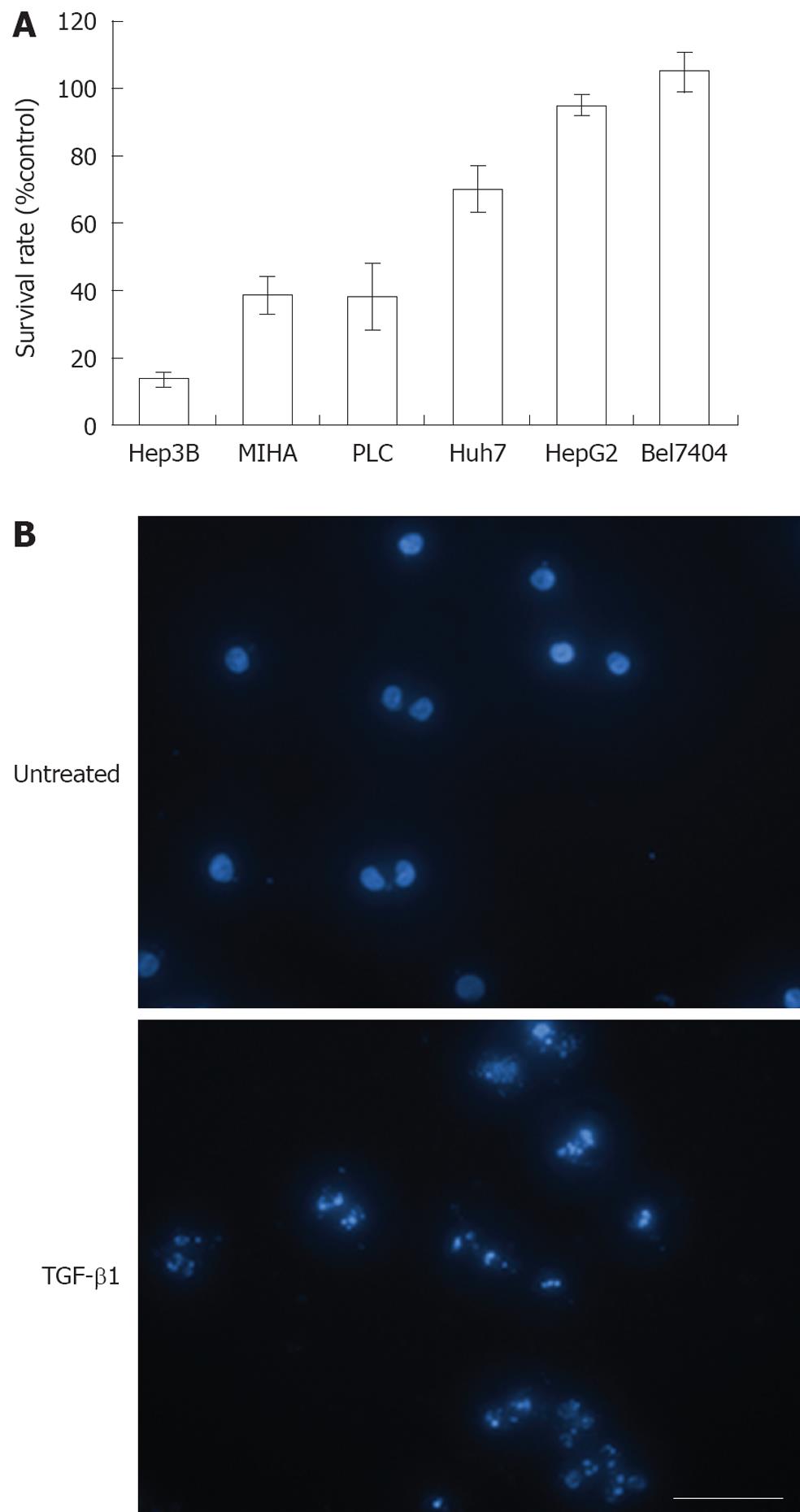
The cell proliferation inhibitory effect of TGF-β1 on various cell lines was evaluated by MTT assay (Figure 1A). TGF-β1 exerted the highest inhibitory effect on Hep3B (> 80%), a known TGF-β-sensitive hepatoma cell line, exhibited a moderate inhibition (approximately 60%) in MIHA cells, a TGF-β-sensitive hepatocyte cell line, and an inhibition rate of approximately 60% was observed in HCC PLC cells. In addition, TGF-β1 exerted only marginal inhibitory effects on Huh7 cells (approximately 30%). Lastly, HepG2 and Bel7404 cells exhibited resistance to growth inhibition by TGF-β1 and completely lost their sensitivity to TGF-β. Using DAPI staining and subsequent fluorescence it was revealed by microscopic examination that the growth inhibitory effect of TGF-β1 on Hep3B cells was via the induction of apoptosis (Figure 1B).
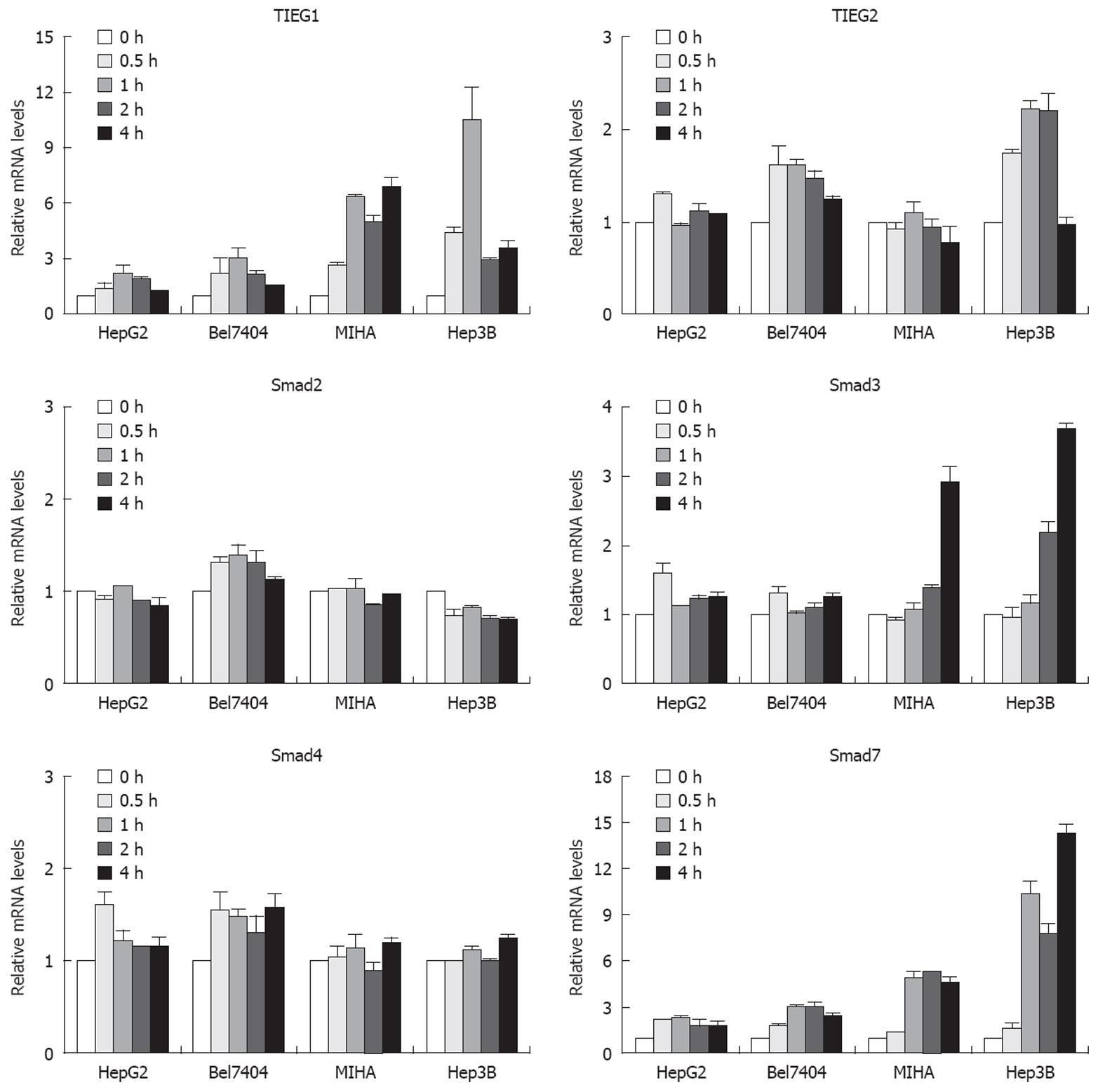
Firstly, we employed semi-quantitative PCR analysis to determine the basal mRNA level of eight different genes, namely, the TGF-β1 receptor 1, the TGF-β1 receptor 2, Smad2, Smad3, Smad4, Smad7, TIEG1 and TIEG2, which are involved in the TGF-β signaling pathway in all the cell lines studied. We found no correlation between the expression of TGF-β-related genes and their sensitivity to TGF-β. All interested genes were present in all cell lines at slightly varied levels (data not shown). Because there was no correlation between the static expression of the TGF-β-related genes and the sensitivity of different HCC cells to TGF-β, we next examined the expression changes of these genes after they had been treated with TGF-β1 in a TGF-β-sensitive hepatocyte cell line (MIHA), a TGF-β-sensitive hepatoma cell line (Hep3B) and two TGF-β-insensitive hepatoma cell lines (HepG2 and Bel7404). Figure 2 shows the relative changes in expression of various genes in response to TGF-β1 over time. The TIEG1 was sharply upregulated as early as 30 min after TGF-β1 treatment in MIHA and Hep3B cells, which were sensitive to TGF-β. One hour after treatment, TIEG1 was upregulated seven-fold in MIHA and more than ten-fold in Hep3B cells, respectively. However, there was a slight increase in TGF-β-insensitive cells (HepG2 and Bel7404). TIEG1 mRNA levels were more potently induced by TGF-β1 in TGF-β1-sensitive cell lines (Hep3B and MIHA) than in TGF-β1-insensitive cell lines (HepG2 and Bel7404).
We also observed that the Smad3 gene was upregulated in TGF-β-sensitive cells after TGF-β1 treatment for 4 h, and that there was no significant change in insensitive hepatoma cells. However, the upregulation of TIEG1 expression appeared earlier than upregulation of the Smad3 gene in TGF-β-sensitive cells. In the case of another gene, namely Smad7, the expression was sharply upregulated after treatment in TGF-β-sensitive cells and weakly upregulated in insensitive hepatoma cells. There was no significant change in TIEG2, Smad2 and Smad4 gene expression after TGF-β1 treatment. Smad7 is thought to be a TGF-β-inducible antagonist of TGF-β signaling[16], and there are autoregulatory negative-feedback signals in the signal transduction of the TGF-β superfamily[17]. These data imply that the TIEG1 gene might play a critical role in TGF-β-mediated growth inhibition of HCC cells.
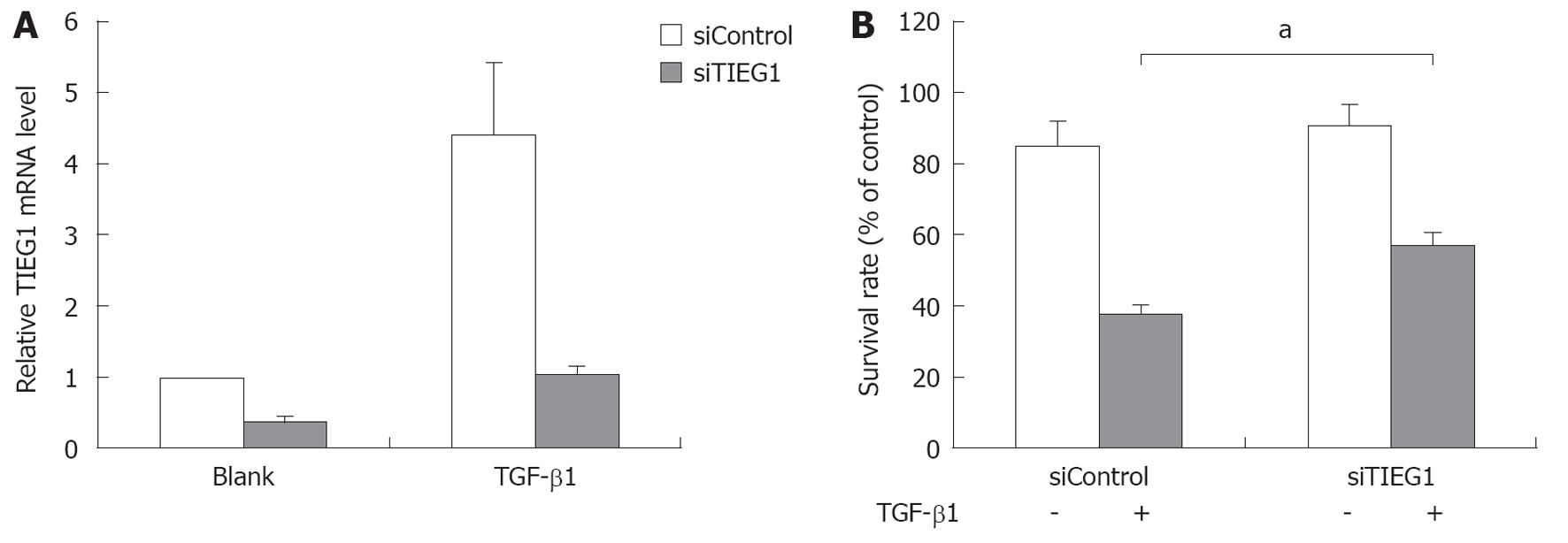
To study the role of TIEG1 in TGF-β induced growth inhibition in HCC cells, we used siRNA to target TIEG1 in the TGF-β-sensitive hepatoma cell line Hep3B. The siRNA targeting significantly decreased the mRNA expression of TIEG1 with or without TGF-β1 treatment (Figure 3A), and consequently increased the survival rate of the cells after treatment with TGF-β1 for 72 h (Figure 3B).
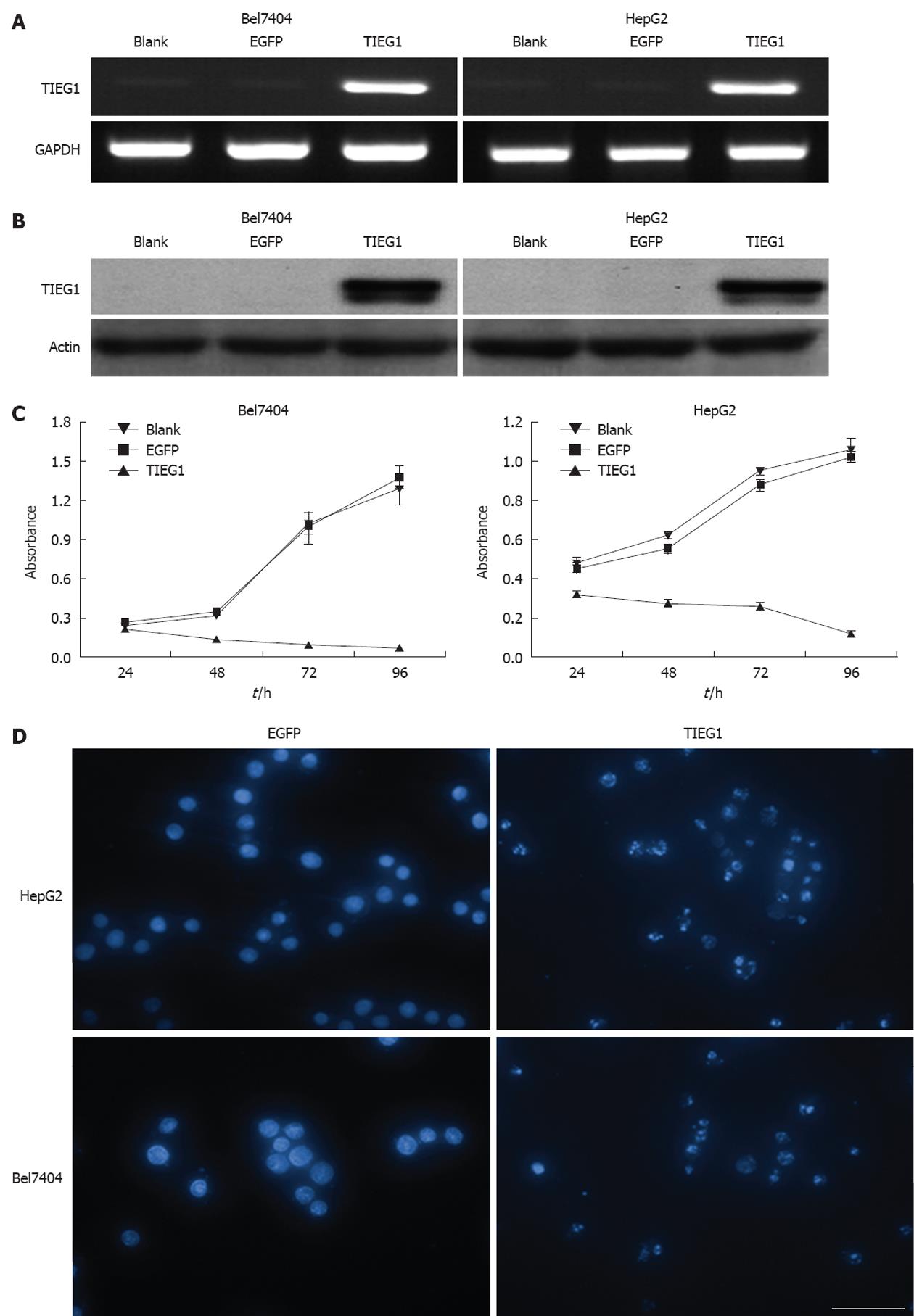
As seen in Figure 4, the overexpression of TIEG1 was successfully induced by lentiviral-mediated transduction in the TGF-β1-resistant hepatoma cell lines (Bel7404 and HepG2) both at transcription (Figure 4A) and translational levels (Figure 4B). MTT assay revealed a very significant inhibitory effect on growth of the two cell lines after the induction of TIEG1 by the lentivirus (Figure 4C). DAPI staining demonstrated a significantly higher amount of apoptotic cells in the two HCC cell lines after the overexpression of TIEG1 (Figure 4D).
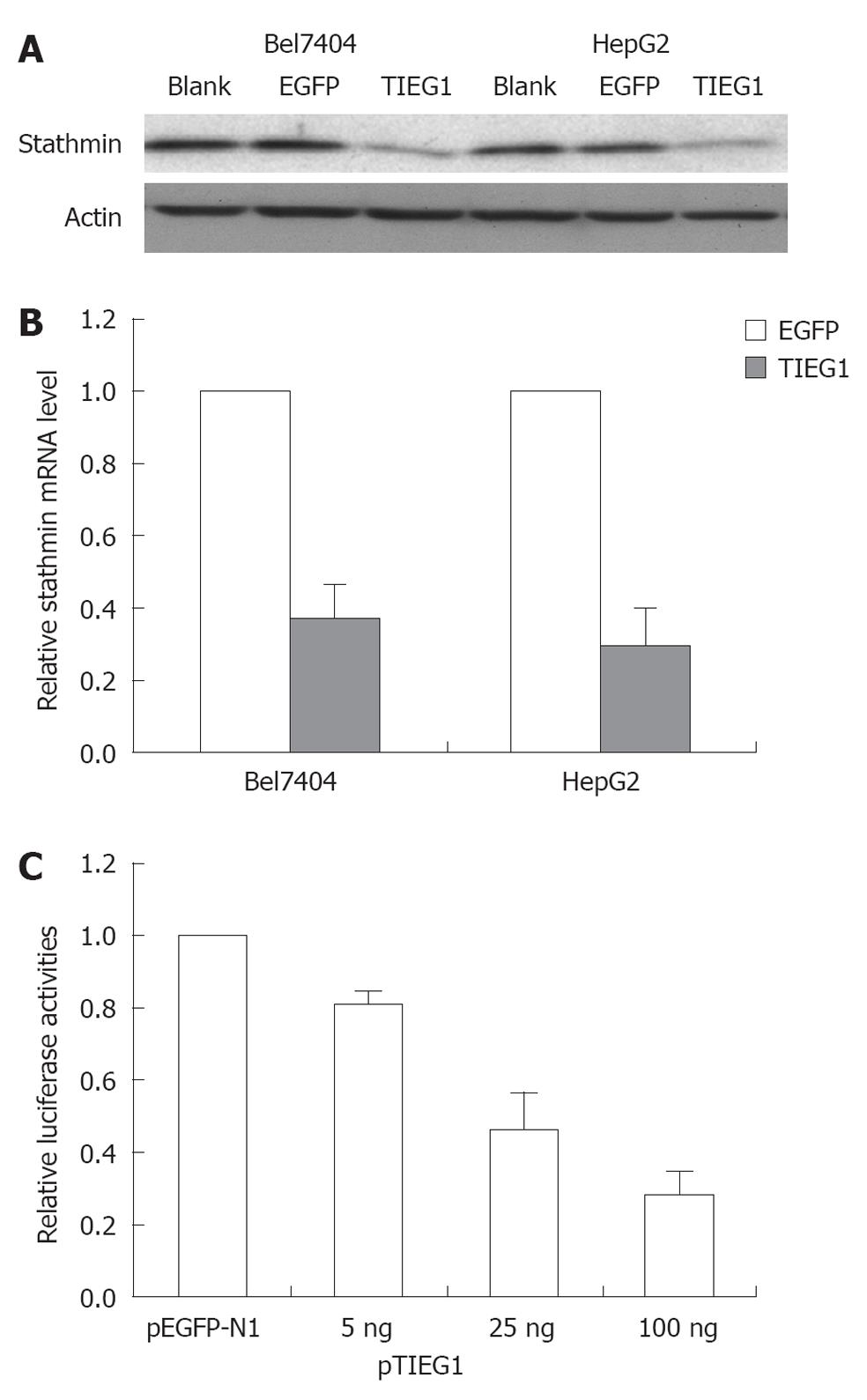
In the present study, the correlation between TIEG1 and STMN was also evaluated. Overexpression of TIEG1 was found to decrease STMN expression at both the transcription and translational levels (Figure 5A and B). STMN promoter activity was reduced in a dose-dependent manner with the induction of TIEG1 gene (Figure 5C).
The present study determined the molecular mechanism underlying the differential susceptibilities of HCC cells to TGF-β treatment. In this study, one TGF-β-sensitive hepatocyte cell line (MIHA) and five TGF-β-various-sensitive hepatoma cell lines were investigated. In general, treatment with TGF-β1 significantly inhibited the growth of the two TGF-β-sensitive cell lines (Hep3B and MIHA). However, HepG2 and Bel7404 cells did not respond to TGF-β1. Taken together, treatment with TGF-β1 caused varied levels of inhibition in different cell lines. Some HCC cell lines were sensitive to TGF-β, whereas others were resistant. Recently, studies on the relationship between HCC and the TGF-β signaling pathway have been extensive. One recent study has reported a negative relationship between interleukin-6, a major stem cell signaling pathway, and the TGF-β signaling pathway in human HCC[18]. TGF-β signaling and Smad adaptor embryonic liver fodrin could suppress HCC via cyclin D1 deregulation[8]. Another study showed that human HCC cells could be protected by interleukin-4 from TGF-β-induced apoptosis, which indicated another therapeutic option by the targeting of interleukin-4[19]. Lost sensitivity to TGF-β has been postulated to be an early event in HCC development[18,20].
In the present study, the role of TIEG1 in TGF-β-induced growth inhibition was analyzed in established TGF-β-sensitive and -insensitive cell systems. Our studies revealed that TIEG1 mRNA was dramatically upregulated by TGF-β1 in TGF-β1-sensitive cell lines but not in resistant cell lines. However, expression of the endogenous TIEG1 protein was not detected in all cell lines (namely, Hep3B, MIHA, Bel7404, and HepG2) before and after the TGF-β1 treatment. One of the reasons for this could be that the amounts of endogenous TIEG1 protein in the cells were too low to be detected. We found that the induction of TIEG1 was transient and occurred before the phenomenon of cell growth inhibition. The time course for the induction of TIEG1 expression was similar to that found in human osteoblast cells[21] and pancreatic epithelial cells[22] following TGF-β1 treatment. Although TIEG1 induction was transient following TGF-β1 treatment, it might participate in the TGF-β1 signaling processes or amplify the TGF-β1 signaling events that inhibited cell growth.
The suppression of TIEG1 by siRNAs decreased the sensitivity of Hep3B cells to TGF-β1, whereas the overexpression of TIEG1 mediated growth inhibition and apoptosis in TGF-β1-resistant HCC cell lines (HepG2 and Bel7404), which resembled those of TGF-β1-sensitive HCC cells treated with TGF-β1. This indicated the pivotal role of TIEG1 in TGF-β1-induced growth inhibition in HCC. In a previous study, the overexpression of TIEG1 was shown to inhibit cell proliferation and growth in TGF-β-sensitive Hep3B cells[15] and in pancreatic carcinoma cell lines[13,22]. Also, TIEG1 plays a role in TGF-β-induced inhibition of cell proliferation and apoptosis in human osteoblast cells[12]. Nevertheless, our data indicated that TIEG1 overexpression alone was capable of inducing sufficient inhibition or apoptosis in HCC tumor cells, despite the cells’ sensitivity to TGF-β. We also found that TIEG1 was identified as a transcriptional repressor of STMN, as the mRNA expression and the promoter activity of STMN were significantly reduced in the presence of overexpressed TIEG1. Bioinformatics analysis revealed several Sp1-binding sites in the promoter region of STMN. TIEG1 regulates STMN transcription by binding to the STMN promoter. These findings indicate the pivotal role of STMN in promoting tumor cell survival which has also been reported elsewhere[23-25]. Various strategies have been suggested to target the expression of STMN for treating tumors, including prostate, cervical[26] and breast cancer[27]. In pancreatic carcinoma cells, we have revealed that overexpression of TIEG1 could induce cell growth inhibition and promote gemcitabine chemosensitivity through downregulation of STMN[13]. In this study, the data again suggest a pivotal role for STMN (i.e., downregulation) in diminishing HCC proliferation, or in facilitating tumor cell apoptosis.
Taken together, these results demonstrate that TIEG1 is involved in TGF-β1-mediated growth inhibition. Transactivation of TIEG1 conferred growth inhibition of TGF-β-susceptible human HCC cells. However, it should be noted that this study was based on in vitro investigations and in vivo models should be explored.
Transforming growth factor-β (TGF-β) has been shown to inhibit cell proliferation and to induce apoptosis to control excessive growth of hepatocytes and maintain liver size, and is considered a liver tumor suppressor. However, many hepatocellular carcinoma (HCC) cells are thought to have lost their sensitivity to TGF-β, and thus escape the antiproliferative effect of TGF-β. Lost sensitivity to TGF-β has been postulated to be an early event in HCC development. The reasons why some HCC cells are sensitive yet others are resistant to TGF-β mediated growth inhibition are still poorly understood.
The TGF-β-inducible early gene 1 (TIEG1) can be activated at the initial stage of the TGF-β pathway. Recent reports have highlighted the importance of TIEG1 in the TGF-β signaling pathway and in regulating cell growth. In the present study, the authors demonstrated the role of the TIEG1 gene in TGF-β-induced growth inhibition in HCC cells.
This study indicated the TIEG1 was significantly upregulated by TGF-β1 in the TGF-β1-sensitive HCC cell line, Hep3B, but not in the resistant cell lines. The suppression of TIEG1 by siRNAs decreased the sensitivity of Hep3B cells to TGF-β1, whereas the overexpression of TIEG1 mediated growth inhibition and apoptosis in TGF-β1-resistant HCC cell lines, which resembled those of TGF-β1-sensitive HCC cells treated with TGF-β1. The studies suggest that transactivation of TIEG1 conferred growth inhibition of TGF-β-susceptible human HCC cells.
By understanding the molecular mechanism underlying the differential susceptibility of HCC cells to TGF-β, this study may provide new molecular targets for therapeutic intervention in HCC.
This paper deals with the transactivation of the TIEG1 in growth inhibition of TGF-β-susceptible HCC cells. The authors aimed to investigate the role of TIEG1 in TGF-β-induced growth inhibition in HCC. They found that transactivation of the TIEG1 conferred growth inhibition of the TGF-β-susceptible human HCC cells. The results are interesting and the design of this study is appropriate.
Peer reviewers: Arezoo Aghakhani, MD, PhD, Assistant Professor, Clinical Research Department Pasteur Institute of Iran, No 69, Pasteur Ave., Tehran 13164, Iran; Dr. Andreas Hillenbrand, Department of General Surgery, University of Ulm, 89075 Ulm, Germany
S- Editor Gou SX L- Editor Webster JR E- Editor Zheng XM
| 1. | Lin L, Amin R, Gallicano GI, Glasgow E, Jogunoori W, Jessup JM, Zasloff M, Marshall JL, Shetty K, Johnson L. The STAT3 inhibitor NSC 74859 is effective in hepatocellular cancers with disrupted TGF-beta signaling. Oncogene. 2009;28:961-972. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 181] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 2. | Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47:2059-2067. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 276] [Cited by in RCA: 283] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 3. | Massagué J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1828] [Cited by in RCA: 1865] [Article Influence: 74.6] [Reference Citation Analysis (0)] |
| 4. | Caja L, Sancho P, Bertran E, Iglesias-Serret D, Gil J, Fabregat I. Overactivation of the MEK/ERK pathway in liver tumor cells confers resistance to TGF-{beta}-induced cell death through impairing up-regulation of the NADPH oxidase NOX4. Cancer Res. 2009;69:7595-7602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 91] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 5. | Sohn BH, Park IY, Lee JJ, Yang SJ, Jang YJ, Park KC, Kim DJ, Lee DC, Sohn HA, Kim TW. Functional switching of TGF-beta1 signaling in liver cancer via epigenetic modulation of a single CpG site in TTP promoter. Gastroenterology. 2010;138:1898-1908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 80] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 6. | Dooley S, Weng H, Mertens PR. Hypotheses on the role of transforming growth factor-beta in the onset and progression of hepatocellular carcinoma. Dig Dis. 2009;27:93-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 7. | Wakefield LM, Roberts AB. TGF-beta signaling: positive and negative effects on tumorigenesis. Curr Opin Genet Dev. 2002;12:22-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 650] [Cited by in RCA: 633] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 8. | Kitisin K, Ganesan N, Tang Y, Jogunoori W, Volpe EA, Kim SS, Katuri V, Kallakury B, Pishvaian M, Albanese C. Disruption of transforming growth factor-beta signaling through beta-spectrin ELF leads to hepatocellular cancer through cyclin D1 activation. Oncogene. 2007;26:7103-7110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 100] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 9. | Senturk S, Mumcuoglu M, Gursoy-Yuzugullu O, Cingoz B, Akcali KC, Ozturk M. Transforming growth factor-beta induces senescence in hepatocellular carcinoma cells and inhibits tumor growth. Hepatology. 2010;52:966-974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 183] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 10. | Hjelmeland AB, Hjelmeland MD, Shi Q, Hart JL, Bigner DD, Wang XF, Kontos CD, Rich JN. Loss of phosphatase and tensin homologue increases transforming growth factor beta-mediated invasion with enhanced SMAD3 transcriptional activity. Cancer Res. 2005;65:11276-11281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Blok LJ, Grossmann ME, Perry JE, Tindall DJ. Characterization of an early growth response gene, which encodes a zinc finger transcription factor, potentially involved in cell cycle regulation. Mol Endocrinol. 1995;9:1610-1620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 12. | Subramaniam M, Hawse JR, Johnsen SA, Spelsberg TC. Role of TIEG1 in biological processes and disease states. J Cell Biochem. 2007;102:539-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 13. | Jiang L, Chen Y, Chan CY, Wang X, Lin L, He ML, Lin MC, Yew DT, Sung JJ, Li JC. Down-regulation of stathmin is required for TGF-beta inducible early gene 1 induced growth inhibition of pancreatic cancer cells. Cancer Lett. 2009;274:101-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 14. | Chalaux E, López-Rovira T, Rosa JL, Pons G, Boxer LM, Bartrons R, Ventura F. A zinc-finger transcription factor induced by TGF-beta promotes apoptotic cell death in epithelial Mv1Lu cells. FEBS Lett. 1999;457:478-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 86] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 15. | Ribeiro A, Bronk SF, Roberts PJ, Urrutia R, Gores GJ. The transforming growth factor beta(1)-inducible transcription factor TIEG1, mediates apoptosis through oxidative stress. Hepatology. 1999;30:1490-1497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 130] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 16. | Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2875] [Cited by in RCA: 2918] [Article Influence: 104.2] [Reference Citation Analysis (0)] |
| 17. | Nakao A, Afrakhte M, Morén A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1433] [Cited by in RCA: 1474] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 18. | Tang Y, Kitisin K, Jogunoori W, Li C, Deng CX, Mueller SC, Ressom HW, Rashid A, He AR, Mendelson JS. Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc Natl Acad Sci USA. 2008;105:2445-2450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 254] [Cited by in RCA: 266] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 19. | Lin SJ, Chang C, Ng AK, Wang SH, Li JJ, Hu CP. Prevention of TGF-beta-induced apoptosis by interlukin-4 through Akt activation and p70S6K survival signaling pathways. Apoptosis. 2007;12:1659-1670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Ding W, Mouzaki M, You H, Laird JC, Mato J, Lu SC, Rountree CB. CD133+ liver cancer stem cells from methionine adenosyl transferase 1A-deficient mice demonstrate resistance to transforming growth factor (TGF)-beta-induced apoptosis. Hepatology. 2009;49:1277-1286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 21. | Hefferan TE, Reinholz GG, Rickard DJ, Johnsen SA, Waters KM, Subramaniam M, Spelsberg TC. Overexpression of a nuclear protein, TIEG, mimics transforming growth factor-beta action in human osteoblast cells. J Biol Chem. 2000;275:20255-20259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 22. | Tachibana I, Imoto M, Adjei PN, Gores GJ, Subramaniam M, Spelsberg TC, Urrutia R. Overexpression of the TGFbeta-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J Clin Invest. 1997;99:2365-2374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 188] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 23. | Yuan RH, Jeng YM, Chen HL, Lai PL, Pan HW, Hsieh FJ, Lin CY, Lee PH, Hsu HC. Stathmin overexpression cooperates with p53 mutation and osteopontin overexpression, and is associated with tumour progression, early recurrence, and poor prognosis in hepatocellular carcinoma. J Pathol. 2006;209:549-558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 126] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 24. | Baldassarre G, Belletti B, Nicoloso MS, Schiappacassi M, Vecchione A, Spessotto P, Morrione A, Canzonieri V, Colombatti A. p27(Kip1)-stathmin interaction influences sarcoma cell migration and invasion. Cancer Cell. 2005;7:51-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 231] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 25. | Singer S, Ehemann V, Brauckhoff A, Keith M, Vreden S, Schirmacher P, Breuhahn K. Protumorigenic overexpression of stathmin/Op18 by gain-of-function mutation in p53 in human hepatocarcinogenesis. Hepatology. 2007;46:759-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 96] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 26. | Mistry SJ, Bank A, Atweh GF. Targeting stathmin in prostate cancer. Mol Cancer Ther. 2005;4:1821-1829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Alli E, Yang JM, Hait WN. Silencing of stathmin induces tumor-suppressor function in breast cancer cell lines harboring mutant p53. Oncogene. 2007;26:1003-1012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 82] [Article Influence: 4.3] [Reference Citation Analysis (0)] |