Published online Feb 14, 2011. doi: 10.3748/wjg.v17.i6.809
Revised: January 4, 2011
Accepted: January 11, 2011
Published online: February 14, 2011
AIM: To identify and assess mutations in the K-ras and BRAF genes in a cohort of Chinese patients with colorectal cancer (CRC) for their association with various clinicopathological parameters and prognosis.
METHODS: Genomic DNA was isolated from frozen tissues. Pyrosequencing analysis was conducted to detect mutations in the K-ras (codons 12, 13, and 61) and BRAF genes (codon 600). Statistical analysis was carried out using SPSS-15.0 software.
RESULTS: Among the 118 colorectal cancer patients, we detected 41 (34.7%) mutations in the K-ras gene. Mutation frequencies at codon 12 and codon 13 were 23.7% (28/118) and 10.2% (12/118), respectively. Only one patient harbored a point mutation at codon 61 (0.8%, 1/118). Gender was the only factor that showed an obvious relationship with K-ras gene mutation (female 44.7% vs male 28.2%, P = 0.037). Other clinicopathological features, such as age, location of the tumor, tumor differentiation, Tumor, Node and Metastases classification, and the Union for International Cancer Control staging, showed no positive relationship with K-ras gene mutations. No significant correlation was observed between the presence of K-ras mutations (codons 12, 13, and 61) and the survival of the patients. BRAF mutations were rare, and only two patients (1.7%) harbored a detectable mutation at codon 600.
CONCLUSION: K-ras gene mutation is a common event in our 118 Chinese CRC patients, with an obvious relationship with gender. However, it seems not to be an independent prognostic factor in CRC patients. The BRAF gene is rarely mutated in Chinese CRC patients.
- Citation: Shen H, Yuan Y, Hu HG, Zhong X, Ye XX, Li MD, Fang WJ, Zheng S. Clinical significance of K-ras and BRAF mutations in Chinese colorectal cancer patients. World J Gastroenterol 2011; 17(6): 809-816
- URL: https://www.wjgnet.com/1007-9327/full/v17/i6/809.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i6.809
Colorectal cancer (CRC) is one of the most common malignancies in the world. In recent years, the morbidity and mortality of colorectal cancer has risen in the Chinese population. The development of CRC is a multistep process, which can arise due to the cumulative effect of mutations in various proto-oncogenes, tumor suppressor genes, and also from epigenetic changes in DNA. Recent evidence suggests that the extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase signaling pathway, which mediates cellular responses to growth factors and regulates the elements of the cell cycle, apoptosis and differentiation[1], plays a critical role in the pathogenesis of colorectal cancer. Both the K-ras and BRAF genes encode proteins that act in the ERK signaling pathway. The K-ras proto-oncogene encodes a 21 kDa RAS protein, a member of a highly conserved family of GTPases involved in signal transduction processes. Mutations in the K-ras gene render the protein constitutively active in signaling by eliminating the GTPase activity. More recently, mutations in the K-ras gene have proved to be predictors of response to epidermal growth factor receptor-targeted therapies, such as cetuximab and panitumumab, for patients with metastatic colorectal cancer.
In human CRC, mutations in the K-ras gene are very frequent (20%-50%), whereas mutations of the BRAF gene, a downstream molecule of K-ras, occur in only 9%-11% of patients with sporadic diseases. Mutations in the K-ras and BRAF genes are frequently found to be mutually exclusive in colorectal cancer[2,3]. Both genes harbor the majority of mutations in distinct hotspots in the BRAF gene at codons 463-468[4] and 600[3,4], and in the K-ras gene at codons 12, 13[5], and, more infrequently, at codon 61[6]. Approximately 90% of the activating mutations in the K-ras gene are scored at codon 12 (wild-type: GGT) and codon 13 (wild-type: GGC) in exon 1, while only 5% are located at codon 61 (wild-type: CAA) in exon 2[5,7,8]. Some studies have been conducted about the relationship between K-ras gene mutation and various clinicopathological characteristics, but no consistent results were obtained. The prognostic significance of K-ras gene mutations is still controversial. As for the BRAF gene, very few papers regarding its prognostic significance are available in Western countries or China.
For the detection of mutations in the K-ras and BRAF genes, various techniques have been described, including temporal temperature gradient electrophoresis[9], denaturing gradient gel electrophoresis[10], restriction endonuclease-mediated selective polymerase chain reaction (PCR)[11], and direct sequencing of a PCR product[12]. For all non-sequencing methods, it is difficult to independently confirm the existence of any mutations that are identified. In addition, our previous work showed that direct sequencing of a PCR product was not a sensitive method for the detection of K-ras gene mutations[12]. Pyrosequencing has emerged as a sensitive and rapid sequencing method for single-nucleotide polymorphism (SNP)/mutation analysis, which overcomes the above limitations[13]. It is a non-electrophoretic, real-time sequencing technic, and a sequence-by-synthesis method that relies on the luminometric detection of pyrophosphate released upon nucleotide incorporation via a four-enzyme mixture reaction cascade[14,15]. It can analyze multiple samples in a short time, which makes it attractive for clinical use. So far, few papers have reported the use of pyrosequencing for the detection of K-ras gene mutations in colorectal cancer patients. Pyrosequencing studies on the BRAF gene are even fewer.
In this paper, we detected mutations in the K-ras and BRAF genes from 118 Chinese CRC patients using pyrosequencing. Correlations with various clinicopathological characteristics and the prognosis of patients were further analyzed.
Tumor specimens used in this study were obtained from 118 CRC patients who received a radical resection operation in the 2nd Affiliated Hospital of Zhejiang University College of Medicine from February 2001 to January 2005. All patients were followed up by the Cancer Research Institute until September 2010, and the data concerning cancer recurrence and patient survival were collected. Tumor stage was classified according to the 7th edition of the Tumor, Node and Metastases (TNM) classification of the Union for International Cancer Control (UICC) staging. The clinicopathological data of all patients are shown in Table 1. Data and tissue collection was approved by the Ethics Committee of Zhejiang University College of Medicine, following the ethical guidelines of the 1975 Declaration of Helsinki.
| Terms | n (%) |
| No. of patients | 118 |
| Median age (yr) | 61 |
| Gender | |
| Male | 71 (60.2) |
| Female | 47 (39.8) |
| Colorectal segment | |
| Cecum | 5 (4.2) |
| Ascending colon | 23 (19.5) |
| Transversal colon | 8 (6.8) |
| Descending colon | 5 (4.2) |
| Sigmoid | 25 (21.2) |
| Rectum | 52 (44.1) |
| UICC stage | |
| I | 18 (15.3) |
| II | 48 (40.7) |
| IIA | 32 (27.1) |
| IIB | 16 (13.6) |
| III | 37 (31.4) |
| IIIA | 5 (4.2) |
| IIIB | 25 (21.2) |
| IIIC | 7 (5.9) |
| IV | 15 (12.7) |
Tumor tissues were collected from the Zhejiang University Cancer Institute tissue bank. All the tissue samples were confirmed independently by two gastrointestinal pathologists. Genomic DNA was extracted with the QIAamp DNA Mini-Kit (QIAGEN, Mississauga, ON), according to the manufacturer’s recommendations. The primers for the amplification and pyrosequencing assay of K-ras and BRAF gene are listed in Table 2.
| Primer sequence | Product (bp) | |
| K-ras gene codon 12 & 13 (exon 1) | Forward: 5'-GCAGTCAACTGGAATTTTCATG-3' | 431 |
| Reverse: 5'-biotin-GAAACCCAAGGTACATTTCAGA-3' | ||
| Pyrosequencing assay: 5'-TGTGGTAGTTGGAGCT- 3' | ||
| K-ras gene codon 61 (exon 2) | Forward:5'-ATCCAGACTGTGTTTCTCCCTTC-3' | 378 |
| Reverse: 5'-biotin-ACTGCTCTAATCCCCCAAGAACT-3' | ||
| Pyrosequencing assay: 5'-TATTCACGACACAGCAGGT-3' | ||
| BRAF gene codon 600 (exon 15) | Forward: 5'-ACAAGCCTTCAAAAATGAAGTAG-3' | 362 |
| Reverse: 5'-biotin-ATCCAGACAACTGTTCAAACTGA-3' | ||
| Pyrosequencing assay: 5'-GGTGATTTTGGTCTAACTACA-3' |
PCR was performed using 100 ng genomic DNA as template. Each mixture contained 10 pmol of each primer. The reactions were performed in 1 × reaction buffer, 0.2 μmol/L dNTPs, 2 mmol/L MgCl2, and 1.25 U Blend Taq polymerase (ToYoBo) in a total volume of 50 μL. The amplification reactions were as follows: an initial denaturing cycle of 94°C for 2 min; 35 cycles of 94°C for 30 s, 55°C for 25 s, and 72°C for 30 s; and a final extension cycle at 72°C for 2 min.
The PCR products were directly subjected to the sequencing analysis using the pyrosequencing PyroMark ID system (PSQ 96 MA, Biotage AB, Sweden). For pyrosequencing, ssDNA was prepared from 40 μL biotinylated PCR product using streptavidin-coated sepharose, and 0.5 mmol/L sequencing primer was used for analysis (Table 2). Sequencing was performed with the SNP Reagent Kit (Biotage AB, Sweden) according to the manufacturer’s instructions.
The Mann-Whitney t test, the Kruskal-Wallis test, and Fisher’ test were used to evaluate the associations between the K-ras wild-type/mutation type and the clinicopathological variables of patients [Mann-Whitney t test for dichotomous variables (gender, metastasis vs no metastasis); Kruskal-Wallis test for no dichotomous variables (age, tumor region, differentiation and TNM stage). Kaplan-Meier survival analysis was performed to evaluate the relationship between K-ras wild-type/mutation type and survival of CRC patients. Calculations were carried out using the SPSS-15.0 software (SPSS Inc., Chicago, IL). P value less than 0.05 was regarded as statistically significant. All statistical tests were two-sided.
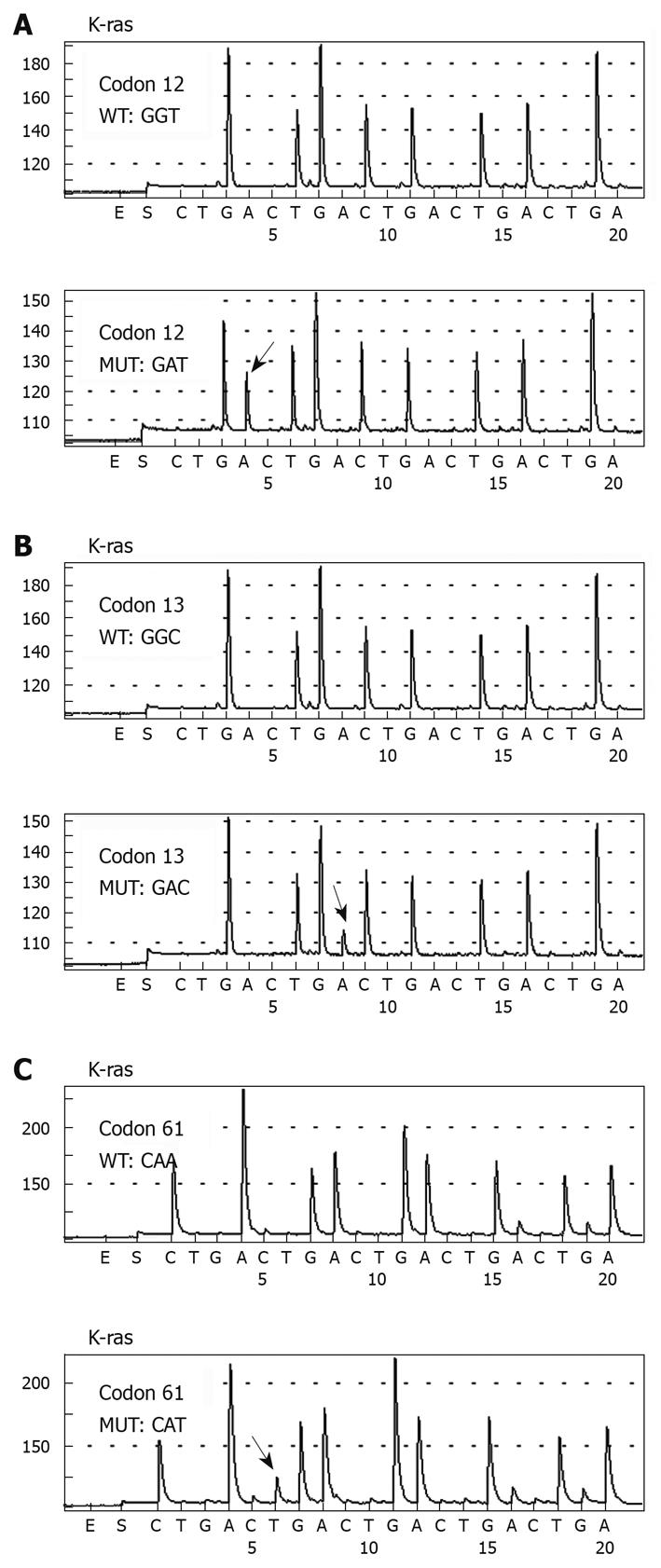
A total of 41 mutations of K-ras gene were detected in the 118 patients, with a mutation rate of 34.7% (41/118). And 23.7% (28/118) of mutations were at codon 12 and 10.2% (12/118) at codon 13. Only one patient harbored a point mutation at codon 61 (0.8%, 1/118). The mutations in the K-ras gene are summarized in Table 3. Figure 1 shows an example of pyrosequencing analysis of mutations located at codon 12, 13, and 61. Compared with the wild-type sequence of codons 12, 13, and 61 (GGT/GGC/CAA), one mutation was detected at codon 12 (Figure 1A) (GGT > GAT), one at codon 13 (Figure 1B) (GGC > GAC), and one at codon 61 (Figure 1A) (CAA > CAT).
| Wild type (AA) | Point mutation (AA) | No. of mutations (%) | |
| K-ras codon 12 | GGT (Gly) | AGT (Ser) | 2 (1.7) |
| GGT (Gly) | GAT (Asp) | 16 (13.6) | |
| GGT (Gly) | GCT (Ala) | 2 (1.7) | |
| GGT (Gly) | GTT (Val) | 8 (6.8) | |
| K-ras codon 13 | GGC (Gly) | GAC (Asp) | 12 (10.2) |
| K-ras codon 61 | CAA (Gln) | CAT (His) | 1 (0.8) |
The distribution of all the detected mutations is shown in Figure 2. The most frequently observed mutations were G-A transitions (30/41, 73.2%), followed by G-T transversions (8/41, 19.5%), two G-C transversions (2/41, 4.9%), and one A-T transversion (1/41, 2.4%). A total of 28 mutations were detected at codon 12 (wild-type GGT), representing four different mutational types. Two G-A transitions (2/28, 7.1%) were located at the first nucleotide of codon 12, resulting in an amino acid change from glycine (Gly) to serine, while the other base substitutions were all located at the second nucleotide of codon 12. The mutated GAT leading to an amino acid change from Gly to aspartic acid (Asp) (16/28, 57.1%) was the most frequently observed mutation. Eight patients were found to harbor a GTT mutation leading to an amino acid change from Gly to valine (Val) (8/28, 28.6%), while the other two patients harbored a GCT mutation (2/28, 7.1%), changing Gly to alanine. Twelve patients (12/118, 10.2%) had detectable mutations at codon 13. The G-A transition was the only mutational type found, resulting in an amino acid change from Gly to Asp. Among all the 118 CRC patients, only one (1/118, 0.8%) had a detectable point mutation (A-T transversion) at codon 61, which resulted in an amino acid change from glutamine to histidine.
As shown in Table 4, differences in the categorical variables, including age, gender, anatomical location of the tumor, tumor differentiation, TNM classification and UICC staging, between patients with and without K-ras mutations were evaluated for significance with χ2 tests. Gender was the only variable that showed a significant relationship with K-ras gene mutation status. The prevalence of gene mutations was higher in female patients than in male patients (44.7% vs 28.2%, P = 0.037). The K-ras mutation frequency in the rectum was higher in female than in male patients (34.0% vs 11.2%, P < 0.05). Male patients had a higher mutation rate in the colon than female patients (16.9% vs 10.6%, P < 0.05) (Table 5).
| Terms | All | Wild type | Mutation type | P value |
| No. of patients | 118 | 77 (65.3) | 41 (34.7) | |
| Gender | 0.037 | |||
| Male | 71 | 51 (71.8) | 20 (28.2) | |
| Female | 47 | 26 (55.3) | 21 (44.7) | |
| Median age (yr) | 61.0 | 64.0 | 60.0 | 0.728 |
| Males | 65.0 | 65.0 | 65.5 | |
| Females | 60.0 | 60.5 | 58.0 | |
| Colorectal segment | 0.559 | |||
| Cecum | 5 | 1 (20.0) | 4 (80.0) | |
| Ascending colon | 23 | 17 (73.9) | 6 (26.1) | |
| Transversal colon | 8 | 4 (50.0) | 4 (50.0) | |
| Descending colon | 5 | 4 (80.0) | 1 (20.0) | |
| Sigmoid | 25 | 16 (64.0) | 9 (36.0) | |
| Rectum | 52 | 35 (67.3) | 17 (32.7) | |
| Differentiation | 0.761 | |||
| Poor | 17 | 12 (70.6) | 5 (29.4) | |
| Moderate | 42 | 25 (59.5) | 17 (40.5) | |
| Well | 59 | 40 (67.8) | 19 (32.2) | |
| UICC classification | 0.631 | |||
| I | 18 | 9 (50.0) | 9 (50.0) | |
| II | 48 | 37 (77.1) | 11 (22.9) | |
| III | 37 | 23 (62.2) | 14 (37.8) | |
| IV | 15 | 8 (53.3) | 7 (46.7) | |
| Bowel wall invasion (pT) | 0.120 | |||
| pT1 | 2 | 1 (50.0) | 1 (50.0) | |
| pT2 | 21 | 11 (52.4) | 10 (47.6) | |
| pT3 | 65 | 43 (66.2) | 22 (33.8) | |
| pT4 | 30 | 22 (73.3) | 8 (26.7) | |
| Lymph node metastasis (pN) | 0.585 | |||
| pN0 | 69 | 47 (68.1) | 22 (31.9) | |
| pN1-2 | 49 | 31 (63.3) | 18 (36.7) | |
| Distant metastasis (pM) | 0.301 | |||
| pM0 | 103 | 70 (68.0) | 33 (32.0) | |
| pM1 | 15 | 8 (53.3) | 7 (46.7) | |
| No. of K-ras mutated patients (%) | Total No. of patients | ||
| Colon | Rectum | ||
| Male | 12 (16.9) | 8 (11.2) | 71 |
| Female | 5 (10.6) | 16 (34) | 47 |
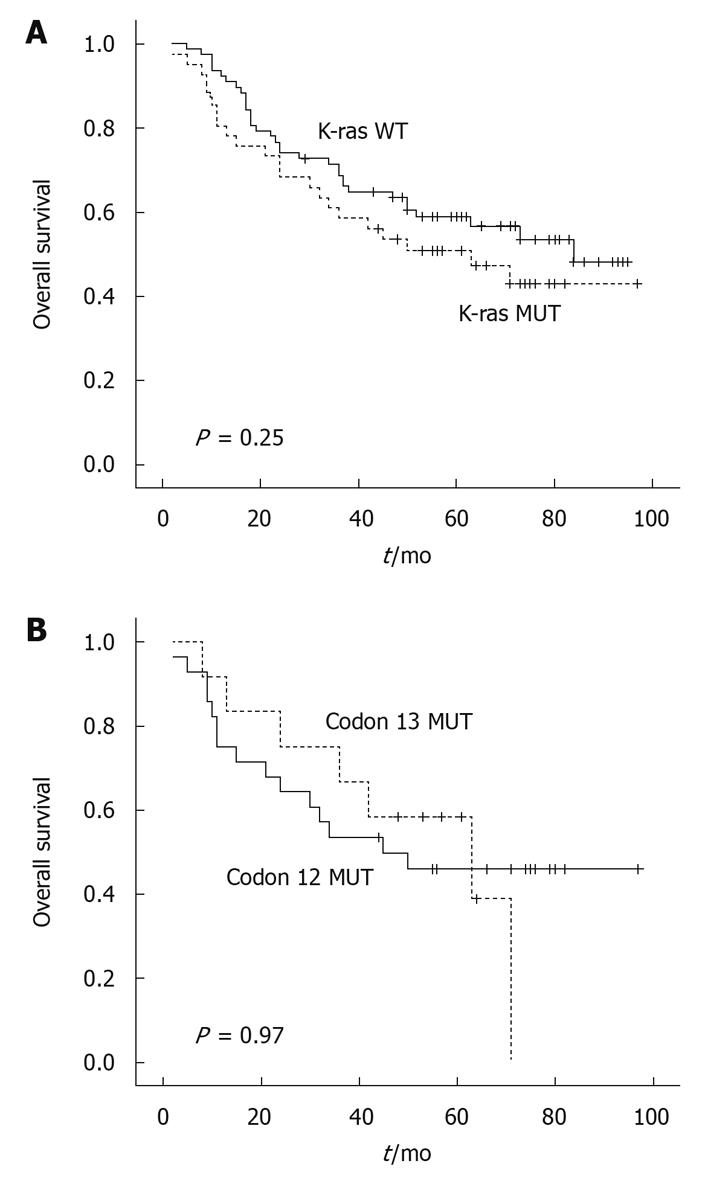
As shown in Figure 3, patients with the wild-type K-ras gene had a median survival of 84.0 mo, which was a little longer than the patients with a mutated K-ras gene, whose median survival was 63.0 mo. However, this difference was not statistically significant (Figure 3A) (P = 0.25). The patients with a mutation at codon 12 and codon 13 had a median survival of 45.0 mo and 63.0 mo, respectively. The patient harboring a mutation at codon 61 had a rather long survival of 73.0 mo. There were no significant correlations between the presence of K-ras mutations at codons 12, 13 and 61) and patient survival (Figure 3B).
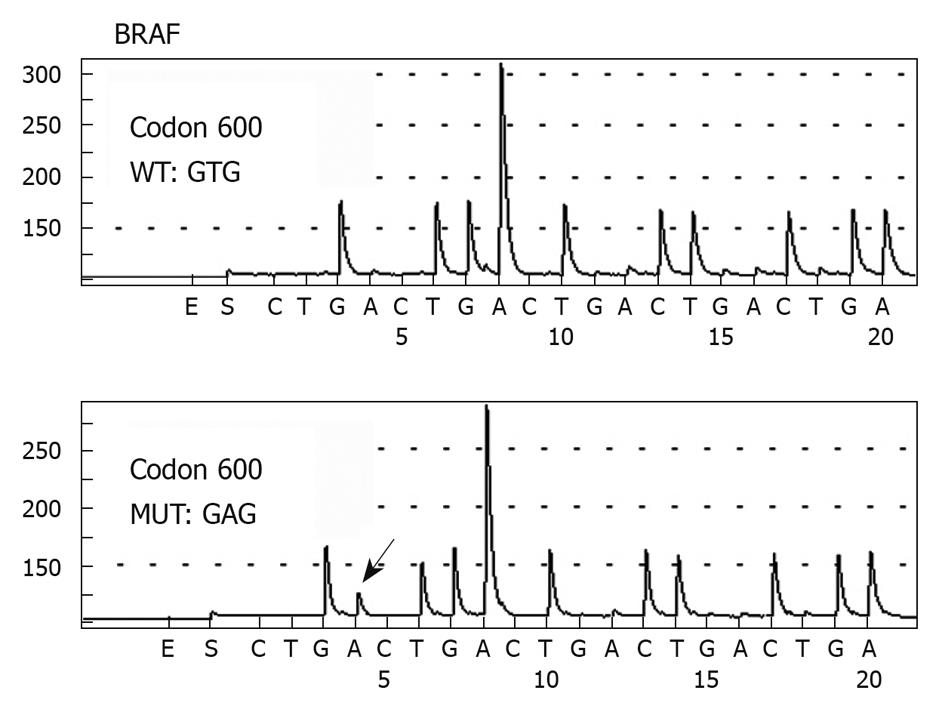
Of the 118 CRC patients analyzed, only two patients (1.7%, 2/118) harbored a detectable mutation at codon 600 in exon 15 of the BRAF gene (Figure 4). Both mutations were T > A transitions (GTG > GAG) at codon 600, which resulted in an amino acid change from Val to glutamic acid (Glu). Both patients had a wild-type K-ras gene. One patient was a 70-year-old man with a moderately differentiated adenocarcinoma in the ascending colon at pathological stage of T3N1M0. The patient survived for 13 mo after the operation, but died of other diseases without evidence of tumor recurrence and metastasis. The other patient was a 56-year-old man. He was operated on for a well-differentiated adenocarcinoma in the transverse colon also at pathological stage of T3N1M0. Sixty-three months after operation, the patient died of tumor recurrence.
In this study, we detected various mutations of the K-ras and BRAF gene in 118 Chinese CRC patients using pyrosequencing. Mutations in the K-ras gene occurred very frequently (20%-50%) in CRC. It was reported in Western countries that approximately 90% of the activating K-ras mutations were found at codon 12 and codon 13 of exon 1, and about 5% at codon 61 located in exon 2. In domestic reports, with a limited number of samples, there was a similar mutation rate at codon 12 and 13, and a rather lower rate at codon 61 (0%-4.8%).
In our previous studies published in the 1990s, K-ras gene mutations were detected in 12 out of 35 (12/35, 34.3%) CRC patients by the PCR-RFLP method[16]. Eleven out of the 12 detected mutations (11/35, 31.4%) were located at codon 12, while the other one was at codon 61 (1/35, 2.9%). None of the patients had a mutation at codon 13. Mutations were also found in the pericancerous mucosa in some cases[17], indicating that the K-ras gene might play an important role in the early stage of colorectal carcinogenesis. In the present study, a similar mutation rate was observed. Among the 118 patients in this study, the rates of mutated (41/118, 34.7%) and non-mutated (77/118, 65.3%) K-ras genes were similar to those reported by other countries. The K-ras mutation rates at codon 12, codon 13 and codon 61 were 23.7% (28/118), 10.2% (12/118) and 0.8% (1/118), respectively. Among all 41 mutations detected in K-ras gene in this study, 68.3% (28/41) were located at codon 12, 29.3% (12/41) at codon 13, and 2.4% (1/41) at codon 61. Similar to the previously reported data, G-A transitions were the most frequently found type of K-ras gene mutations in our study, followed by G-T transversions. Among the four different mutations detected at codon 12, Gly12Asp was the most frequent point mutation, accounting for about 40% of all mutations detected in the K-ras gene and 60% of all mutations at codon 12. All 12 point mutations detected at codon 13 were Gly13Asp. This base substitution accounted for 30% of all mutations detected in the K-ras gene. The distribution of the six different mutations (Figure 1) among the mutated patients was in concordance with the published data[18]. These point mutations resulting in amino acid substitution would activate RAS proteins, produce an alteration in the transduction of signals in the RAS pathway and ultimately lead to increased mitogenic signaling.
It is widely accepted that mutations in the K-ras gene are early events in colorectal carcinogenesis. As described by Vogelstein et al[5], a K-ras gene mutation might happen in the progression from adenoma to carcinoma. Some studies have investigated the relationship between K-ras mutation and various clinicopathological parameters, but the results remain controversial. For example, it was reported by Zlobec et al[19] that K-ras mutations were associated with neither clinicopathological parameters, such as gender, age, tumor location, histological type, tumor T and N stage, tumor grade and vascular invasion nor survival time of patients. Naguib et al[20] reported a mutation rate of 22% in the K-ras gene (codon 12 and 13), and a positive relationship with more advanced Dukes’ stage and microsatellite stable status. In the present study, we did not find any significant correlations between the presence of K-ras mutations at codons 12, 13, and 61 or mutation type and various clinicopathological features, such as age, anatomical location of the tumor, tumor differentiation, TNM classification, and UICC staging. Gender was the only variable that showed an obvious relationship with K-ras gene mutation, and suggested that female patients had a higher prevalence of gene mutation than male patients (44.7% vs 28.2%, P = 0.037). Breivik et al[7] demonstrated that K-ras mutations were much less frequent in colon samples from male patients compared with female patients at codon 12 and 13 in Western populations. However, we found that male patients had a higher mutation rate in colon samples than female patients in the Chinese population (16.9% vs 10.6%, P < 0.05). Ethnicity, environment, and lifestyle differences may explain the difference in K-ras mutation frequency in the colon between Chinese and Western population. However, a larger population of Chinese patients is needed to confirm our findings.
Some studies have indicated the importance of K-ras alterations in predicting long-term outcome, while others have failed to show such a relationship. The collaborative RASCAL study reported a correlation between the specific K-ras mutation Gly12Val and poor prognosis, especially in Dukes’ C CRC patients[21]. The Gly12Val mutation at codon 12 reported by Al-Mulla et al[22] and the G > A mutation at codon 13 reported by Samowitz et al[18] both might be related to the poor survival of the patients. On the other hand, Tortola et al[23] and Dix et al[24] failed to prove a positive relationship between K-ras mutation and shorter survival. In our study, prognostic analysis for K-ras mutations showed that none of the K-ras mutations were predictive of patient survival. Patients with a wild-type K-ras gene had a median survival of 84.0 mo, which was slightly longer than patients with a mutated K-ras gene (63.0 mo); however, this difference was not statistically significant (P = 0.25). There was also no significant difference in survival between patients harboring a mutation at codon 12 and those at codon 13 (45.0 mo vs 63.0 mo, P = 0.97). The shorter survival in the patients reported by Al-Mulla et al[22] (Gly12Val mutations at codon 12) and by Samowitz et al[18] (G > A mutation at codon 13), was not observed in our study. A further study with a larger number of samples will hopefully confirm the results in this study.
The K-ras/BRAF/ERK signaling pathway plays an important role in colorectal carcinogenesis. Encoding a downstream molecule of K-ras, the BRAF gene (codon 600) is mutated in 12%-15.6% (45/374) of colorectal carcinomas[19,20]. It was reported by Zlobec et al[19] that BRAF gene mutations are strongly associated with right-sided tumor location, higher tumor grade, absence of peritumoral lymphocytic inflammation, and microsatellite instability (MSI-H)[19]. It was also reported that a mutated BRAF gene is an adverse prognostic factor in right-sided colon cancer patients independent of MSI status, and in patients with lymph node-negative disease[19]. More recently, Richman[25] showed that mutations in either KRAS or BRAF are factors for poor prognosis and the overall survival (OS), and have minimal impact on progression-free survival (PFS). The mutation status of either gene does not affect the impact of irinotecan or oxaliplatin on PFS or OS. To our knowledge, this is the first paper reporting mutations in the BRAF gene in a large population of Chinese CRC patients. In our study, only two mutations (1.7%, 2/118) at codon 600 in exon 15 of the BRAF gene were detected in 118 Chinese CRC patients. Both patients had a wild-type K-ras gene. The mutation rate was very low compared with that reported in Western studies. Both mutations were T to A transversions (GTG > GAG), resulting in an amino acid change from Val to Glu. Both patients were male, with a Dukes’ C (T3N1M0) carcinoma at the right-side of the colon. Due to the limited mutations detected, it was impossible to analyze the correlation of BRAF gene mutations with various clinicopathological features or prognosis. There might be a few explanations for the low incidence of BRAF gene mutations in our study. First, we only analyzed mutations at codon 600 in exon 15 of the BRAF gene. Therefore, mutations at other sites (e.g. codons 463-468) might affect the result. Second, ethnic differences might exist in CRC between Chinese and Western populations. Liao et al[26] found that the BRAF mutation frequency was only 4.9% in 61 Chinese colorectal tissues, which is also lower than that in Western populations. Wójcik et al[27] screened for mutations in exons 11 and 15 of the BRAF gene in 163 resected adenocarcinomas in a Polish population and only six (3.7%) tumors had a missense point mutation (G469A, D594G, G596R, K601N, and two V600E). Future studies containing a larger number of Chinese samples are expected to further clarify the result.
In conclusion, the mutation of the K-ras gene was a common event in our 118 Chinese CRC patients, with an obvious relationship with gender. Female patients had a higher prevalence of gene mutation than male patients. K-ras gene mutation seemed not to be an independent prognostic factor in CRC patients. The BRAF gene was rarely mutated in Chinese CRC patients.
In recent years, the morbidity and mortality of colorectal cancer (CRC) has risen in the Chinese population. The K-ras and BRAF genes encode proteins that act in the extracellular signal-regulated kinase signaling pathway, which mediates cellular responses to growth factors and regulates the elements of the cell cycle, apoptosis and differentiation. Both K-ras and BRAF are prone to mutations in sporadic CRC. Mutations in K-ras could lead to constitutive activation of this pathway, resulting in cancer progression. Several recent studies have shown a strong correlation between K-ras and BRAF mutations and response to panitumumab and cetuximab.
Results were in consistent among studies of the relationship between K-ras gene mutations and various clinicopathological characteristics. The prognostic significance of K-ras gene mutations is also controversial. With regard to the prognostic significance of the BRAF gene, less information is available in both Western and Chinese populations.
Recent reports have highlighted the importance of K-ras and BRAF gene mutations, the clinicopathological characteristics of CRC and its response to epidermal growth factor receptor-targeted therapies. Pyrosequencing is a powerful, sensitive and rapid sequencing method for single-nucleotide polymorphism (SNP)/mutation analysis. This is the first study to report mutations of K-ras and BRAF genes in a large population of Chinese CRC patients using pyrosequencing.
Using pyrosequencing technology, the authors found that K-ras gene mutations were common in Chinese CRC patients, with an obvious relationship with gender. The BRAF gene was rarely mutated in Chinese CRC patients.
Pyrosequencing: Pyrosequencing is a method of determining the order of nucleotides in DNA based on the “sequencing by synthesis” principle. It relies on the detection of pyrophosphate release on nucleotide incorporation, rather than chain termination with dideoxynucleotides. It is a powerful, sensitive and rapid sequencing method and can be used for DNA SNP/mutation analysis.
This paper presents new results on the frequency of K-ras and BRAF mutations in colorectal carcinomas of Chinese patients.
Peer reviewer: Daniel S Straus, PhD, Professor, Biomedical Sciences Division, University of California, Riverside, CA 9252, United States
S- Editor Tian L L- Editor Ma JY E- Editor Lin YP
| 1. | Marshall CJ. Small GTPases and cell cycle regulation. Biochem Soc Trans. 1999;27:363-370. |
| 2. | Li WQ, Kawakami K, Ruszkiewicz A, Bennett G, Moore J, Iacopetta B. BRAF mutations are associated with distinctive clinical, pathological and molecular features of colorectal cancer independently of microsatellite instability status. Mol Cancer. 2006;5:2. |
| 3. | Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. |
| 4. | Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949-954. |
| 5. | Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM, Bos JL. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525-532. |
| 6. | Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682-4689. |
| 7. | Breivik J, Meling GI, Spurkland A, Rognum TO, Gaudernack G. K-ras mutation in colorectal cancer: relations to patient age, sex and tumour location. Br J Cancer. 1994;69:367-371. |
| 8. | Kislitsin D, Lerner A, Rennert G, Lev Z. K-ras mutations in sporadic colorectal tumors in Israel: unusual high frequency of codon 13 mutations and evidence for nonhomogeneous representation of mutation subtypes. Dig Dis Sci. 2002;47:1073-1079. |
| 9. | Kressner U, Bjørheim J, Westring S, Wahlberg SS, Påhlman L, Glimelius B, Lindmark G, Lindblom A, Børresen-Dale AL. Ki-ras mutations and prognosis in colorectal cancer. Eur J Cancer. 1998;34:518-521. |
| 10. | Hayes VM, Westra JL, Verlind E, Bleeker W, Plukker JT, Hofstra RM, Buys CH. New comprehensive denaturing-gradient-gel- electrophoresis assay for KRAS mutation detection applied to paraffin-embedded tumours. Genes Chromosomes Cancer. 2000;29:309-314. |
| 11. | Martinez-Garza SG, Núñez-Salazar A, Calderon-Garcidueñas AL, Bosques-Padilla FJ, Niderhauser-García A, Barrera-Saldaña HA. Frequency and clinicopathology associations of K-ras mutations in colorectal cancer in a northeast Mexican population. Dig Dis. 1999;17:225-229. |
| 12. | Yuan Y, Hu HG, Ye XX, Shen H, Zheng S. [K-ras gene mutation in colorectal cancer and its clinicopathologic significance]. Zhonghua Waike Zazhi. 2010;48:1247-1251. |
| 13. | Gao J, Li YY, Sun PN, Shen L. Comparative analysis of dideoxy sequencing, the KRAS StripAssay and pyrosequencing for detection of KRAS mutation. World J Gastroenterol. 2010;16:4858-4864. |
| 14. | Ronaghi M, Karamohamed S, Pettersson B, Uhlén M, Nyrén P. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem. 1996;242:84-89. |
| 15. | Ronaghi M, Uhlén M, Nyrén P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363, 365. |
| 16. | Gan YB, Cai XH, Zheng S. Detection of Ki-ras gene mutations in tumor tissues and stools of patients with colorectal carcinoma. J Zhejiang Univ (Medicine Edition). 1995;24:241-247. |
| 17. | Gan YB, Cai XH, Zheng S. The ras gene mutations in mucosa adjacent to colorectal cancer. Zhongliu Yanzhi Yanjiu. 1994;21:350-351. |
| 18. | Samowitz WS, Curtin K, Schaffer D, Robertson M, Leppert M, Slattery ML. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: a population-based study. Cancer Epidemiol Biomarkers Prev. 2000;9:1193-1197. |
| 19. | Zlobec I, Bihl MP, Schwarb H, Terracciano L, Lugli A. Clinicopathological and protein characterization of BRAF- and K-RAS-mutated colorectal cancer and implications for prognosis. Int J Cancer. 2010;127:367-380. |
| 20. | Naguib A, Mitrou PN, Gay LJ, Cooke JC, Luben RN, Ball RY, McTaggart A, Arends MJ, Rodwell SA. Dietary, lifestyle and clinicopathological factors associated with BRAF and K-ras mutations arising in distinct subsets of colorectal cancers in the EPIC Norfolk study. BMC Cancer. 2010;10:99. |
| 21. | Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N. Kirsten ras mutations in patients with colorectal cancer: the 'RASCAL II' study. Br J Cancer. 2001;85:692-696. |
| 22. | Al-Mulla F, Going JJ, Sowden ET, Winter A, Pickford IR, Birnie GD. Heterogeneity of mutant versus wild-type Ki-ras in primary and metastatic colorectal carcinomas, and association of codon-12 valine with early mortality. J Pathol. 1998;185:130-138. |
| 23. | Tortola S, Marcuello E, González I, Reyes G, Arribas R, Aiza G, Sancho FJ, Peinado MA, Capella G. p53 and K-ras gene mutations correlate with tumor aggressiveness but are not of routine prognostic value in colorectal cancer. J Clin Oncol. 1999;17:1375-1381. |
| 24. | Dix BR, Robbins P, Soong R, Jenner D, House AK, Iacopetta BJ. The common molecular genetic alterations in Dukes' B and C colorectal carcinomas are not short-term prognostic indicators of survival. Int J Cancer. 1994;59:747-751. |
| 25. | Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931-5937. |
| 26. | Liao W, Liao Y, Zhou JX, Xie J, Chen J, Huang W, Luo R. Gene mutations in epidermal growth factor receptor signaling network and their association with survival in Chinese patients with metastatic colorectal cancers. Anat Rec (Hoboken). 2010;293:1506-1511. |
| 27. | Wójcik P, Okoń K, Osuch C, Klimkowska A, Tomaszewska R. BRAF mutations in sporadic colorectal carcinoma from polish patients. Pol J Pathol. 2010;61:23-26. |