Published online Dec 28, 2011. doi: 10.3748/wjg.v17.i48.5317
Revised: October 28, 2011
Accepted: November 5, 2011
Published online: December 28, 2011
AIM: To explore mutations around the interferon sensitivity-determining region (ISDR) which are associated with the resistance of hepatitis C virus 1b (HCV-1b) to interferon-α treatment.
METHODS: Thirty-seven HCV-1b samples were obtained from Hong Kong patients who had completed the combined interferon-α/ribavirin treatment for more than one year with available response data. Nineteen of them were sustained virological responders, while 18 were non-responders. The amino acid sequences of the extended ISDR (eISDR) covering 64 amino acids upstream and 67 amino acids downstream from the previously reported ISDR were analyzed.
RESULTS: One amino acid variation (I2268V, P = 0.023) was significantly correlated with treatment outcome in this pilot study with a limited number of patients, while two amino acid variations (R2260H, P = 0.05 and S2278T, P = 0.05) were weakly associated with treatment outcome. The extent of amino acid variations within the ISDR or eISDR was not correlated with treatment outcome as previously reported.
CONCLUSION: Three amino acid mutations near but outside of ISDR may associate with interferon treatment resistance of HCV-1b patients in Hong Kong.
- Citation: Zhou XM, Chan PK, Tam JS. Mutations around interferon sensitivity-determining region: A pilot resistance report of hepatitis C virus 1b in a Hong Kong population. World J Gastroenterol 2011; 17(48): 5317-5323
- URL: https://www.wjgnet.com/1007-9327/full/v17/i48/5317.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i48.5317
The interferon (IFN)-α/ribavirin combination treatment for hepatitis C virus (HCV) can achieve a sustained virological response rate of 20%-50%. The effectiveness of the combination treatment is partly correlated with HCV genotypes[1]. HCV genotype 6a shows a more favorable response with a sustained virological response rate around 50%-70%[2,3], while HCV genotype 1 is hard to be treated with a low sustained virological response rate between 20% and 40%[4]. Pegylated (PEG)-IFN-α can improve the sustained virological response. But HCV genotype 1 is still more difficult to treat than other HCV genotypes, i.e., 2 and 3 by PEG-IFN-α[4,5].
The interferon sensitivity-determine region (ISDR, amino acid position 2209-2248, refers to HCV-J, GenBank accession no: D90208) for HCV 1b was previously identified to be associated with resistance to IFN-α-based treatment[6,7]. However, this observation was not reproducible in the studies in Europe, China and United States[8-12]. The host genetic background may be one of the reasons accounting for the discrepancies[13]. Furthermore, isolates of HCV 1b circulated in Japan may have substantial genetic difference compared with those circulating in Europe or United States[14]. The genetic difference of virus strains may be another reason for the discrepancies[15].
Several functional domains were identified within NS5A encoding region[16]. One of them can bind to Protein Kinase-R (PKR)[17,18]. This domain encloses the entire ISDR, with additional 26 amino acids extended downstream. PKR can be activated by IFN-α, then phosphorylates elongation-initiation-factor-2, which is an essential component of cellular protein translation complex, resulting in stopping the synthesis of intracellular proteins[19]. When NS5A binds to PKR, the synthesis of proteins will be restored, then the replication of HCV within host cells can be rescued[18]. This hypothesis suggests that the PKR binding domain is still a good candidate to look for variations that are associated with outcomes of IFN-α-based treatment [20].
No study on the IFN resistance-associated mutation in HCV-1b has yet been carried out in the Hong Kong population[21]. Although result from this population may conform to other geographic regions, HCV-1b strains circulated in Hong Kong had a more conservative genomic pattern than its counterpart in any other regions due to its specific geographic location and political history. Thus we carried out this study to explore amino acid variations within extended ISDR (eISDR) associated with the IFN-α/ribavirin combination treatment[22].
This work was conducted in accordance with the Declaration of Helsinki (2000) of the World Medical Association. Recruited patients all submitted written consent to take part in the study. The study was approved by the local Joint Chinese University of Hong Kong-New Territories East Cluster Clinical Research Ethics Committee (ref. CRE-2006.405).
Samples were collected from Hong Kong patients between 1998 and 2004. Their HCV genotypes were determined by the type-primer-specific reverse transcription-polymerase chain reaction (RT-PCR) genotyping method as described before.
All patients included in this study were previously naive to HCV treatment. After HCV-1b genotyping determination, they received 5 million international units IFN-α three times per week and ribavirin (1000 mg for those weighing less than 75 kg and 1200 mg for those weighing more than 75 kg) for one year. If HCV-6a genotyping was determined, they would receive a short-term treatment for 6 mo. The HCV RNA load was measured once every six months during treatment and then every year after the completion of treatment. Patients with HCV RNA detected in the serum six months after treatment were defined as non-response/resistant (NR). Patients with negative HCV RNA results for at least one year after treatment were defined as having sustained virological response (SVR).
Altogether 19 NR and 18 SVR patients with determined endpoint of treatment were recruited in this pilot study. These patients had no recorded history of injecting drug use or liver transplantation.
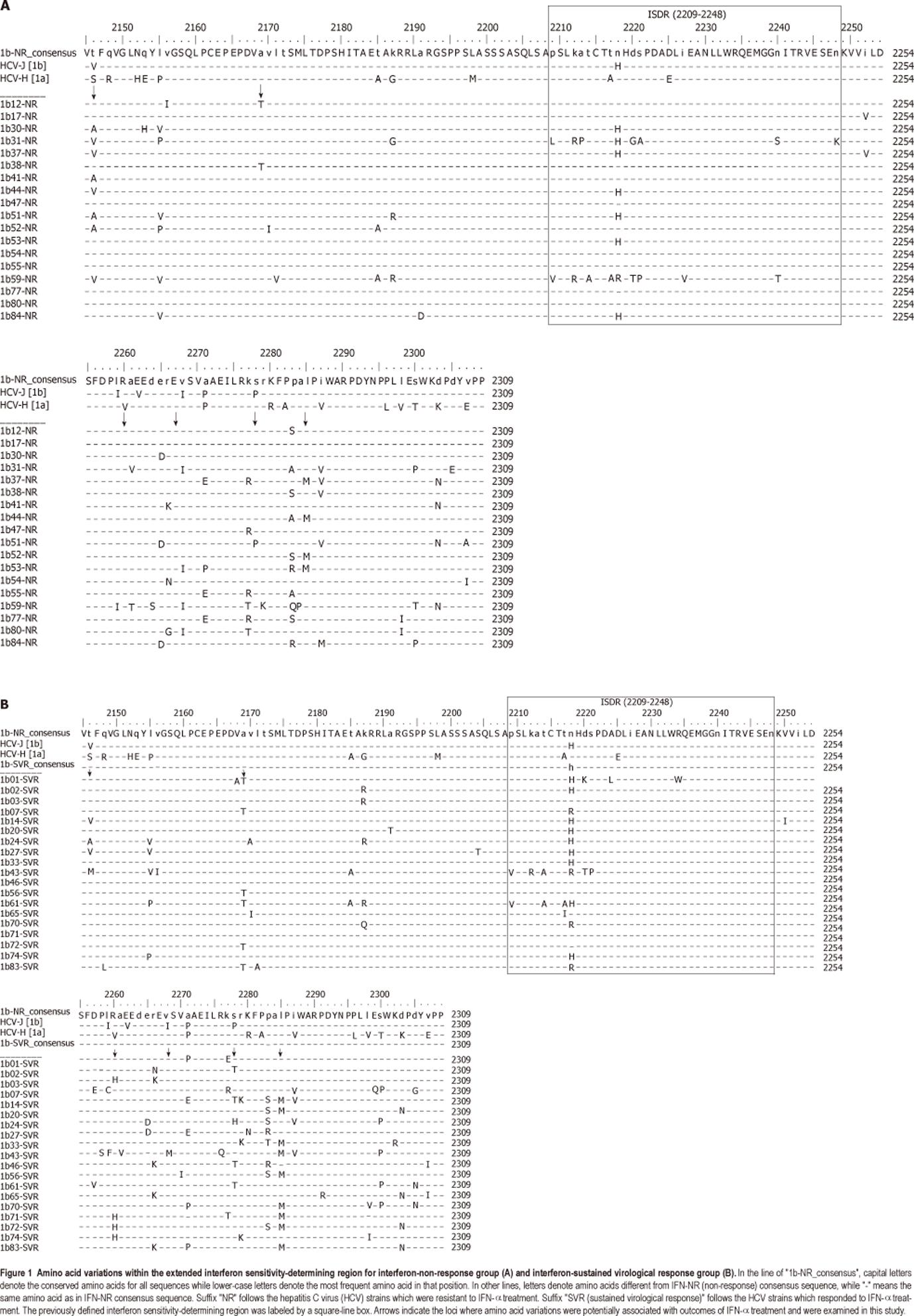
The eISDR within HCV 1b genome starts from amino acid loci 2145 to 2315 (refers to HCV-J, GenBank accession no: D90208). This region extends 64 amino acids upstream and 67 amino acids downstream to the ISDR (2209-2248 in HCV-J). With the downstream extension, this eISDR includes the entire PKR-binding domain.
This region was amplified by a nested RT-PCR reaction. Primers used were: outer forward primer (NS5A-1b-1F): 5’-TGGATGGAGTGCGGTTGCACAGGTA-3’; outer reverse primer (NS5A-1b-1R): 5’-TCT TTC TCC GTG GAG GTG GTA TTG G-3’; inner forward primer (NS5A-1b-2F): 5’-TGTAAAACGACGGCCAGTCAGGTACGCTCCGGCGTGCA-3’; inner reverse primer (NS5A-1b-2R): 5’-CAGGAAACAGCTATGACCGGGGCCTTGGTAGGTGGCAA-3’. The underlined inner-primer sequences represent the M13 sequencing primers.
HCV RNA was extracted from each serum sample using the High Pure Viral Nucleic Acid Kit (Roche Diagnostics GmbH, Mannheim, Germany) following the manufacturer’s protocol. Five microliters of viral RNA were used in a final 50-μL single-tube single-step RT-PCR reaction using SuperScript™ One-Step RT-PCR system with Platinum®Taq kit (Invitrogen, Life Technologies, Carlsbad, CA). Cycling conditions were: reverse transcription at 50 °C for 30 min, 94 °C for 2 min followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min, extension at 68 °C for 1 min. Second round nested PCR reactions were carried out using 1 μL of the first round RT-PCR product as template and using HotStar Taq polymerase (Qiagen, Hilden, Germany) in a final 50-μL reaction with 1 × Q-solution. The HotStar Taq polymerase was used at a working concentration of 5 U per 50-μL reaction. Cycling conditions were: 95 °C for 15 min to activate the HotStar Taq polymerase followed by 30 cycles of denaturation at 95 °C for 1 min, annealing at 55 °C for 1 min, extension at 72 °C for 1 min, and a final extension cycle at 72 °C for 5 min.
The 609-bp PCR amplicons were purified from agarose gel using the GFX™ PCR DNA and Gel Band Purification kit (Amersham Bioscience UK limited, Little Chalfont, United Kingdom), and directly used for BigDye® Terminator v3.1 Cycle Sequencing reaction (Applied Biosystems, Foster City, CA). The nucleotide sequences were translated to amino acid sequences following the right open-reading-frame. All the sequences were deposited into GenBank.
The amino acid sequences were aligned by the multiple-alignment algorithm under Clustal-X (version 1.83) using a protein weight matrix of BLOSUM 62.
Consensus amino acid sequences were deduced from all the eISDR sequences. The number of amino acids that were different between each query sequence and the consensus eISDR sequence was added up as the degree of amino acid variations for this query sequence in the eISDR (1b-VAR2). The number of amino acids that were different within the ISDR was added up as the degree of amino acid variations for this query sequence in the ISDR (1b-VAR1).
The difference in patient age between the IFN-NR group and the IFN-SVR group was assessed by two-tailed Student's t test. The sex distribution between patients of the IFN-NR group and the IFN-SVR group was assessed by two-tailed Fisher’s exact test (mid-P approach). The association of the amino acid variations within eISDR with treatment outcomes was evaluated by two-tailed Fisher's exact test. Statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS 11.1.0, SPSS Inc., Chicago, IL) and Two-by-two (http://www.med. uio.no/imb/stat/two-by-two/manual.html) for two-tailed Fisher's exact test.
The mean age of the 37 patients was 48.4 years (SD = 9.3 years, range 28-67 years). There was no significant difference in age between the IFN-NR and the IFN-SVR patients (47.9 years vs 49.0 years, P = 0.73). There was no significant difference in sex distribution between the IFN-NR (male/female: 11/7) and the IFN-SVR (male/female: 11/8) patients (P = 1.00).
The sequence differences between the IFN-SVR and the IFN-NR patient groups are illustrated in Figure 1. Amino acid variations were found distributed over 56 different loci. Six loci, at which more variations existed between the two groups, were examined by two-tailed Fisher’s exact test (Table 1). One amino acid variation (I2268V, P = 0.023) was significantly correlated with the treatment outcome, while the two amino acid variations (R2260H, P = 0.05 and S2278T, P = 0.05) were weakly associated with treatment outcomes.
| Locus | Amino acid variations | mid-P | Prediction | ||
| IFN-NR (n = 18) | IFN-SVR (n = 19) | Sensitivity (%) | Specificity (%) | ||
| 2146 | T (10/18, 55.6%) | T (15/19, 78.9%) | 0.085 | ||
| V (4/18, 22.2%) | V (2/19, 10.5%) | 0.20 | |||
| A (4/18, 22.2%) | A (1/19, 5.3%) | 0.09 | |||
| M (1/19, 5.3%) | - | ||||
| 2169 | A (16/18, 88.9%) | A (13/19, 68.9%) | 0.12 | ||
| T (2/18, 11.1%) | T (6/19, 31.6%) | 0.12 | |||
| 2260 | R (18/18, 100%) | R (15/19, 78.9%) | 0.05 | ||
| H (0/18, 0%) | H (4/19, 21.1%) | 0.05 | 100 | 21.1 | |
| 2268 | V (14/18, 77.8%) | V (18/19, 100%) | 0.09 | ||
| I (4/18, 22.2%) | I (0/19, 0%) | 0.023 | 22.2 | 100 | |
| M (1/19, 5.3%) | - | ||||
| 2278 | S (17/18, 94.4%) | S (14/19, 73.7%) | 0.09 | ||
| T (0/18, 0%) | T (4/19, 21.1%) | 0.05 | 100 | 21.1 | |
| P (1/18, 5.6%) | H (1/19, 5.3%) | - | |||
| 2285 | L (14/18, 77.8%) | L (10/19, 52.6%) | 0.085 | ||
| M (4/18, 22.2%) | M (9/19, 47.4%) | 0.085 | |||
Prediction of the treatment outcome by I2268V has a specificity of 100%, but a low sensitivity of 22.2%. Prediction by R2260H or by S2278T has a sensitivity of 100%, but a low specificity 21.1%.
Previous studies have suggested that the degree of amino acid variations within the ISDR was associated with resistance to IFN-α treatment. We examined the correlation between the number of amino acid variations within the ISDR and the eISDR and the treatment outcome. A consensus resistant amino acid sequence was deduced from the conserved sequence of the IFN-NR group. This sequence has one amino acid different from reference strain HCV-J within the ISDR and seven amino acids within the entire eISDR. The degree of amino acid variations within the ISDR (1b-VAR1) and the eISDR (1b-VAR2) was calculated and compared. 1b-VAR1 ranged from 0 to 9 and 1b-VAR2 ranged from 1 to 24, both showing no correlation with the treatment outcome (P = 0.958, P = 0.563) with a previously defined cutoff of four amino acid variations.
In this pilot study with a limited number of cases, amino acid variations in three loci within the eISDR but outside the ISDR were identified to be possibly correlated with the resistance to the IFN-α/ribavirin combination treatment. R2260H (P = 0.05) and I2268V (P = 0.023) were located within PKR-binding domain[23,24]. S2278T (P = 0.05) was located within a hyperphosphorylation signal motif “PPALP”[25]. All these three loci were located within the putative NS5A transcriptional activation domain[16].
Based on these preliminary results, R2260H and I2268V suggested that PKR binding ability of NS5A may still play a major role in determining the IFN-α resistance of HCV-1b[20]. S2278T may imply that modification of the NS5A protein, which eventually alters the interaction of NS5A with other proteins like PKR, can contribute to the resistance to treatment[26,27].
This trans-activating activity of the NS5A transcriptional activation domain was identified in an in vitro GAL4 system, and had not been demonstrated in vivo[28,29]. Some studies have shown that this NS5A trans-activating activity can stimulate the expression of IL8, an antagonist of IFN-αin vivo[30]. Whether the variations in these three loci can affect the expression of IL8 remains to be studied.
In contrast to previous reports, no correlation was found in the current study between the degree of amino acid variations within the ISDR or eISDR and IFN-α resistance of HCV-1b.
The association of IFN-α resistance and the amino acid variations was different from the reports from Europe and United States[15]. Previous reports did not find any amino acid variations within the ISDR that were correlated with treatment resistance. This difference may be due to the concurrent circulation of multiple subtypes of HCV-1b. The similarities between HCV-1b strains in Europe or United States were 90.3%, while in Hong Kong were 94.3% in average. Thus, HCV-1b strains distributed in Europe or United states were more diversified than those in Hong Kong. The convergency of HCV-1b population in Hong Kong introduces less genomic confounding factor. This may lead to different observations among studies involving a small number of samples.
Therefore, an expanded study is needed to further clarify the observations acquired from the Hong Kong population.
The association can be further ascertained in future studies by a larger group of registered HCV-1b patients, for those patients infected with IFN-resistant type of HCV-1b would be pursuable to abandon IFN treatment as early as possible. Alternative treatment, i.e., traditional Chinese medicine may be used instead, which may improve patient living status without the side-effects caused by IFN simultaneously. For the patients infected with IFN-sensitive type of HCV-1b, less IFN dosage should be administered to achieve the same efficiency of treatment.
Currently, an interferon-α (IFN-α) based regimen is the most effective and widely accepted therapy for hepatitis C virus (HCV) infections. When combined with ribavirin, IFN-α typically can achieve a 20% to 50% sustained virological response (SVR) in HCV-infected patients. The effect of HCV sequence variation in response to therapy is less clear.
The association of a region of 40 amino acids in length within HCV 1b genome, termed as the interferon sensitivity-determining region (ISDR). However, similar studies performed with cohorts of American, European and Chinese patients reached discrepancy results. This discrepancy may resulted from high divergence of HCV 1b strains distributed worldwide.
The authors had early found that mutations within eISDR but outside of ISDR are in association with interferon treatment failure for HCV genotype 6a. This exploration were carried out in scope of full genomewide analysis. Thus, the authors explored mutations within eISDR in HCV-1b patients from Hong Kong, wish to reach a relative firmly report.
The study results suggest that we also can predict the prognosis of interferon treatment to HCV-1b patients. Interferon dosage can be regulated based on HCV-1b sequences, so reach to a balance between treatment effectiveness and tolerance of the side-effect of this drug.
ISDR: Interferon sensitive-determine region, located within HCV NS5A. HCV NS5A had been firmly demonstrated that it can interact with various signal proteins induced by interferon. The relation of NS5A with interferon resistance were firmly established in HCV patients. But certain mutations had not ever been determined for predicting the prognosis of interferon treatment.
This is a good study in which authors analyze the association of mutations within eISDR of HCV-1b with the resistance of treatment by IFN-α in Hong Kong. The results are interesting and suggest that three mutations may associated with treatment success/failure. These mutations could be used for predicting prognosis before the start of treatment. Thus, prescription of IFN-α may be improved for future treatments to HCV-1b patients.
Peer reviewer: Dr. Hisato Nakajima, The Jikei University School of Medicine, Department of Gastroenterology and Hepatology, 3-25-8, Nishi-Shinbashi, Minato-ku, Tokyo 105-8461, Japan
S- Editor Tian L L- Editor Ma JY E- Editor Xiong L
| 1. | Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345:41-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2042] [Cited by in RCA: 2025] [Article Influence: 84.4] [Reference Citation Analysis (0)] |
| 2. | Hui CK, Yuen MF, Sablon E, Chan AO, Wong BC, Lai CL. Interferon and ribavirin therapy for chronic hepatitis C virus genotype 6: a comparison with genotype 1. J Infect Dis. 2003;187:1071-1074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 3. | Yuen MF, Lai CL. Response to combined interferon and ribavirin is better in patients infected with hepatitis C virus genotype 6 than genotype 1 in Hong Kong. Intervirology. 2006;49:96-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 4. | Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958-965. [PubMed] |
| 5. | Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL, Häussinger D, Diago M, Carosi G, Dhumeaux D. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975-982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4847] [Cited by in RCA: 4748] [Article Influence: 206.4] [Reference Citation Analysis (0)] |
| 6. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Invest. 1995;96:224-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 452] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 7. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 752] [Cited by in RCA: 734] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 8. | Duverlie G, Khorsi H, Castelain S, Jaillon O, Izopet J, Lunel F, Eb F, Penin F, Wychowski C. Sequence analysis of the NS5A protein of European hepatitis C virus 1b isolates and relation to interferon sensitivity. J Gen Virol. 1998;79:1373-1381. [PubMed] |
| 9. | Hofgärtner WT, Polyak SJ, Sullivan DG, Carithers RL, Gretch DR. Mutations in the NS5A gene of hepatitis C virus in North American patients infected with HCV genotype 1a or 1b. J Med Virol. 1997;53:118-126. [PubMed] |
| 10. | Hu Y, Tang M, Jiang W, Wu Y, Yuan Z, Wen Y. [Association between NS5A gene sequence and response to interferon therapy in chronic hepatitis C patients in Shanghai]. Zhonghua Shiyan He Linchuang Bingduxue Zazhi. 2002;16:114-118. [PubMed] |
| 11. | Veillon P, Payan C, Gaudy C, Goudeau A, Lunel F. [Mutation analysis of ISDR and V3 domains of hepatitis C virus NS5A region before interferon therapy with or without ribavirin]. Pathol Biol (Paris). 2004;52:505-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 12. | Zeuzem S, Lee JH, Roth WK. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon alfa. Hepatology. 1997;25:740-744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 146] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 13. | Gao B, Hong F, Radaeva S. Host factors and failure of interferon-alpha treatment in hepatitis C virus. Hepatology. 2004;39:880-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 131] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 14. | Herion D, Hoofnagle JH. The interferon sensitivity determining region: all hepatitis C virus isolates are not the same. Hepatology. 1997;25:769-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 63] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 15. | Schinkel J, Spaan WJ, Kroes AC. Meta-analysis of mutations in the NS5A gene and hepatitis C virus resistance to interferon therapy: uniting discordant conclusions. Antivir Ther. 2004;9:275-286. [PubMed] |
| 16. | Pawlotsky JM, Germanidis G. The non-structural 5A protein of hepatitis C virus. J Viral Hepat. 1999;6:343-356. [PubMed] |
| 17. | Gale M, Blakely CM, Kwieciszewski B, Tan SL, Dossett M, Tang NM, Korth MJ, Polyak SJ, Gretch DR, Katze MG. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208-5218. [PubMed] |
| 18. | Gale M, Korth MJ, Katze MG. Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance. Clin Diagn Virol. 1998;10:157-162. [PubMed] |
| 19. | Gale M. Effector genes of interferon action against hepatitis C virus. Hepatology. 2003;37:975-978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 20. | Boulestin A, Sandres-Saune K, Payen JL, Rostaing L, Pasquier C, Izopet J. Genetic heterogeneity of the NS5A gene of hepatitis C virus and early response to interferon-alpha. J Infect Dis. 2003;188:1367-1370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 21. | Zhou DX, Tang JW, Chu IM, Cheung JL, Tang NL, Tam JS, Chan PK. Hepatitis C virus genotype distribution among intravenous drug user and the general population in Hong Kong. J Med Virol. 2006;78:574-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 22. | Zhou DX, Chan PK, Zhang T, Tully DC, Tam JS. Sequence diversity of hepatitis C virus 6a within the extended interferon sensitivity-determining region correlates with interferon-alpha/ribavirin treatment outcomes. Virus Res. 2010;153:44-49. [PubMed] |
| 23. | Korth MJ, Katze MG. Evading the interferon response: hepatitis C virus and the interferon-induced protein kinase, PKR. Curr Top Microbiol Immunol. 2000;242:197-224. [PubMed] |
| 24. | Tan SL, Katze MG. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology. 2001;284:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 153] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 25. | Reed KE, Rice CM. Identification of the major phosphorylation site of the hepatitis C virus H strain NS5A protein as serine 2321. J Biol Chem. 1999;274:28011-28018. [PubMed] |
| 26. | Frese M, Pietschmann T, Moradpour D, Haller O, Bartenschlager R. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J Gen Virol. 2001;82:723-733. [PubMed] |
| 27. | Taguchi T, Nagano-Fujii M, Akutsu M, Kadoya H, Ohgimoto S, Ishido S, Hotta H. Hepatitis C virus NS5A protein interacts with 2',5'-oligoadenylate synthetase and inhibits antiviral activity of IFN in an IFN sensitivity-determining region-independent manner. J Gen Virol. 2004;85:959-969. [PubMed] |
| 28. | Kato N, Lan KH, Ono-Nita SK, Shiratori Y, Omata M. Hepatitis C virus nonstructural region 5A protein is a potent transcriptional activator. J Virol. 1997;71:8856-8859. [PubMed] |
| 29. | Tanimoto A, Ide Y, Arima N, Sasaguri Y, Padmanabhan R. The amino terminal deletion mutants of hepatitis C virus nonstructural protein NS5A function as transcriptional activators in yeast. Biochem Biophys Res Commun. 1997;236:360-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 57] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 30. | Girard S, Shalhoub P, Lescure P, Sabile A, Misek DE, Hanash S, Bréchot C, Beretta L. An altered cellular response to interferon and up-regulation of interleukin-8 induced by the hepatitis C viral protein NS5A uncovered by microarray analysis. Virology. 2002;295:272-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 59] [Article Influence: 2.6] [Reference Citation Analysis (0)] |