Published online Jan 7, 2012. doi: 10.3748/wjg.v18.i1.70
Revised: July 11, 2011
Accepted: July 18, 2011
Published online: January 7, 2012
AIM: To explore germline hypermethylation of the tumor suppressor genes MLH1, CDH1 and P16INK4a in suspected cases of hereditary gastric cancer (GC).
METHODS: A group of 140 Chinese GC patients in whom the primary cancer had developed before the age of 60 or who had a familial history of cancer were screened for germline hypermethylation of the MLH1, CDH1 and P16INK4a tumor suppressor genes. Genomic DNA was extracted from peripheral blood leukocytes and modified by sodium bisulfite. The treated DNA was then subjected to bisulfite DNA sequencing for a specific region of the MLH1 promoter. The methylation status of CDH1 or P16INK4a was assayed using methylation-specific PCR. Clonal bisulfite allelic sequencing in positive samples was performed to obtain a comprehensive analysis of the CpG island methylation status of these promoter regions.
RESULTS: Methylation of the MLH1 gene promoter was detected in the peripheral blood DNA of only 1/140 (0.7%) of the GC patient group. However, this methylation pattern was mosaic rather than the allelic pattern which has previously been reported for MLH1 in hereditary non-polyposis colorectal cancer (HNPCC) patients. We found that 10% of the MLH1 alleles in the peripheral blood DNA of this patient were methylated, consistent with 20% of cells having one methylated allele. No germline promoter methylation of the CDH1 or P16INK4a genes was detected.
CONCLUSION: Mosaic germline epimutation of the MLH1 gene is present in suspected hereditary GC patients in China but at a very low level. Germline epimutation of the CDH1 or P16INK4a gene is not a frequent event.
- Citation: Wu PY, Zhang Z, Wang JM, Guo WW, Xiao N, He Q, Wang YP, Fan YM. Germline promoter hypermethylation of tumor suppressor genes in gastric cancer. World J Gastroenterol 2012; 18(1): 70-78
- URL: https://www.wjgnet.com/1007-9327/full/v18/i1/70.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i1.70
Gastric cancer (GC) is one of the most common malignancies worldwide and is the most frequent form of cancer in East Asian countries. Point mutations or deletions within large genomic segments of tumor suppressor genes have previously been detected in about 30% of individuals with a genetic predisposition to GC[1,2]. However, the underlying genetic abnormalities in more than 70% of GC patients thus remain unknown. The methylation of cytosines in CpG dinucleotides within the promoter regions of tumor suppressor genes can result in transcriptional silencing and this occurs frequently in GC tumors. As one of the major genes in the mismatch repair (MMR) system, MLH1 is considered to be one of the key causative genes for GC. Hypermethylation of the MLH1 gene promoter is extremely frequent and often correlates well with loss of the MLH1 protein in tumor cells[3-6]. The epithelial cadherin gene CDH1 is also associated with GC and shows decreased expression in GC tumors. However, germline mutations in CDH1 have been found in only some patients with hereditary diffuse gastric cancer (HDGC). A high rate of promoter methylation of CDH1 is observed in GC tumors and is associated with the inactivation of this gene[7-11]. As an important protein in the cell cycle regulatory pathways, the inactivation of P16INK4a is one of the most commonly observed abnormalities in human cancer. This can occur via the hypermethylation of CpG islands within its promoter in many tumor types including gastric cancers and germline mutations in this gene in GC are relatively rare[10,11].
Recent reports have shown that promoter hypermethylation of the MLH1 gene is not limited to tumor cells but might also occur in the peripheral blood and other tissues of patients with early-onset colorectal cancer (CRC), suggesting a germline origin[12-18]. Importantly however, although the promoters of MLH1, CDH1 and P16INK4a are hypermethylated in GC tumors, it is currently not known whether these epigenetic events exist in the germline and cause a GC predisposition in affected individuals.
In our current study, we characterize the germline promoter methylation of the MLH1, CDH1 and P16INK4a tumor suppressor genes in GC. We screened a selected cohort of Chinese GC cases in whom the cancer had developed before the age of 60 years or in which there was familial history of MLH1, CDH1 and P16INK4a germline epimutations.
A cohort of 140 GC patients from the Jiangsu, Anhui and Zhejiang provinces of China was assembled for this study. The selection of patients was based on the GC family history and onset ages i.e., (1) individuals with GC and two or more first-degree relatives with GC, denoted as high familial cancer history (HF) cases; (2) individuals with GC and one first- or second-degree relative with GC, designated low familial cancer history (LF) cases; and (3) individuals diagnosed with GC prior to 60 years of age, but no family history of this disease, referred to as young onset (Y) patients. Of the 140 GC patients in our selected cohort, we identified 18 HF, 43 LF and 79 Y cases. Thirty age-matched normal controls were also included in the analysis. Informed consent was obtained from each subject who underwent genetic testing, in accordance with the guidelines of the Ethics Committee of the Medical School of Nanjing University.
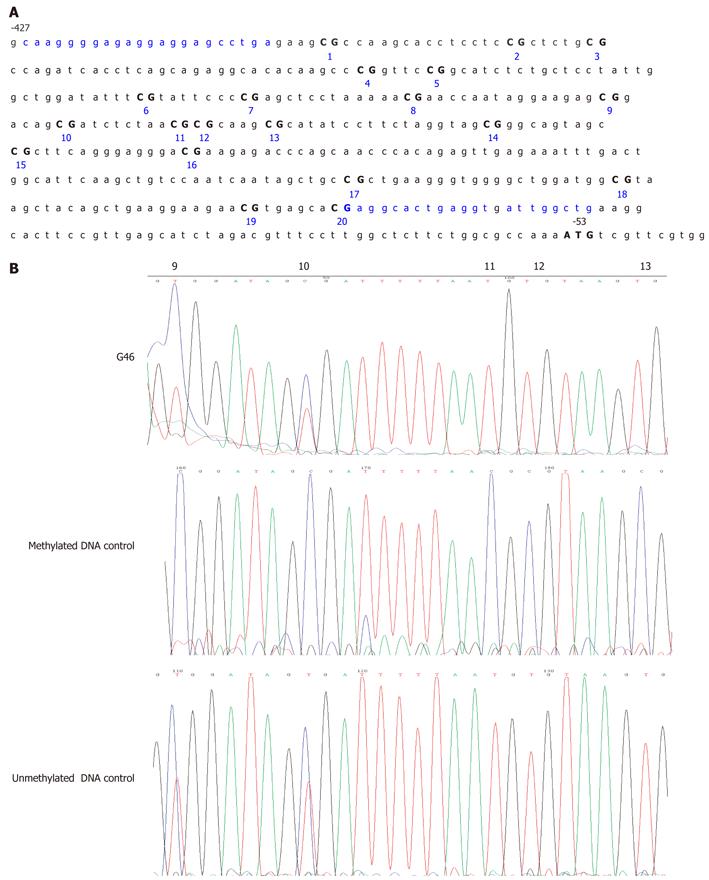
Peripheral blood leukocyte DNA was bisulfite modified using the CpGenome™ DNA Modification Kit (Chemicon International, Temecula, CA, United States) in accordance with the manufacturer’s instructions. A specific CpG-rich sequence in the MLH1 promoter region (from -427 to -53 bp relative to the translation start site for human MLH1, a 375 bp fragment containing 20 CpG sites; Figure 1A) was selected. This region is purported to be strongly associated with MLH1 silencing[19-22]. Bisulfite DNA sequencing (BSP) of this region was performed to determine the comprehensive CpG island methylation status of the MLH1 gene promoter using an ABI 3100-Avant automated sequencer (Applied Biosystems, Foster City, CA, United States). The primers used for BSP were MLH1-BF, 5’- TAAGGGGAGAGGAGGAGTTTGA-3’ (sense) and MLH1-BR, 5’-CAACCAATCACCTCAATACCTC-3’ (antisense). The obtained PCR products displaying methylated CpG sites were subcloned into the PMD18-T vector [TaKaRa Biotechnology (Dalian) Co., Ltd., Dalian, China] and at least 10 clones were selected and sequenced for each sample using universal M13 primers to determine the level and extent of gene promoter methylation.
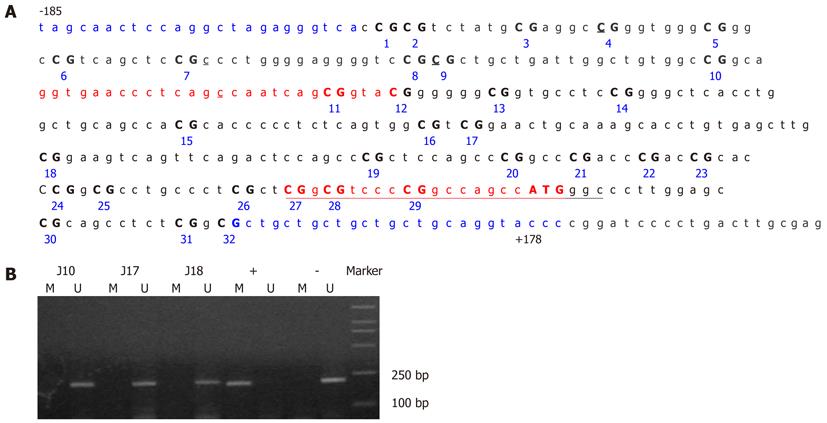
The methylation status of CDH1 and P16 was determined by methylation-specific PCR (MSP) after treatment of the DNA with sodium bisulfite. The primer sequences used have been reported previously for CDH1[8,11] and P16[7,23] and are as follows: CDH1-M-F, 5’-GGTGAATTTTTAGTTAATTAGCGGTAC-3’ (sense) and CDH1-M-R, 5’-CATAACTAACCGAAAACGCCG-3’ (antisense) for methylated CDH1; CDH1-U-F, 5’-GGTAGGTGAATTTTTAGTTAATTAGTGGTA-3’ (sense); and CDH1-U-R, 5’-ACCCATAACTAACCAAAAACACCA-3’ (antisense) for unmethylated CDH1; P16-M-F, 5’-TTATTAGAGGGTGGGGCGGATCGC-3’ (sense) and P16-M-R, 5’-GACCCCGAACCGCGACCGTAA-3’ (antisense) for methylated P16; P16-U-F, 5’-TTATTAGAGGGTGGGGTGGATTGT-3’ (sense), and P16-U-R, 5’- CAACCCCAAACCACAACCATAA-3’ (antisense) for unmethylated P16. The PCR-amplified regions for the methylated (from -78 to +127 bp relative to the transcription initiation site for human CDH1, 205 bp) and unmethylated (from -78 to +130 bp relative to the transcription initiation site, 208 bp) CDH1 alleles contained 19 CpG dinucleotides, including five CpGs at the primer annealing sites (Figure 3A). The primer sets spanned the transcriptional start site and were designed to include methylation sites that best corresponded with the transcriptional silencing of CDH1 in the published literature[24].
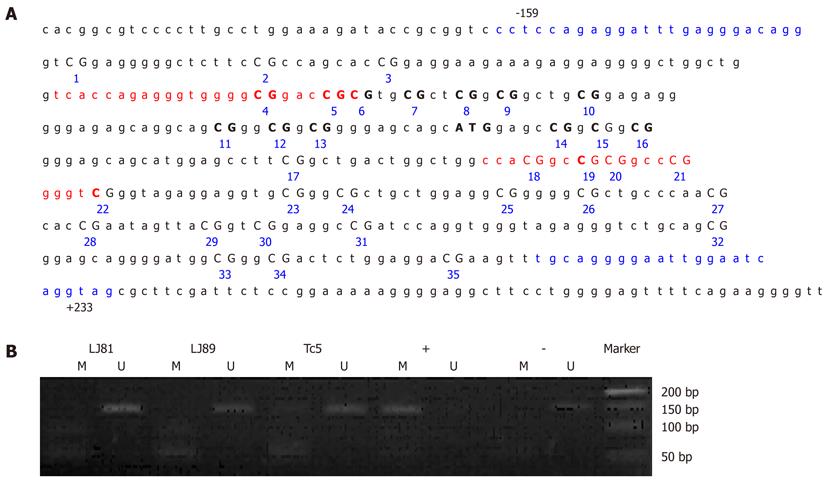
The PCR-amplified region for the methylated (from -80 to +70 bp relative to the translation initiation site for human P16, 150 bp) and unmethylated (from -80 to +71 bp relative to the translation initiation site, 151 bp) p16 alleles contained 19 CpG dinucleotides, including eight CpGs at the primer annealing sites (Figure 6A).
The positive samples were further amplified using CDH1 or p16 gene-specific primers CDH1-BF-5’-TAGTAATTTTAGGTTAGAGGGTTA-3’ (sense) and CDH1-BR-5’-AAATACCTACAACAACAACAACAAC-3’ (anti-sense)[25]; P16-BF-5’-TTTTTAGAGGATTTGAGGGATAGG-3’ (sense) and P16-BR-5’-CTACCTAATTCCAATTCCCCTACA-3’ (anti-sense)[19]. The amplified fragments (363 bp, from -185 to +178 bp relative to the transcription initiation site of CDH1, Figure 3A; 392 bp, from -159 to +233 bp relative to the translation initiation site of P16, Figure 6A), were sequenced using an ABI 3100-Avant automated sequencer (Applied Biosystems, Foster City, CA, United States). CpGenome universal methylated and unmethylated DNA (Chemicon International, Temecula, CA, United States) served as a positive control for gene promoter hypermethylation and hypomethylation, respectively.
A stretch of 375 bases incorporating 20 CpG sites in the MLH1 proximal promoter was analyzed (Figure 1A). In one of the 140 suspected hereditary GC patients in our cohort (patient G46), partial methylation was detected by BSP through the 6th to 20th CpG sites of the MLH1 promoter from blood DNA (Figure 1B). This individual was a 60-year-old female with no family history of GC. For the remaining samples, partial methylation was evident at the 9th and 10th CpG sites (-269 and -262 bp from the translation initiation site, respectively), but no other CpGs in this region were found to be methylated. The peripheral blood leukocyte DNA of 30 age-matched normal controls was examined for MLH1 methylation in comparison with the GC patients. None of the control samples showed detectable MLH1 promoter methylation by bisulfite DNA sequencing apart from the 9th and 10th CpG sites which seemed to have methylated alleles more than 50% for all the patients. However, because this alteration was also found in unmethylated controls and in healthy blood donor samples, it is far less likely to be linked to GC (Figure 1B).
To determine the extent of MLH1 allele methylation in patient G46, clonal bisulfite allelic sequencing of the MLH1 promoter in peripheral blood from this individual was performed. Upon analysis of the peripheral blood DNA from patient G46, one of ten clones had a cytosine present through the 6th to 20th CpG sites (Figure 2). All other non-CpG cytosines were converted, indicating that this corresponded to a methylated allele. For one other clone, only one CpG (the 12th) was found to be methylated and this may have been due to an incomplete conversion. Four of these ten clones had a cytosine present at the 9th and 10th CpGs, confirming the methylation pattern evidenced by bisulfite DNA sequencing. This result supported the possibility that mosaic germline MLH1 methylation could be occurring in this patient. Indeed, 10% of the MLH1 allele was methylated in this individual, equating to 20% of the cells found with one methylated MLH1 allele in the peripheral blood. Unfortunately, tumor material was not available from patient G46 to evaluate the methylation profile of the MLH1 gene in the GC cancer cells.
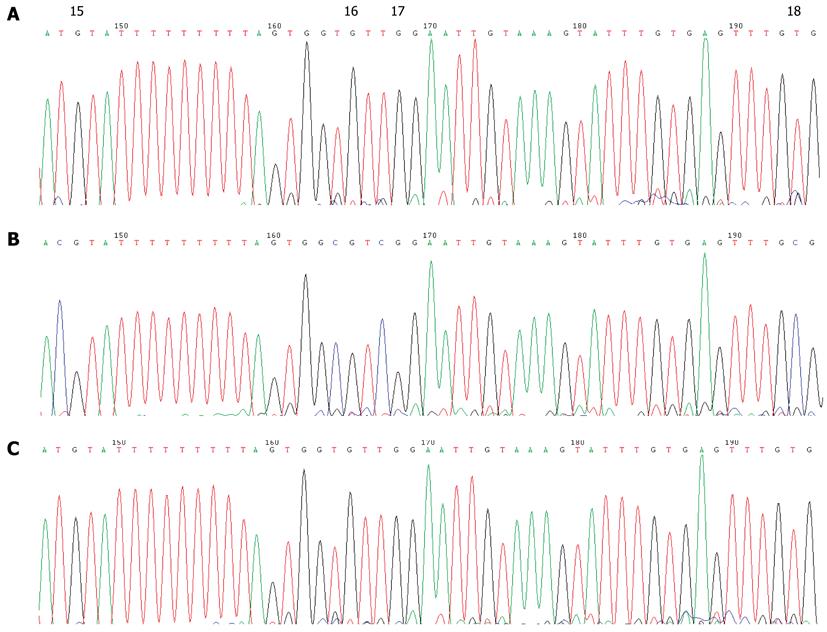
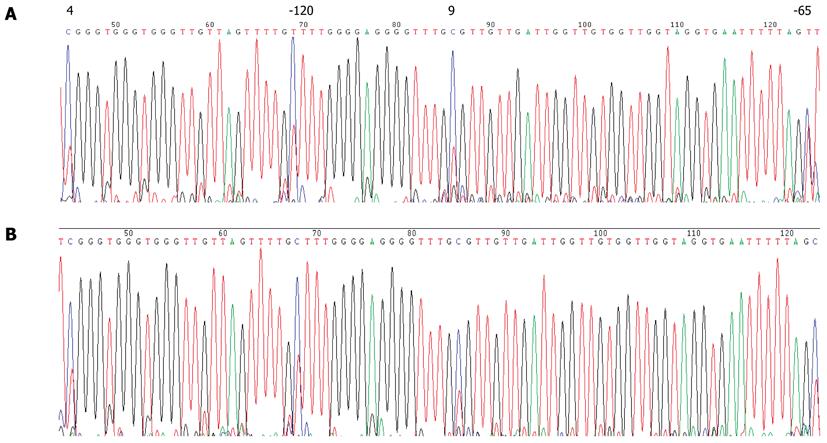
None of the GC cases under study showed detectable CDH1 promoter methylation in their peripheral blood leukocytes by MSP assay (Figure 3B). Direct sequencing of the CDH1 promoter region from -185 to +178 bp relative to the transcription initiation site confirmed this result (Figure 4). Several C bases (including the 4th and 9th CpG sites, and at positions -120 and -65 bp relative to the transcription initiation site) were evident (more than 50%) in all patients. However, because this variation was also found in the healthy blood donor samples, it is unlikely to be linked to GC (Figure 5). This result also suggested that the incidence of germline hypermethylation of the CDH1 promoter is low in Chinese GC patients.
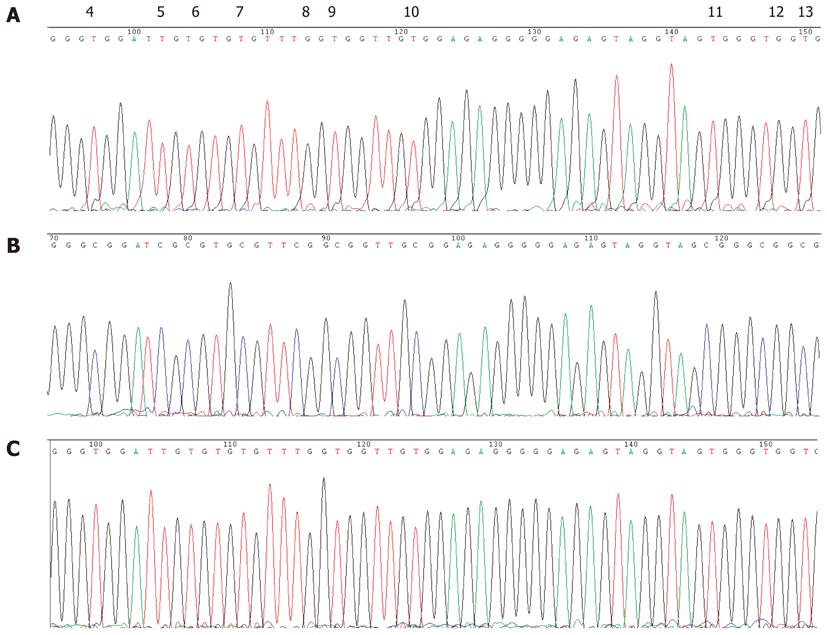
None of the GC cases under study showed detectable P16 promoter methylation of the DNA from their peripheral blood leukocytes by MSP assay (Figure 6B). Direct sequencing of the P16 promoter region from -159 to +233 bp relative to the translation initiation site confirmed that all of the 35 CpG dinucleotides present in the fragment were unmethylated (Figure 7). This result suggested that germline hypermethylation of the P16 promoter is also rare in Chinese GC patients.
In our current study, we demonstrated that a mosaic MLH1 promoter methylation pattern existed in the peripheral blood of a Chinese GC patient. Deng et al[19] previously divided the MLH1 promoter area into four regions, A–D, and proposed that methylation in region C plays an important role in silencing hMLH1 expression. In our current experiments, we carried out methylation analysis on region C of the MLH1 promoter. We excluded the 9th and 10th CpG sites of the MLH1 promoter region as they were found to be methylated in the control samples (Figure 1B). Our analyses also showed that rather than an allelic methylation pattern as has previously been reported for MLH1 in CRC patients, a mosaic level of MLH1 methylation was found from the 6th to 20th CpG sites in this promoter region in the germline genomic DNA of GC patient G46 in our study cohort. Clonal sequencing further revealed a 10% methylation level at each of these CpG sites. Taken together, our data suggest that a mosaic germline MLH1 epimutation might be present in some patients who develop gastric cancer. This mosaic methylation of MLH1 might not be due to disseminated GC cells in the blood because these do not occur at sufficiently high levels to be detected by our assay. In patient G46, aberrant DNA methylation may have occurred after fertilization, when the maternal CpG methylation pattern is established[26]. It is also possible that CpG methylation is not always faithfully replicated during early embryogenesis, and the degree of mosaicism might be influenced by the genetic or epigenetic background. Our results raise the further possibility that silencing of MLH1 by promoter methylation could occur as a germline or an early somatic event that generates a predisposition to GC.
We did not detect germline CDH1 or P16 promoter CpG methylation in any of the 140 GC patients investigated (Figures 3, 4, 5, 6 and 7), suggesting that germline hypermethylation in the CDH1 or P16 promoter is not a common mechanism of CDH1 or P16 inactivation leading to GC. A detailed inspection of the bisulfate transformed sequence corresponding to the promoter region of CDH1 revealed two methylated CpGs (the 4th and 9th), and also two methylated Cs at positions -120 and -65 bp relative to the transcription start site of the CDH1 gene. However, given that this methylation was found in all of the patients and normal controls, it is unlikely to be associated with GC.
In summary, we report here that germline mosaic hypermethylation of the CpG islands in the MLH1 promoter region may be the underlying cause of some gastric cancers, while germline hypermethylation of CDH1 or P16 gene is not a common mechanism responsible for GC in familial patients.
Gastric cancer is considered to be one of the leading cancers in East Asia but the underlying genetic abnormalities that lead to more than 70% of these cases remain unknown.
Epigenetic methylation-associated inactivation of genes is not limited to tumor cells. Monoallelic promoter hypermethylation of the MLH1 gene in the peripheral blood of patients with early-onset colorectal cancer has recently been reported in the literature. However, it is uncertain whether germline epimutations are responsible for gastric cancer onset in hereditary cases.
In this report, we provide a comprehensive description of germline epimutations of the tumor suppressor genes MLH1, CDH1 and P16INK4a in Chinese gastric cancer cases. We report the rare occurrence of germline mosaic hypermethylation of the CpG islands in the MLH1 promoter region in gastric cancer. We also demonstrate that germline epimutation of the CDH1 or P16INK4a gene is not a frequent occurrence in this disease.
The finding of human MLH1 gene germline epimutations is of some significance as it not only reveals a possible new mechanism in the tumorigenesis of gastric cancers, but also indicates that germline epimutations may be associated with a wide range of human diseases. On the other hand, germline epimutations show both mosaic and non-Mendelian characteristics, which may explain the phenotypic variability and changes in the genetic penetrance of some complex diseases, and thereby provide new avenues for studying the etiology of such disorders.
Germline epimutations not only arise in tumor cells but in nontumor cells of a germline origin.
The manuscript is well written and the methods are adequate. The results justify the conclusions drawn.
Peer reviewer: Mario M D’Elios, Professor, University of Florence, viale Morgagni 85, Florence 50134, Italy
S- Editor Sun H L- Editor Webster JR E- Editor Zhang DN
| 1. | Fan Y, Liu X, Zhang H, Dai J, Zhang X, Zhu M, Gao X, Wang Y. Variations in exon 7 of the MSH2 gene and susceptibility to gastrointestinal cancer in a Chinese population. Cancer Genet Cytogenet. 2006;170:121-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 2. | Zhang Y, Liu X, Fan Y, Ding J, Xu A, Zhou X, Hu X, Zhu M, Zhang X, Li S. Germline mutations and polymorphic variants in MMR, E-cadherin and MYH genes associated with familial gastric cancer in Jiangsu of China. Int J Cancer. 2006;119:2592-2596. [PubMed] |
| 3. | Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870-6875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1343] [Cited by in RCA: 1370] [Article Influence: 50.7] [Reference Citation Analysis (0)] |
| 4. | Fleisher AS, Esteller M, Wang S, Tamura G, Suzuki H, Yin J, Zou TT, Abraham JM, Kong D, Smolinski KN. Hypermethylation of the hMLH1 gene promoter in human gastric cancers with microsatellite instability. Cancer Res. 1999;59:1090-1095. [PubMed] |
| 5. | Fleisher AS, Esteller M, Tamura G, Rashid A, Stine OC, Yin J, Zou TT, Abraham JM, Kong D, Nishizuka S. Hypermethylation of the hMLH1 gene promoter is associated with microsatellite instability in early human gastric neoplasia. Oncogene. 2001;20:329-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 93] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 6. | Miyakura Y, Sugano K, Konishi F, Ichikawa A, Maekawa M, Shitoh K, Igarashi S, Kotake K, Koyama Y, Nagai H. Extensive methylation of hMLH1 promoter region predominates in proximal colon cancer with microsatellite instability. Gastroenterology. 2001;121:1300-1309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 144] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 7. | Toyota M, Ahuja N, Suzuki H, Itoh F, Ohe-Toyota M, Imai K, Baylin SB, Issa JP. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999;59:5438-5442. [PubMed] |
| 8. | Tamura G, Yin J, Wang S, Fleisher AS, Zou T, Abraham JM, Kong D, Smolinski KN, Wilson KT, James SP. E-Cadherin gene promoter hypermethylation in primary human gastric carcinomas. J Natl Cancer Inst. 2000;92:569-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 261] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 9. | Lee TL, Leung WK, Chan MW, Ng EK, Tong JH, Lo KW, Chung SC, Sung JJ, To KF. Detection of gene promoter hypermethylation in the tumor and serum of patients with gastric carcinoma. Clin Cancer Res. 2002;8:1761-1766. [PubMed] |
| 10. | Oue N, Motoshita J, Yokozaki H, Hayashi K, Tahara E, Taniyama K, Matsusaki K, Yasui W. Distinct promoter hypermethylation of p16INK4a, CDH1, and RAR-beta in intestinal, diffuse-adherent, and diffuse-scattered type gastric carcinomas. J Pathol. 2002;198:55-59. [PubMed] |
| 11. | Waki T, Tamura G, Tsuchiya T, Sato K, Nishizuka S, Motoyama T. Promoter methylation status of E-cadherin, hMLH1, and p16 genes in nonneoplastic gastric epithelia. Am J Pathol. 2002;161:399-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 114] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 12. | Gazzoli I, Loda M, Garber J, Syngal S, Kolodner RD. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res. 2002;62:3925-3928. [PubMed] |
| 13. | Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36:497-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 14. | Miyakura Y, Sugano K, Akasu T, Yoshida T, Maekawa M, Saitoh S, Sasaki H, Nomizu T, Konishi F, Fujita S. Extensive but hemiallelic methylation of the hMLH1 promoter region in early-onset sporadic colon cancers with microsatellite instability. Clin Gastroenterol Hepatol. 2004;2:147-156. [PubMed] |
| 15. | Hitchins M, Williams R, Cheong K, Halani N, Lin VA, Packham D, Ku S, Buckle A, Hawkins N, Burn J. MLH1 germline epimutations as a factor in hereditary nonpolyposis colorectal cancer. Gastroenterology. 2005;129:1392-1399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 125] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 16. | Valle L, Carbonell P, Fernandez V, Dotor AM, Sanz M, Benitez J, Urioste M. MLH1 germline epimutations in selected patients with early-onset non-polyposis colorectal cancer. Clin Genet. 2007;71:232-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 17. | Hitchins MP, Wong JJ, Suthers G, Suter CM, Martin DI, Hawkins NJ, Ward RL. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356:697-705. [PubMed] |
| 18. | Morak M, Schackert HK, Rahner N, Betz B, Ebert M, Walldorf C, Royer-Pokora B, Schulmann K, von Knebel-Doeberitz M, Dietmaier W. Further evidence for heritability of an epimutation in one of 12 cases with MLH1 promoter methylation in blood cells clinically displaying HNPCC. Eur J Hum Genet. 2008;16:804-811. [PubMed] |
| 19. | Deng G, Chen A, Hong J, Chae HS, Kim YS. Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res. 1999;59:2029-2033. [PubMed] |
| 20. | Deng G, Peng E, Gum J, Terdiman J, Sleisenger M, Kim YS. Methylation of hMLH1 promoter correlates with the gene silencing with a region-specific manner in colorectal cancer. Br J Cancer. 2002;86:574-579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 82] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 21. | Park SJ, Rashid A, Lee JH, Kim SG, Hamilton SR, Wu TT. Frequent CpG island methylation in serrated adenomas of the colorectum. Am J Pathol. 2003;162:815-822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 183] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 22. | Capel E, Fléjou JF, Hamelin R. Assessment of MLH1 promoter methylation in relation to gene expression requires specific analysis. Oncogene. 2007;26:7596-7600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 23. | Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4183] [Cited by in RCA: 4247] [Article Influence: 146.4] [Reference Citation Analysis (0)] |
| 24. | Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1307] [Cited by in RCA: 1210] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 25. | Leung WK, Man EP, Yu J, Go MY, To KF, Yamaoka Y, Cheng VY, Ng EK, Sung JJ. Effects of Helicobacter pylori eradication on methylation status of E-cadherin gene in noncancerous stomach. Clin Cancer Res. 2006;12:3216-3221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 98] [Article Influence: 5.2] [Reference Citation Analysis (0)] |